SUMMARY
Heterochromatin impacts various nuclear processes by providing a recruiting platform for diverse chromosomal proteins. In fission yeast, HP1 proteins Chp2 and Swi6, which bind to methylated histone H3 lysine 9, associate with SHREC (Snf2/HDAC repressor complex) and Clr6 histone deacetylases (HDACs) involved in heterochromatic silencing. However, heterochromatic silencing machinery is not fully defined. We describe a histone chaperone complex containing Asf1 and HIRA that spreads across silenced domains via its association with Swi6 to enforce transcriptional silencing. Asf1 function in concert with a Clr6 HDAC complex to silence heterochromatic repeats, and it suppresses antisense transcription by promoting histone deacetylation. Furthermore, we demonstrate that Asf1 and SHREC facilitate nucleosome occupancy at heterochromatic regions but TFIIIC transcription factor binding sites within boundary elements are refractory to these factors. These analyses uncover a role for Asf1 in global histone deacetylation and suggest that HP1-associated histone chaperone promote nucleosome occupancy to assemble repressive heterochromatin.
INTRODUCTION
Eukaryotic genomes are in general organized into two types of chromatin: euchromatin and heterochromatin. Whereas euchromatin is transcriptionally competent, heterochromatin is typically transcriptionally repressed. The factors involved in heterochromatin assembly are found broadly distributed across genomes but their main targets are chromosomal regions containing high density of repetitive DNA such as transposons and their remnants found at centromeres and telomeres (Grewal and Jia, 2007). Heterochromatin promotes genomic stability by exerting repressive influence on the expression of parasitic transposable elements and by prohibiting the illegitimate recombination between dispersed repetitive DNA elements (Peng and Karpen, 2008).
Heterochromatin assembly involves posttranslational modifications of histones and a common set of structural proteins. With the exception of budding yeast, heterochromatin assembly requires methylation of histone H3 at lysine 9 (H3K9me) that provides binding sites for HP1 family of chromodomain proteins (Jenuwein and Allis, 2001). In the fission yeast Schizosaccharomyces pombe, Clr4 not only methylates H3K9 to create binding sites for the localization of chromodomain proteins Chp1, Chp2 and Swi6 (Bannister et al., 2001; Nakayama et al., 2001; Sadaie et al., 2004; Schalch et al., 2009), but it also binds to H3K9me via its own chromodomain to promote the spreading of heterochromatin (Zhang et al., 2008). Heterochromatin factors are preferentially enriched across the chromosomal domains containing specific repeats, referred to as dg and dh repeats, that are present at pericentromeric regions, subtelomeres and the silent mating-type (mat) locus (Cam et al., 2005). RNA polymerase II (RNAPII) transcribes the dg/dh repeats but their expression is repressed by heterochromatin. Chp1, a subunit of the RITS (RNA-induced transcriptional silencing) complex containing Ago1 and Tas3 proteins docks RNAi machinery to heterochromatin, where RITS and its associated factors degrade repeat transcripts, thus causing posttranscriptional silencing in cis (cis-PTGS) (Noma et al., 2004; Schalch et al., 2009; Verdel et al., 2004). Similarly, Chp2 and Swi6 provide recruiting platform for factors involved in transcriptional gene silencing (TGS). The localization of SHREC, which contains a class II HDAC Clr3 and an Snf2 family protein Mit1, across heterochromatin domains requires Chp2 and Swi6 (Sugiyama et al., 2007; Yamada et al., 2005). Swi6 also associates with class I HDAC Clr6 that acts broadly to mediate the global deacetylation of histones, including at RNAPII transcribed regions (Nicolas et al., 2007). Clr3 and Clr6 as well as their interacting HP1 proteins act in an overlapping manner to mediate heterochromatic TGS (Fischer et al., 2009). Mutations in Clr3 and Mit1 subunits of SHREC affect nucleosome positioning that correlates with the TGS defects (Sugiyama et al., 2007).
Heterochromatin assembly also requires histone chaperones (Eitoku et al., 2008). Among the chaperones that deliver histones to DNA, CAF-1 (chromatin assembly factor 1) and HIRA (histone regulatory homolog A) mediate DNA replication-dependent and –independent chromatin assembly, respectively (Groth et al., 2007b; Ransom et al., 2010). CAF-1 interacts with HP1 and is required for the replication and the maintenance of heterochromatin (Murzina et al., 1999; Quivy et al., 2004). HIRA is involved in silencing heterochromatic loci (Greenall et al., 2006; Kaufman et al., 1998; Sharp et al., 2001; Ye et al., 2007). Both CAF-1 and HIRA cooperate with a ubiquitous histone chaperone Asf1 (anti-silencing factor 1), which is believed to deliver histones H3 and H4 heterodimer for nucleosome assembly (Ransom et al., 2010). Loss of Asf1 causes sensitivity to genotoxic agents (Tyler et al., 1999). However, the exact cause of this phenotype is not fully understood.
In this study, we define Asf1 functions in heterochromatic silencing and global protective functions of chromatin in S. pombe. Asf1 forms complex with histones H3 and H4 as well as HIRA that spreads across heterochromatin domains in a manner dependent upon Swi6/HP1. Asf1 associates with a Clr6 complex and these factors act in concert to mediate large-scale deacetylation of histones, including at euchromatic loci. This function of Asf1 is essential to suppress the intragenic antisense transcripts and to protect the DNA from damage. We demonstrate that Asf1 and SHREC function in overlapping pathways impacting nucleosome occupancy at heterochromatic loci. Thus, Asf1 contributes to chromatin structural modulations by facilitating histone deacetylation and governing nucleosome occupancy, which has important implications for the assembly and the propagation of repressive heterochromatin.
