We review the molecular etiology of poor prognosis basal-like breast cancer and efforts to translate these pathogenic insights into improved therapies.
Abstract
The classification of breast cancer into molecular subtypes with distinctive gene expression signatures that predict treatment response and prognosis has ushered in a new era of personalized medicine for this remarkably heterogeneous and deadly disease. Basal-like breast cancer (BLBC) is a particularly aggressive molecular subtype defined by a robust cluster of genes expressed by epithelial cells in the basal or outer layer of the adult mammary gland. BLBC is a major clinical challenge because these tumors are prevalent in young woman, often relapsing rapidly. Additionally, most (but not all) basal-like tumors lack expression of steroid hormone receptors (estrogen receptor and progesterone receptor) and human epidermal growth factor receptor 2, limiting targeted therapeutic options for these predominantly triple-negative breast cancers. This minireview will focus on new insights into the molecular etiology of these poor-prognosis tumors that underlie their intrinsic genomic instability, deregulated cell proliferation and apoptosis, and invasive tumor biology. We will also review ongoing efforts to translate these fundamental insights into improved therapies for women with BLBC.
Breast cancer is the most common noncutaneous malignancy in women and second only to lung carcinoma in cancer mortality (1). In the United States, women have an estimated 12.0% lifetime risk of being diagnosed with breast cancer; the risk of breast cancer-related death is estimated at 2.82% (2). One of the genuine triumphs of personalized medicine in the last decade has been the molecular classification of breast cancer based on gene expression profiles. Transcriptome analyses of human breast tumors have revealed remarkably robust molecular subtypes with distinctive gene signatures (differential expression of an ∼500 intrinsic gene subset) and clinical outcomes (3–6). These intrinsic subtypes include luminal A and B, defined by the expression of genes in the luminal epithelial layer of the mammary gland, such as the estrogen receptor (ER) and its targets; human epidermal growth factor receptor 2 (HER2/ErbB2), characterized by high expression of the HER2 oncogene and neighboring genes on its 17q12–21 amplicon; basal-like, defined by expression of genes characteristic of the outer or basally located epithelial layer of the mammary gland, such as cytokeratins 5 and 17 and the epidermal growth factor receptor (EGFR/HER1); and normal-like, which express adipose and other nonepithelial genes and have high basal-like and low luminal gene expression. Strikingly, these molecular subtypes are strongly associated with survival: luminal A tumors have the most favorable prognosis, normal-like tumors have an intermediate prognosis; luminal B, HER2-positive, and basal-like tumors are associated with the shortest relapse-free and overall survival (4–6). Molecular subtypes also predict treatment response, with HER2-positive and basal-like tumors paradoxically having higher rates of complete response to presurgery chemotherapy than luminal and normal-like tumors (7). Overall, gene profiling has radically altered our conceptualization of breast cancer and provided a myriad of translational opportunities to improve (i.e. personalize) prognostic and therapeutic approaches to this disease.
Of all the molecular subtypes, basal-like breast cancer (BLBC) remains the greatest challenge because of its clinically aggressive nature and poorly characterized molecular pathogenesis. Unlike ER-positive luminal tumors and HER2-positive tumors, the basal-like subtype typically lacks expression of the molecular targets that confer responsiveness to highly effective targeted therapies such as tamoxifen and aromatase inhibitors (ER) or trastuzumab (HER2 amplification) (8, 9). Indeed, identification of the relevant molecular targets in BLBC remains a formidable challenge. Although there are several excellent reviews on various aspects of BLBC (8–12), the present review will highlight recent discoveries that have led to fundamentally new insights into the molecular etiology of these tumors and emerging efforts to translate these discoveries into improved therapies.