RESULTS
Purification of S. pombe Asf1
We previously showed that amino-terminal TAP-tagged Swi6 co-purifies with factors involved in chromosome dynamics and heterochromatic silencing (Fischer et al., 2009). Mass spectrometry of purified Swi6 samples also identified peptides corresponding to HIRA subunit Hip3 (Figure S1A). These results indicated that HIRA might directly participate in heterochromatic silencing. Considering that Asf1 synergizes with HIRA and CAF-1 to assemble nucleosomes (Tagami et al., 2004; Tyler et al., 1999), we wondered whether these factors act together in heterochromatin assembly. To test this, we purified carboxy-terminal TAP-tagged Asf1 (Asf1-TAP) using tandem affinity purification (TAP). Mass spectrometry of the purified samples showed that Asf1 associate with histones H3 and H4 as well as HIRA comprising Hip1, Slm9, Hip3 and Hpc2 (Figure 1A and S1B). This analysis did not identify CAF-1 in the Asf1 purified fraction (Figure 1A). To address this further, we purified carboxy-terminal TAP-tagged Pcf3 (p48) subunit of CAF-1. In addition to Pcf3, we identified Pcf1 (p150) and Pcf2 (p60) subunits of CAF-1, as well as few peptides of Pcn1 (PCNA) (Figure 1A and S1C), which associates with CAF-1 (Shibahara and Stillman, 1999), but no Asf1 peptides. It is possible that Asf1 interacts with CAF-1 in a cell cycle stage-specific manner (e.g. during S-phase) that is not detected in our assays. Regardless, Asf1 interactions with histones H3/H4 and HIRA are conserved in S. pombe; thus, raising the possibility of these factors acting together, in association with HP1, to enforce heterochromatic silencing.
Figure 1. Asf1 Affects Heterochromatic Silencing in S. pombe.
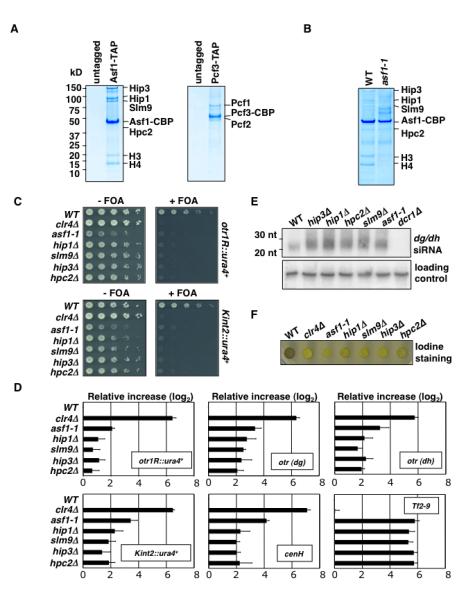
(A) Purification of Asf1 and CAF-1. SDS-PAGE followed by coomassie blue staining of untagged control, Asf1-TAP and Pcf3-TAP purification. Purified fractions were subjected to tandem MS (LC-MS/MS). (B) Coomassie blue staining of Asf1-TAP purification from wild type and asf1-1 mutant. (C) Heterochromatic silencing of reporter genes inserted at pericentromeric repeats (otr1R::ura4+) and silent mat locus (Kint2::ura4+). Serial dilution plating on counter-selective fluoroorotic acid (FOA) medium used to assay ura4+ expression. (D) asf1-1 affects silencing of heterochromatic repeats and Tf2. qPCR was used to assay expression of indicated loci. Values shown were normalized to act1+ expression. Error bars indicate standard deviations from three independent experiments. (E) Defective TGS in mutants results in elevated levels of dg/dh siRNAs. siRNAs levels were analyzed by northern blot. (F) Mutations in Asf1/HIRA impair mating-type switching. In contrast to colonies formed by wild-type homothallic (h90) cells, which switch mating-type efficiently and stain dark with iodine vapors, colonies formed by h90 asf1 and HIRA mutants stain light, owing to defective mating-type switching. Δclr4 is shown as a control.
Asf1 Histone Chaperone is an Effector for Heterochromatic Silencing
We next investigated whether Asf1 affects heterochromatic silencing. Since Asf1 is essential for growth in S. pombe (Umehara et al., 2002), we constructed a mutant allele of asf1 (asf1-1). Mutant Asf1 shows a considerable reduction in its ability to bind histones and HIRA (Figure 1B). Sequencing of asf1-1 showed one mutation mapping to a residue involved in histone-binding, while another mapped close to HIRA binding site (Figure S2A). We tested the effects of asf1-1 on silencing at heterochromatic loci (Figure S2B). For comparison, we included deletion mutants of genes encoding HIRA subunits. asf1-1 alleviated silencing of the ura4+ inserted at the outer centromeric repeat region (otr1R::ura4+) and within a centromere-homologous (cenH) element at the silent mat locus (Kint2::ura4+), in a manner similar to HIRA null mutants (Figure 1C).
Since the growth-based silencing assay is not quantitative, we evaluated silencing defects by quantitative real-time RT-PCR (qPCR). Effects of asf1-1 and HIRA null mutants on expression of ura4+ reporters and heterochromatic repeats at centromeres (dg/dh) and mat locus (cenH) were examined (Figure 1D). Also included in this analysis was Tf2 retrotransposons. Tf2 silencing requires HIRA, SHREC and Clr6 but not Clr4 and HP1 (Cam et al., 2008; Greenall et al., 2006; Hansen et al., 2005). The asf1-1 and HIRA mutants showed severe defects in the silencing of Tf2. However, these mutations only partially derepressed heterochromatic repeats or ura4+ reporters, as compared to the clr4Δ (Figure 1D). Clr4 is required for both TGS and cis-PTGS of heterochromatic loci (Noma et al., 2004). Transcripts that escape heterochromatic TGS are processed into siRNAs by RNAi machinery. asf1-1 and HIRA mutants only affect TGS and not cis-PTGS. Indeed, we observed increased levels of siRNAs in asf1-1 and HIRA mutants (Figure 1E), revealing that dg/dh repeats are derepressed in these mutants but transcripts are not accumulating due to their degradation by RNAi.
Heterochromatin also regulates mating-type switching by promoting nonrandom choice of mat2 and mat3 donor loci (Jia et al., 2004). S. pombe spores, but not the vegetative cells, stain dark with iodine, while the switching-defective mutants produce spores inefficiently, resulting in light staining. Defective mating-type switching in heterochromatin mutants results in poor iodine staining of colonies, in contrast to the dark staining of wild-type colonies (Jia et al., 2004). asf1-1 and HIRA mutants were defective in mating-type switching, as indicated by the light iodine staining of the colonies (Figure 1F). Therefore, Asf1 contributes not only to silencing but also other functions that rely on heterochromatin.
Swi6-dependent and -independent Localization of Histone Chaperone
HP1 proteins serve as a recruiting platform to target silencing activities such as SHREC (Sugiyama et al., 2007; Yamada et al., 2005). Asf1/HIRA might affect silencing by facilitating the assembly of H3K9me-HP1 platform or it could act downstream of HP1 loading as an effector involved in repressive chromatin assembly. We used chromatin immunoprecipitation (ChIP) assay to examine the effects asf1-1 and hip1Δ on levels of H3K9me and HP1 proteins at centromeres. Whereas asf1-1 cells showed slight reduction in levels of H3K9me, Swi6 and Chp2, the levels of these factors in hip1Δ appeared comparable to wild type (Figure 2A and 2B). Thus, silencing defects in asf1-1 and hip1Δ were not coupled to complete loss of the heterochromatin platform. The reduction in H3K9me is not due to defective Clr4 recruitment but rather reflects dynamic nature of nucleosomes in mutant cells (see below).
Figure 2. Swi6-dependent and –independent Localization of Histone Chaperone.
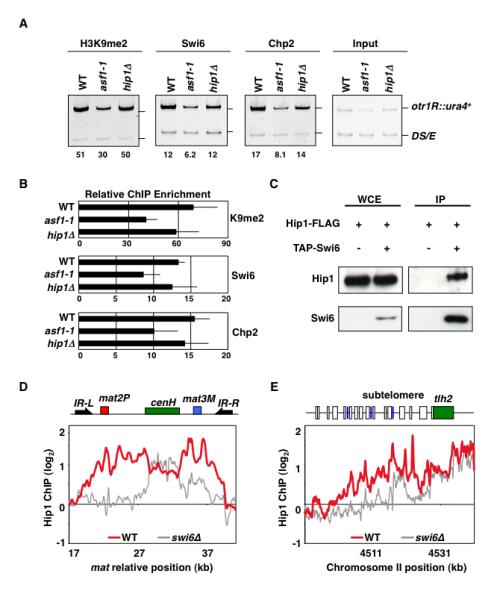
(A) asf1-1 and hip1Δ maintain heterochromatin signatures of H3K9me, Swi6, and Chp2. Strains carrying otr1R::ura4+ and ura4DS/E minigene at the endogenous locus were used to perform ChIP. Intensities of bands representing otr1R::ura4+ and ura4DS/E in ChIP and input lanes were used to calculate relative fold enrichment values shown. (B) qPCR using DNA isolated from either immunoprecipitated chromatin or input DNA was used to calculate relative fold enrichment at centromeres. Error bars indicate standard deviations from three independent experiments. (C) Swi6 interacts with Hip1. Strains expressing functional FLAG-epitope tagged Hip1 (Hip1-FLAG) and/or TAP-tagged Swi6 (TAP-Swi6) were used to perform purification. Purified fractions from untagged or TAP-tagged Swi6 samples were analyzed by western blotting using anti-FLAG antibody. (D and E) Hip1 localization across silent mat locus and subtelomeres requires Swi6. ChIP-chip was used to determine Hip1-FLAG distribution in wild type and swi6Δ cells. At silent mat locus, heterochromatic domain containing mat2 and mat3 loci as well as cenH is surrounded by IR-L and IR-R boundary elements. The subtelomeric region of right arm of chromosome II includes tlh2 gene sharing homology to dh, LTRs (blue boxes) and ORFs (open boxes).
We next investigated whether HP1 proteins contribute to the localization of histone chaperone at heterochromatic loci. Since we detected Hip3 in Swi6 purified fraction (Figure S1A), we wondered if HIRA associates with heterochromatin machinery. Hip1 was readily detected in the affinity-purified Swi6 fraction (Figure 2C). This interaction is functionally important because Swi6 was required for Hip1 distribution across the heterochromatic domains. ChIP-chip showed that Hip1 was enriched throughout heterochromatin domains in the wild-type cells (Figure 2D, 2E, and S3). In the absence of Swi6, Hip1 localization was restricted to transcribed dg/dh repeats, and it failed to spread outward to the surrounding sequences (Figure 2D, 2E, and S3). Defects in spreading of Hip1 in swi6Δ mutant could be seen clearly at silent mat locus and telomeres, which unlike centromeres contain a single copy of repeat element and require Swi6 for spreading heterochromatin across the silenced domain (Cam et al., 2005; Hall et al., 2002; Kanoh et al., 2005). These results suggest that Hip1 can be targeted to dg/dh elements independent of Swi6, but its localization across the heterochromatin domains requires Swi6.
Asf1 Acts in Concert with Clr6 HDAC to Enforce Heterochromatic Silencing
Asf1/HIRA influence heterochromatic silencing directly by associating with Swi6/HP1. To address the underlying mechanism, we explored the effects of asf1 and hip1 mutants in combination with the mutant alleles of other silencing activities such as RNAi RITS (tas3), SHREC (clr3 and mit1) and Clr6 complex (clr6 and alp13). The effects of single and double mutants on the levels of transcripts derived from dg/dh and cenH elements were assayed using qPCR. Consistent with both TGS and cis-PTGS contributing to heterochromatin silencing, combining asf1-1 or hip1Δ with tas3Δ resulted in synergistic defects in heterochromatic silencing (Figure 3A). We also found that double mutants carrying mutations in clr3 or mit1 along with either asf1-1 or hip1Δ showed cumulative derepression of repeat elements (Figure 3A and S4), indicating overlapping functions for Asf1/HIRA and SHREC. However, when asf1-1 or hip1Δ were combined with clr6 or alp13 mutant alleles, double mutants did not show additive defects on silencing as compared to the single mutants (Figure 3A and S4). This result argues that Asf1/HIRA and Clr6 HDAC act in concert to silence heterochromatic repeats.