Basal-Like or Triple-Negative: Capturing the Gene Signature in the Clinic
Although the intrinsic subtypes have robust prognostic and predictive value (4–7), standard microarray-based transcriptional profiling, which requires fresh frozen tissue, is not currently feasible in the clinic. One potential strategy to overcome this translational barrier is a 50-gene subtype predictor that utilizes quantitative RT-PCR analysis of clinically available breast tumor tissue to determine molecular subtype (13); however, this methodology is a research tool at present that needs to be validated in additional cohorts. A more practical strategy is the use of immunohistochemistry to identify protein expression surrogates for the basal-like gene signature. Basal-like tumors are generally ER- and progesterone receptor (PR)-negative and also lack high expression/amplification of HER2 (i.e. triple-negative tumors), but not uniformly so (4, 5, 14). In one series, 71% of triple-negative breast tumors had a basal-like gene profile, whereas 29% did not (14). To address these disparities between basal-like and triple-negative tumors, several biomarker surrogates have been proposed that incorporate basal-like markers in combination with hormone receptor negativity. A four-biomarker panel defined by positive staining for one basal maker [cytokeratin 5/6 and/or EGFR], and negative staining for both ER and HER2 has been shown to be 76% sensitive and 100% specific for BLBC identified by basal-like gene expression profile (15). The four-biomarker panel has also been expanded by adding PR or additional basal cytokeratins (cytokeratin 14 or -17) to identify BLBC (16–18). This distinction between BLBC and triple-negative breast cancer (TNBC) is not merely an academic subtlety: triple-negative tumors that express basal markers have distinct molecular lesions (e.g. p53 stabilization and higher mitotic indices) and are associated with worse survival than triple-negative tumors that lack basal-like markers (Table 1) (17, 18). In this review, BLBC refers to tumors defined by gene expression or biomarker surrogates, whereas TNBC refers to ER, PR, and HER2-negative tumors not otherwise characterized.
Table 1.
Comparison of TNBCs: basal-like vs. nonbasal
| Triplenegative:nonbasal | Triplenegative:basal-like | |
|---|---|---|
| Basal-like markers: | ||
| CK 5/6 and/or EGFR-positive by IHC | No (47.4%) | Yes (52.6%) (17) |
| CK 5/6, CK14, CK17,and/or EGFR-positiveby IHC | No (28.9%) | Yes (71.1%) (18) |
| Basal-like gene profile | No (28.5%) | Yes (71.5%) (14) |
| p53 positive by IHC | 41.0% | 62.0% (18) |
| Distant metastasis, 10-yrfollow-up | 26.0% | 37.0% (18) |
| Breast cancer-specific survival, 10-yr follow-up | 71.6% 75.5% | 62.2% (17) 56.6% (18) |
IHC, Immunohistochemistry; CK, cytokeratin.
Epidemiology and Clinical Presentation
The prevalence of BLBC ranges from 12.3–36.7% of breast cancer cases in different patient cohorts (3–6, 15, 16, 18–25). BLBC is more common in African and African-Americans and in young and premenopausal women (especially among African-Americans) (15, 23, 24). The incidence of BLBC is inversely related to duration of lactation. However, unlike luminal tumors, BLBC is more common in women with increased parity, early age of menarche, and first full-term pregnancy before age 26 (23, 24). Although body mass index has not been shown to be significantly associated with BLBC as it has for the other molecular subtypes, an increased waist-hip ratio is positively associated with BLBC in premenopausal women (23, 24).
Much more epidemiological data exists for TNBC. In addition to the above BLBC risk factors, TNBC is more common in Hispanic women (25–29), in women with lower social economic status (27), in women with the metabolic syndrome (30), and in some studies in women with more than 1 yr of oral contraceptive use or use before age 18 (31, 32). Of note, young African-American women are especially adversely affected by TNBC: not only is their recurrence-free and overall survival rate reduced compared with postmenopausal and non-African-American women with TNBC, those women with stage III/IV disease at diagnosis have only a 14% 5-yr survival rate compared with 37% and 36% survival in Hispanic and Caucasian women, respectively (16, 26).
Despite often presenting as large, advanced stage tumors at diagnosis, TNBC/BLBC tumors may be more sensitive to presurgery chemotherapy as evidenced by higher rates (22–45%) of pathological complete response (i.e. no tumor found at surgery) (7, 33, 34). Meta-analyses by the Early Breast Cancer Trialists' Collaborative Group have examined the impact of combined chemotherapy on breast cancer recurrence and include early data from both treated and untreated controls. Among patients with ER-negative breast cancer (TNBC and HER2 subtypes) under the age of 50, the 5-yr recurrence rate in the untreated cohort is 38.8%, which is reduced to 25.5% by combined chemotherapy (35). Hence, chemotherapy is highly effective in patients with ER-negative breast cancer, including TNBCs (35, 36). The higher response rate to neoadjuvant chemotherapy may reflect the typically high tumor grade and mitotic index of BLBC (16, 24, 37). Importantly, patients who achieve pathological complete response have survival rates similar to non-TNBC/BLBC patients (33, 34). However, the majority of women with TNBC/BLBC do not have a complete response and are at high risk for early relapse within the first 2–5 yr after treatment, resulting in an overall lower 5-yr survival rate (26, 28, 34, 38). TNBC/BLBC has a distinctive pattern of organ-specific distant metastases, with the lungs, liver, and central nervous system as the preferred sites (39–41). A particularly devastating aspect of TNBC is the high frequency of parenchymal central nervous system metastases that are observed in up to 46% of women with metastatic TNBC (6.7%–9.6% of all TNBC cases) and are associated with a median survival of less than 5 months from the time of diagnosis (41–43). The reader is referred to the elegant work of Joan Massagué and colleagues (44–46) who have identified genes regulating organic-specific metastasis in breast cancer.