Figure 3. Asf1 Acts in Concert with Clr6 HDAC to Promote Heterochromatic Silencing.
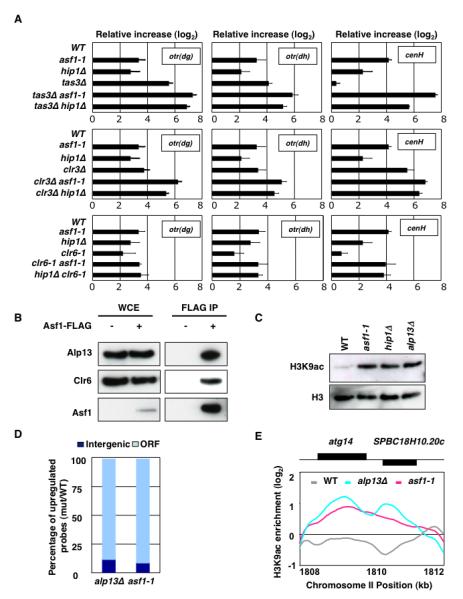
(A) The expression levels of dg [otr(dg)] and dh [otr(dh)] centromeric repeats and cenH in indicated strains were analyzed by qPCR. All values were normalized to act1+ expression. Error bars indicate standard deviations from three independent experiments. (B) Asf1 interacts with Clr6 complex-II. Strain expressing FLAG-tagged Asf1 (Asf1-FLAG) or control untagged strain was used to perform the immunoaffinity purification using anti-FLAG antibody. Purified fractions were analyzed by western blotting with antibodies against Alp13 and Clr6. (C) asf1-1 and hip1Δ show increase in bulk histone acetylation levels, similar to alp13Δ. Histones extracted from indicated strains were analyzed by western blotting using anti-H3K9ac and anti-H3 antibodies. (D and E) ChIP-chip analyses of histone acetylation were performed using anti-H3K9ac antibody. Percentage of upregulated probes (mutant/WT signal intensity ratio > 1.5) corresponding to ORF and intergenic regions are plotted (D). Like alp13Δ, asf1-1 shows increased acetylation in coding regions of genes. A representative region of the genome is shown (E).
Based on the genetic analyses, it was possible that Asf1/HIRA facilitate histone deacetylation by Clr6. Asf1 co-immunoprecipitated with Clr6 complex subunits Alp13 and Clr6 (Figure 3B). Moreover, asf1-1 and hip1Δ exhibited a substantial increase in bulk H3K9ac levels, in a manner similar to alp13Δ (Figure 3C). To confirm this further, we performed ChIP-chip analyses of H3K9ac. Both alp13Δ and asf1-1 mutants showed widespread increase in H3K9ac, as compared to the wild-type cells. Notably, although 30% of the probes in our microarray correspond to intergenic regions, nearly all probes affected by asf1-1 and alp13Δ reside in coding regions (Figure 3D and 3E). These data are in agreement with the previous conclusion that Alp13-containing Clr6 complex (referred to as Clr6 complex-II) deacetylates histones preferentially in transcribed regions of the genome (Nicolas et al., 2007). Closer examination of the data revealed that all genomic sites that showed increased histone acetylation in asf1 mutant were also affected by Clr6 complex-II, except the partial loss of function asf1-1 allele in general showed weaker effects as compared to alp13Δ.
Asf1 Suppresses Intragenic Antisense Transcripts at Loci Targeted by Clr6
Changes in histone acetylation prompted us to analyze transcriptional changes in asf1 mutant. For comparison, we also included alp13Δ and hip1Δ that show upregulation of antisense RNA (Anderson et al., 2009; Nicolas et al., 2007). Expression profiling detected widespread upregulation of transcripts in all three mutant backgrounds. The affected loci included Tf2 retrotransposon and LTRs. Moreover, asf1-1 and hip1Δ showed upregulation of sense and antisense transcripts corresponding to subtelomeric genes located within heterochromatic domains (Figure S5A). Interestingly, asf1-1, but not hip1Δ, also showed substantial increase in the levels of transcripts derived from intergenic portions of rDNA repeat loci (Figure S5B). Notably, asf1-1 produced a disproportionate increase in antisense transcripts – constituting a large proportion of probes upregulated. Detailed expression profiling of individual loci showed that the antisense transcripts upregulated in asf1-1 mutants were also upregulated in hip1Δ and alp13Δ cells (Figure 4A and 4B). The appearance of antisense transcripts in mutant backgrounds was confirmed by strand-specific RT-PCR (Figure 4C).
Figure 4. Asf1 Suppresses Antisense Transcription and Protects Genomic Integrity.

(A) Expression profiling of asf1-1, hip1Δ, and alp13Δ was performed using microarrays containing probes corresponding to both DNA strands. Changes in RNA signal intensities were calculated for each probe. Plotted are number and percentage of upregulated probes (mutant/WT signal intensity ratio > 1.5) corresponding to sense and antisense strands. (B) asf1 and hip1 mutants show antisense upregulation at loci affected by alp13Δ. Ratios of RNA signal intensities for forward and reverse strand probes in mutant versus wild type were converted to color codes and plotted. Black rectangles on top and bottom of solid line indicate genes transcribed in forward and reverse directions, respectively. (C) High resolution view of antisense upregulation at hrp1 gene. Strand-specific RT-PCR performed using RNA prepared from indicated strains is shown. (D) Hierarchical clustering of mutants based on similarities in the distribution of upregulated antisense RNAs. Pearson’s correlation coefficients calculated from pairwise comparisons of mutant expression profiles were converted into color codes. (E) Asf1 and Alp13 protect DNA from damage by genotoxins. Chromosomal DNA samples from indicated strains treated with 0 or 0.5mU/ ml bleomycin for 90 minutes were analyzed by pulse-field gel electrophoresis.
Clr4 complex and RNAi factors facilitate the degradation of readthrough antisense RNA at euchromatic loci via a mechanism involving nuclear exosome (Zofall et al., 2009). This mechanism is distinct from a pathway involving Clr6 and Set2 that suppresses initiation of antisense transcript from cryptic promoters (Nicolas et al., 2007). To gain further insight into Asf1 functions, we compared the distributions of antisense RNA in different mutants. Correlation coefficients from pairwise comparisons were used to cluster mutants based on similarity in their antisense profiles (Figure 4D). Antisense profiles of asf1-1 and hip1Δ mutants closely resembled to that of cells lacking Clr6 complex components Alp13 and Cph1, or Set2 involved in H3K36me at transcribed loci. Thus, in addition to silencing heterochromatic repeats, Asf1 prevents antisense transcription at euchromatic loci.
Asf1 Protects Genome from Damage by Genotoxic Agents
Global deacetylation of histones by Clr6 HDAC is important for protecting the genome from damage by genotoxic agents (Nicolas et al., 2007). To determine whether defects in Asf1 render genome more vulnerable to damage by double-strand break-inducing agents, we exposed asf1-1 to bleomycin for 90 min and then assayed chromosomal breakage by pulse-field gel electrophoresis. Similar to alp13Δ, genomic DNA of asf1-1 cells was considerably more sensitive to bleomycin-induced damage, as indicated by the disappearance of full-length chromosome bands and the appearance of a smear of broken DNA fragments (Figure 4E). We also found that asf1-1 cells were hypersensitive to genotoxic agents such as bleomycin, camptothecin and methylmethane sulfonate (Figure S6), in a manner similar to Clr6 (Anderson et al., 2009; Nicolas et al., 2007). However, both Asf1 and Clr6 complex-II mutants were not sensitive to hydoxyurea (Figure S6) (Nicolas et al., 2007). These results suggest that Asf1 protects genome from damage in part by facilitating global deacetylation of histones by Clr6.
Asf1 and SHREC Affect Nucleosome Occupancy at Heterochromatin
SHREC targeted by H3K9me-bound HP1 proteins positions nucleosomes to promote TGS (Sugiyama et al., 2007). Since Asf1 and SHREC act in an overlapping manner, we wondered whether Asf1 promotes nucleosome occupancy at heterochromatic loci. We addressed this possibility by using a microarray-based approach to measure nucleosome occupancy at heterochromatic loci. The custom microarray used contained probes tiled every 10 bp across the major heterochromatin domains at the silent mat region, centromere 2 (cen2) and left telomere of chromosome 1, as well as a segment of euchromatin. Probes derived from mononucleosomal DNA isolated from micrococcal nuclease (MNase)-digested chromatin were hybridized to the microarray. To validate our MNase-chip method, we compared nucleosome patterns at euchromatic loci to published results (Lantermann et al., 2010). We observed nucleosome free regions at 5′ ends of genes followed by positioned nucleosomes in open reading frames (Figure S7A). Comparison of nucleosome occupancy across heterochromatin domains in asf1, clr3 or asf1clr3 mutant cells to wild-type cells identified several sites showing depletion of nucleosomes in mutant cells. In general, changes observed in clr3 and asf1 single mutants were weaker as compared to asf1clr3 double mutant that showed substantial reduction in the nucleosome occupancy (Figure 5). These results are consistent with cumulative loss of TGS in asf1clr3 double mutants (Figure 3A), and suggest that both Asf1 and SHREC promote nucleosome occupancy to enforce heterochromatic silencing.
Figure 5. Asf1 and SHREC Promote Nucleosome Occupancy at Heterochromatic Loci.
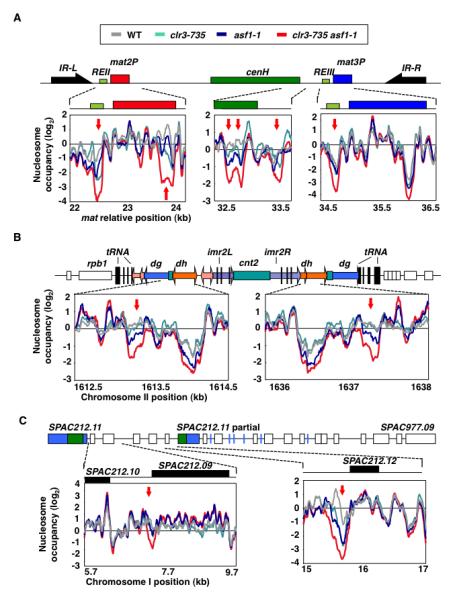
Nucleosome occupancies in wild type, clr3-735 (which carry D232N mutation in Clr3 HDAC domain), asf1-1 and clr3-735 asf1-1 mutants were measured using MNase-chip. Major heterochromatic regions at silent mat locus (A), centromere 2 (B) and subtelomeric region (C) were analyzed for differences in nucleosome occupancy. The regions showing minimum log2 difference of 2 between wild type and clr3-735asf1-1 datasets are highlighted. Red arrows indicate regions showing lower nucleosome occupancy in single and/or double mutants as compared to wild type. Schematic diagrams indicating main features of heterochromatin domains are included (A-C). (A) At silent mat locus, heterochromatic domain containing mat2 and mat3 loci as well as cenH is surrounded by IR-L and IR-R boundary elements. REII and REIII represent cis-acting silencers. (B) At centromeres, heterochromatin coats dg/dh repeats and a portion of inner repeats (imr) that surround central (cnt) core domain. Vertical black lines and boxes indicate individual copies or clusters of tRNAs. (C) Heterochromatin at subtelomeric loci covers several ORFs including SPAC212.11 sharing homology to dh. Blue lines denote LTRs.
The most prominent sites that display decreased nucleosome occupancy corresponded to silencer DNA elements such as REII and REIII located near mat2 and mat3 silent mating-type loci, respectively (Figure 5A). These elements correspond to the major peaks of SHREC binding at mat locus (Cam et al., 2008; Sugiyama et al., 2007). asf1clr3 mutant also showed a hypersensitive site appearing inside mat2 (Figure 5A), suggesting that these factors act outside of the known silencer elements. We also observed nucleosome-depleted regions appearing elsewhere in heterochromatic domains, in particular at repeat loci where RNAPII transcription is repressed by heterochromatin (Fischer et al., 2009). Notably, several changes mapped to dh repeats at centromeres and cenH element at the mat locus (Figure 5A and 5B). Other sites affected included promoters of the genes located in heterochromatin domain at subtelomeric region (Figure 5C). These analyses suggest a mechanism in which HP1-associated Asf1 and SHREC govern nucleosome occupancy at heterochromatic loci to assemble repressive chromatin.