Molecular Pathogenesis
BRCAness of BLBC: the BCRA1 connection
The study of hereditary breast cancer has revealed high-penetrance breast cancer susceptibility genes (BRCA1 and BRCA2) that function as tumor suppressor gene products that preserve genome integrity (47, 48). Women with inactivating germline mutations in BRCA1 or BRCA2 have up to an 85% chance of developing breast cancer in their lifetime. Loss of heterozygosity (LOH) of the second BRCA½ allele in breast epithelium results in disruption of double-strand DNA repair via the high-fidelity homologous recombination repair pathway. Instead, cells rely on nonhomologous end joining, which is error prone and may result in chromosomal translocations because the repair is not templated by the damaged DNA's sister chromatid sequence. Normally, the presence of double-stranded DNA damage leads to cell cycle arrest and cell death, but in the presence of p53 mutations, the checkpoint arrest is abrogated and widespread genomic instability and aneuploidy ensue (47).
One of the earliest insights into the pathogenesis of BLBC was the observation that sporadic BLBC phenocopies many aspects of hereditary breast cancer arising in BRCA1 (but not BRCA2) carriers (49). Specifically, breast tumors in BRCA1 carriers and nonhereditary BLBC share the following features: 1) they are largely triple negative and basal like by gene expression profile and biomarker surrogates (5, 50); 2) they are characterized by high tumor grade, high mitotic indices, p53 mutations, and chromosomal instability (51, 52); 3) they frequently have X chromosome abnormalities, including defects in X chromosome inactivation, a well-established function of BRCA1 (53, 54); and 4) they have similar clinical features, including young age at presentation, poor prognosis, early relapses, and favorable response to DNA-damaging chemotherapy (9, 49). However, mutational inactivation of BRCA1 is uncommon in sporadic breast cancer (54, 55), suggesting that other mechanisms account for the BRCA1 dysfunction phenotype of these tumors. BRCA1 inactivation by promoter CpG island methylation, often in combination with BRCA1 LOH, has been observed in 11–13% of sporadic breast cancers, the majority of which are ER-negative tumors (56, 57). Additionally, the dominant-negative transcriptional regulator ID4 has been shown to regulate BRCA1 expression and to be preferentially expressed in BLBC (58, 59). Nevertheless, many cases of BLBC have normal expression and nuclear localization of BRCA1 (54), suggesting that epigenetic and/or genetic abnormalities in other BRCA1-associated proteins [e.g. Fanconi anemia proteins, ataxia telangiectasia mutated gene product, Bloom syndrome protein, or Rad50 (60, 61)] might underlie the BRCA1 dysfunction phenotype of BLBC. Regardless of the underlying molecular mechanisms, the BRCAness of BLBC has emerged as a promising therapeutic target in these poor-prognosis tumors.
Apoptosis resistance
Defects in the apoptotic cell death machinery play a critical role in the pathogenesis of cancer, and BLBCs are characterized by a distinctive pattern of apoptotic gene abnormalities. As noted, BLBC has a high frequency (44–82%) of TP53 mutations, which impair DNA damage-induced checkpoint activation and apoptosis, thereby promoting genome instability (4, 16, 62). Indeed, loss of one TP53 allele in mice with mammary-specific deletion of BRCA1 dramatically accelerates mammary tumorigenesis (63), suggesting that p53 mutations may act synergistically with functional BRCA1 defects in sporadic BLBC to drive tumor initiation. The receptor tyrosine kinase EGFR is expressed in 39–54% of BLBC and confers resistance to apoptosis by ligand-dependent activation of the phosphatidylinositol-3 (PI3)-kinase/Akt/mTOR pathway (15, 64, 65). Another characteristic apoptotic abnormality in BLBC is expression of the molecular chaperone αB-crystallin, which suppresses apoptosis by inhibiting proteolytic activation of the proapoptotic protease caspase-3 (66, 67). αB-Crystallin is expressed in 45% of BLBC and only rarely (5%) in other molecular subtypes. Notably, αB-crystallin expression is associated with resistance to presurgery chemotherapy and poor survival in breast cancer patients, whereas ectopic expression of this molecular chaperone leads to an invasive tumor phenotype in preclinical models (67, 68). Additionally, loss of the phosphatase and tensin analog (PTEN) tumor suppressor gene, with resultant aberrant activation of the antiapoptotic PI3-kinase/Akt/mTOR pathway, is commonly observed in TNBC (69–71). Intriguingly, PTEN inactivation has also recently been linked to chromosome instability due to defects in Rad51-mediated DNA double-strand break repair, resulting in further genome instability in BLBC (72). Furthermore, mutational inactivation of Fbxw7, a component of an E3 ubiquitin ligase that degrades mTOR and Cyclin E (see next paragraph), has been reported in BLBC and likely results in enhanced levels of these key regulatory molecules (73–75).