Asf1 does not Affect NFRs at a Recombinational Enhancer and Boundary Elements
Previous work has shown that the heterochromatin factors spread to inverted repeat boundary elements, IR-L and IR-R, which flank the heterochromatin domain at mat locus (Noma et al., 2006), but their levels decrease sharply coinciding with a cluster of TFIIIC binding sites (B-boxes) essential for boundary function (Noma et al., 2006). Similarly, the borders of heterochromatin domains at centromeres are demarcated by clusters of tRNAs, which also contain B-boxes (Noma et al., 2006; Scott et al., 2006). Interestingly, nucleosomes were depleted at tRNAs, and B-box-containing region of IRs (Figure 6A and S7B). These NFRs were not an artifact of the MNase-ChIP method as existence of hypersensitive sites could be confirmed by the conventional DNase I- and MNase-hypersensitivity assays (Figure 6B). Our analyses also showed NFRs overlapping with a recombinational enhancer (referred to as SRE) (Figure 6A), which is involved in proper choice of mat2 or mat3 donor loci during mating-type switching (Jia et al., 2004). Considering Asf1 functions in nucleosome disassembly (Adkins et al., 2004; Schwabish and Struhl, 2006) and asf1-1 mutant is defective in mating-type switching, we wondered whether the formation of NFRs require Asf1. Mutation in asf1 alone or in combination with clr3 had no effect on NFRs associated with SRE or TFIIIC-binding sites at boundary elements (Figure 6A), although asf1clr3 mutant showed an NFR appearing in IR element (Figure 6A).
Figure 6. Nucleosome Occupancy at a Recombinational Enhancer and Boundary Elements.
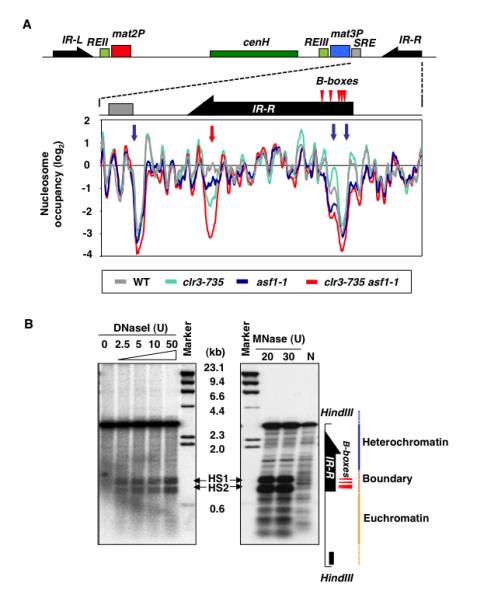
(A) Nucleosome occupancies in indicated mutants, as determined by MNase-chip, are plotted for a region containing mat3M, SRE element containing Swi2-binding site, and IR-R boundary element. Blue arrows indicate NFRs unaffected by mutations in Asf1 or SHREC. Red arrow indicates location of an NFR in IR-R most prominent in asf1clr3 double mutant. (B) DNaseI- or MNase-treated chromatin fractions from wild-type cells were digested with HindIII and then analyzed by Southern blotting with a mat-specific probe (black rectangle). Lane N indicates naked DNA treated with MNase. HS1 and HS2 indicate the positions of DNaseI and MNase hypersensitive sites.
DISCUSSION
Heterochromatin assembled at repetitive DNA elements not only prevents the accumulation of potentially deleterious RNAs but it has also evolved as a powerful mechanism for regulating other chromosomal processes (Grewal and Jia, 2007). HDAC activities (such as SHREC) associated with HP1 proteins spread across heterochromatin domains to restrict the access of RNAPII to underlying DNA (Sugiyama et al., 2007; Yamada et al., 2005). Here we show that Asf1, which exists in a complex containing histone H3 and H4 as well as HIRA, functions in the silencing of repeat elements embedded within heterochromatin domains and various genetic elements in other parts of the genome. These analyses reveal a critical aspect of heterochromatin silencing in which histone chaperone and the HDAC activities associated with H3K9me-HP1 docking platform promote histone deacetylation and nucleosome occupancy to assemble repressive chromatin (Figure 7). Asf1 also facilitates large-scale histone deacetylation that is essential for global protective functions of chromatin.
Figure 7. Model Showing Involvement of HP1-associated Asf1/HIRA, Clr6 and SHREC in Histone Deacetylation and Nucleosome Occupancy at Heterochromatic Regions.
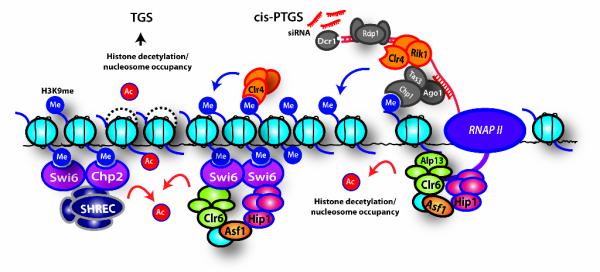
RNAPII transcription of dg/dh repeats generates transcripts that are processed by RNAi machinery (RITS, RNA-dependent RNA polymerase, Rdp1, and Dicer, Dcr1) into siRNAs. Asf1 facilitates histone deacetylation by Clr6 complex-II after the passage of RNAP II to reassemble repressive chromatin. Clr4 binding to H3K9me via its chromodomain facilitates heterochromatin spreading. H3K9me also recruits Chp1, Chp2 and Swi6. Whereas Chp1 tethers RNAi machinery to chromatin for cis-PTGS, Chp2 and Swi6 associate with Asf1/HIRA, Clr6-complex-II and/or SHREC involved in deacetylation of histones and transcriptional silencing. Asf1/HIRA and SHREC promote nucleosome occupancy and positioning to exclude NFRs, which is presumably critical for preventing access to transcriptional and recombinational machineries. The prevention of nucleosome turnover by these effectors may have important implications for the maintenance of heterochromatin.
HP1-Dependent Spreading of Histone Chaperone and Heterochromatic Silencing
Mutations in Asf1 that affect its binding to histones H3/H4 and HIRA cause defects in silencing of reporter genes artificially inserted into heterochromatin domains and naturally repressed repeats. The results presented argue that Asf1/HIRA directly contribute to TGS associated with heterochromatin formation. Notably, Hip1 interacts with Swi6/HP1. Moreover, Hip1 is enriched across heterochromatin domains in a manner dependent upon Swi6. However, deletion of swi6 does not completely abolish Hip1 localization. Hip1 can be detected at transcribed dg/dh repeats in swi6Δ cells. HIRA and presumably its associated Asf1 are likely recruited to RNAPII-transcribed repeat loci independent of Swi6 but their localization across entire silenced domain requires the heterochromatin machinery. This situation resembles Clr6 complex-II that is targeted to transcribed loci by factors associated with elongating RNAPII (Li et al., 2007) but also interacts with Swi6 to enforce heterochromatic silencing (Fischer et al., 2009). Considering that Asf1 interacts with Clr6 and that Asf1/HIRA are required for histone deacetylation, it is possible that the chaperone complex is recruited to transcribed loci by the elongating RNAPII apparatus either directly and/or through interactions with Clr6. Once recruited, however, Asf1/HIRA induces the silencing by promoting nucleosome occupancy and deacetylation of histones (see below).
It is significant that Swi6/HP1 targeted by RNAi and other mechanisms mediate Asf1/HIRA localization across heterochromatin domains. This allows the histone chaperone to act beyond transcribed loci to assemble chromatin throughout the heterochromatin domains. In other words, H3K9me-HP1 permits Asf1/HIRA normally involved in transcription-coupled chromatin assembly to serve as a general-purpose chromatin assembly factor capable of influencing a broad spectrum of genetic elements.
Histone Deacetylation, Nucleosome Occupancy and Maintenance of Heterochromatin
HP1-associated SHREC restricts RNAPII occupancy and this depends upon Clr3 and Mit1 activities that impact the positioning of nucleosomes (Sugiyama et al., 2007). This work suggested that the nucleosome positioning by HP1-associated factors is a critical mechanism for heterochromatin silencing. Further supporting this view, the loss of SHREC or factors (Clr4 and HP1) required for its localization affects nucleosome occupancy at heterochromatic loci (Garcia et al., 2010). Our work significantly enhances the understanding of pathways governing heterochromatic TGS and suggests a critical role for Asf1/HIRA in repressive chromatin assembly coordinated by HP1 proteins. Swi6-asociated Asf1/HIRA is an effector for heterochromatic TGS that acts in an overlapping manner with SHREC to promote nucleosome occupancy. Indeed, double mutant cells carrying mutations in SHREC and Asf1/HIRA show cumulative defects in heterochromatic silencing that correlates with the loss of nucleosomes. These findings, together with previous studies (Sugiyama et al., 2007), suggest a scenario in which the chromatin assembly factors bound to HP1 proteins assemble the repressive structures in part by maintaining nucleosomes at target loci.
How do SHREC and Asf1/HIRA orchestrate the assembly of repressive heterochromatin? Current evidence suggests dual functions for these effectors. SHREC mediates deacetylation of histone H3 via its Clr3 HDAC activity, but its function also requires ATPase activity of Mit1 (Sugiyama et al., 2007). Mit1 might have a role in deposition of histones, similar to its mammalian homolog ATRX required for localization of histone H3.3 at telomeres (Goldberg et al., 2010). However, Mit1 activity might also be required for the deacetylation of nucleosomal histones by Clr3. Similarly, Asf1 plays an important role in histone deacetylation, in addition to its conserved functions in depositing histones H3/H4 (Ransom et al., 2010). Genetic epistasis and biochemical experiments suggest that Asf1/HIRA function together with Clr6 to silence centromeric repeats. Clr6 homologs including mammalian HDAC1/2 deacetylate core histones more efficiently than nucleosomal histones (Zhang et al., 1998). Asf1 could be required to present H3/H4 dimer to Clr6 for deacetylaton. Analogous to Asf1/HIRA, Clr6 complex-II also interacts with Swi6, and Clr6 function is partially redundant to the function of SHREC (Fischer et al., 2009; Nicolas et al., 2007). We propose that deacetylation of histones by SHREC as well as Asf1 and Clr6 complexes bound to HP1, is a critical step in the assembly of silenced chromatin. Deacetylation of histones, which is a hallmark of heterochromatin in most eukaryotes (Ekwall, 2005), may prevent nucleosome disassembly/turnover by chromatin remodeling factors (Kasten et al., 2004), ultimately promoting the nucleosome occupancy and heterochromatic silencing.
Clr4 and Swi6 affect chromatin dynamics by precluding replication-independent exchange of nucleosomes (Choi et al., 2005). HP1-associated SHREC and Asf1/HIRA might act broadly to prevent nucleosome turnover, in addition to eliminating NFRs at specific sites. Such stabilization of nucleosomes marked with heterochromatin-specific modifications is critical for the inheritance of heterochromatin in cis (Hall et al., 2002). Indeed, in cells defective in de novo heterochromatin assembly, stable propagation of preassembled heterochromatic structures requires Swi6 and Clr3 (Hall et al., 2002; Yamada et al., 2005). Mammalian Asf1 engaged in replication coupled chromatin assembly binds to histones carrying parental histone modifications (Groth et al., 2007a). Therefore, Asf1 may transfer parental H3/H4 to the newly replicated DNA to promote heterochromatin maintenance. Asf1 bound to replication factors might also facilitate the histone deacetylation to promote chromatin maturation.
Asf1 has no effect on naturally occurring NFRs at SRE recombinational enhancer at mat locus and boundary elements. These NFRs are also resistant to heterochromatin factors (Garcia et al., 2010). The existence of NFRs at boundary elements in principle could block the spread of heterochromatin. However, heterochromatin spreads beyond NFR at SRE. These NFRs likely signify binding of factors involved in mating-type switching and boundary function such as TFIIIC (Jia et al., 2004; Noma et al., 2006).
Asf1-Facilitated Histone Deacetylation and Antisense Suppression at Euchromatin
A “toolkit” of effectors associated with heterochromatin is also targeted to euchromatic locations by either site-specific recruitment mechanisms or via their association with the elongating RNAPII apparatus (Cam et al., 2008; Nicolas et al., 2007; Zofall et al., 2009). Clr6 deacetylates histones at coding regions to suppress cryptic transcripts (Nicolas et al., 2007). Similarly, Asf1 suppresses the cryptic transcripts. S. cerevisiae Asf1 contributes to restoring chromatin after elongation of RNAPII (Schwabish and Struhl, 2006). Besides providing histones for chromatin reassembly, our results suggest that Asf1 facilitates histone deacetylation by Clr6 that is critical for the suppression of cryptic transcripts. Notably, asf1-1 and clr6 complex-II mutants show upregulation of intragenic transcripts at same loci. Both of these factors also affect histone acetylation in RNAPII-transcribed regions. In addition to affecting nucleosome turnover that could also affect acetylation status, Asf1 might bind to histones displaced during the transcription to facilitate their deacetylation by presenting them to Clr6. The removal of acetyl moieties from histones assembles repressive chromatin, such that transcription initiation from cryptic promoters within coding regions is inhibited. The global histone deacetylation by Clr6 and Asf1 also protects genome integrity. Defects in histone deacetylation, along with the roles of histone chaperones in chromatin assembly during the processes of replication and repair (Groth et al., 2007b; Ransom et al., 2010), may help to explain the sensitivity of asf1 mutant to genotoxic agents.
In contrast to their homologs in S. cerevisiae, both Asf1 and Clr6 are essential in S. pombe (Nicolas et al., 2007; Umehara et al., 2002). However, cells lacking components of HIRA can grow under standard growth conditions. These observations indicate that Asf1 has additional HIRA-independent function(s), which may or may not rely on its partnership with Clr6. We note that Asf1, but not HIRA, is required for inhibiting transcription in the intergenic spacer region of rDNA repeats. Future studies will address whether Asf1 associates with other chromatin modifying activities. Regardless, the widespread involvement of Asf1 in suppressing potentially deleterious RNAs, including transcripts derived from repeat elements and cryptic promoters, is expected to have important implications for maintaining the genome homeostasis.
EXPERIMENTAL PROCEDURES
Strain Constructions
Strains carrying deletion alleles and carboxy-terminal-tagged Asf1-TAP, Asf1-FLAG, and Hip1-FLAG under the control of native promoters were constructed according to a PCR-based method. For constructing asf1-1 allele, DNA fragment containing FLAG-tagged asf1 gene (asf1-FLAG) and KANMX6 was mutagenised using Gene morph II Random mutagenesis kit (Stratagene), and transformed into a wild type strain. Transformants were selected on medium containing G418 and colonies showing temperature-sensitive growth defects at 37 °C were subjected to further characterization by performing backcrosses and sequencing of mutant allele. asf1-1 cells were grown at 30 °C.
ChIP and Expression Profiling
ChIP and ChIP-chip were carried out as previously described (Cam et al., 2005) using anti-FLAG agarose (M2, Sigma), or anti-histone H3K9me2 (ab1220, Abcam) antibody. For expression profiling, 15 ug DNase-treated total RNA was reverse transcribed using Invitrogen Superscript Indirect cDNA Labeling System (Invitrogen). Labeled samples from mutant (Cy5) and wild type (Cy3) were mixed and hybridized to the microarray.
Quantitative Real-time PCR
1 ug of DNase-treated total RNA isolated by MasterPure yeast RNA purification kit (Epicentre) was reverse transcribed using ImProm-IITM Reverse Transcription System (Promega). Complementary DNA (1/50 RT reaction) was used as a template for SYBR green (0.1× dilution) qPCR analysis. Relative fold expressions relative to act1 were calculated.
Descriptions of methods used to perform tandem affinity purification as well as micrococcal nuclease digestion and mapping of nucleosomes can be found in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Zhang and T. Fischer for their helpful contributions, F. Reyes-Turcu and O. Aygun for critical reading of the manuscript, M. Zhou and T. Veenstra for mass spectrometry, and members of the Grewal laboratory for discussions. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Accession numbers Microarray data are available at the GEO repository of the NCBI under accession numbers GSE25597, GSE25598 and GSE25600.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Anderson HE, Wardle J, Korkut SV, Murton HE, Lopez-Maury L, Bahler J, Whitehall SK. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol. Cell. Biol. 2009;29:5158–5167. doi: 10.1128/MCB.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SIS. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SIS. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Choi ES, Shin JA, Kim HS, Jang YK. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucl. Acid. Res. 2005;33:7102–7110. doi: 10.1093/nar/gki1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol. Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. Genome-wide analysis of HDAC function. Trends Genet. 2005;21:608–615. doi: 10.1016/j.tig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SIS. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Dumesic PA, Hartley PD, El-Samad H, Madhani HD. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 2010;24:1758–1771. doi: 10.1101/gad.1946410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenall A, Williams ES, Martin KA, Palmer JM, Gray J, Liu C, Whitehall SK. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J. Biol. Chem. 2006;281:8732–8739. doi: 10.1074/jbc.M512170200. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007a;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007b;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SIS. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bahler J, Thon G. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jia S, Yamada T, Grewal SIS. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell. 2004;119:469–480. doi: 10.1016/j.cell.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SIS. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SIS. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SIS. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SIS. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol. Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SIS. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Umehara T, Chimura T, Ichikawa N, Horikoshi M. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells. 2002;7:59–73. doi: 10.1046/j.1356-9597.2001.00493.x. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SIS. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 2007;27:2452–2465. doi: 10.1128/MCB.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SIS. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