Proliferation
As noted, BLBC is characterized by high mitotic indices and rates of proliferation (76). EGFR is commonly expressed in these tumors and promotes cell proliferation via activation of the Ras/MAPK/MAPK kinase (MEK) pathway (64). BLBC is also characterized by low expression of the RB and Cyclin D1 genes and high expression of E2F3 and Cyclin E genes (77). Cell proliferation requires progression through the G1 to S cell cycle transition that is negatively regulated by the RB tumor suppressor gene product (78). Cyclin D-CDK4/CDK6 complexes phosphorylate RB and promote S-phase entry by releasing E2F family transcription factors, which induce Cyclin E expression. Cyclin E-CDK2 complexes induce additional phosphorylation of RB and ensure S-phase entry. A 59-gene expression signature reflecting RB pathway dysregulation and a distinct RB LOH signature were more prevalent in TNBC than other subtypes (79, 80). Cyclin E1 is present in higher copy number in BLBC than other molecular subtypes, and its expression correlates with poor survival in breast cancer (81–83). Taken together, these studies suggest a specific role of RB loss and/or Cyclin E overexpression in the highly proliferative phenotype of BLBC.
Epithelial-mesenchymal transition
A key step in the metastatic cascade of epithelial tumors is the epithelial-mesenchymal transition (EMT), a carefully orchestrated program whereby carcinoma cells lose epithelial characteristics, such as cell-cell adhesion and polarity, and acquire mesenchymal features, facilitating invasion of the extracellular matrix (84). Developmental EMT pathways may be co-opted by BLBC or stimulated by environmental pressures such as hypoxia (85, 86). EMT markers such as N-cadherin and vimentin are frequently highly expressed in BLBC, whereas epithelial markers such as E-cadherin are often lost (87, 88). Down-regulation of E-cadherin expression and promotion of EMT have shown to be achieved in BLBC through the activation of TGF-β, Wnt, and Notch pathways leading to expression of EMT-associated transcription factors such as FOXC2, Twist, Slug, Snail, and LBX1 (89–91). EGFR also promotes EMT by inducing expression of Twist and plays a key role in cell motility and invasion (92, 93). Moreover, the Src family tyrosine kinase LYN is an EMT mediator that is commonly expressed in BLBC and is associated with poor survival (94). Of note, the recently defined claudin-low gene expression signature is characterized by low expression of cell-cell adhesion genes (e.g. Claudins and E-cadherin) leading to an EMT phenotype (95). Most closely linked with the basal-like subtype, claudin-low tumors are generally ER and HER2 negative; however, claudin-low tumors have variable expression of basal-like markers and so constitute a distinct intrinsic gene expression subtype (95, 96). Overall, these findings point to multiple mechanisms promoting EMT and an invasive tumor phenotype in BLBC.
Angiogenesis
Vascular endothelial growth factor A (VEGFA/VEGF) is a potent mitogen for endothelial cells and regulates tumor angiogenesis and vascular permeability, thereby promoting primary tumor growth and metastasis (97). The actions of VEGF on endothelial cells are mediated by the receptor tyrosine kinases VEGF receptor (VEGFR)1/Flt1 and VEGFR2/KDR/Flk1 as well as Neuropilin coreceptors. VEGF is expressed at approximately 3-fold higher levels in TNBC compared with non-TNBC as determined by ELISA (98). Moreover, the VEGF gene is located on a chromosomal region (6p21.2–6p12.3) characterized by frequent copy number gain in TNBC, and specific probes for VEGF have confirmed VEGF gene copy gain and increased mRNA expression in approximately one third of TNBCs (99). A 13-gene VEGF signature was recently reported to predict distant metastases in breast cancer (100), underscoring the link between VEGF and metastasis. Breast tumors with p53 mutations have higher VEGF levels, suggesting a mechanism by which p53 mutation may promote angiogenesis (101). Additionally, high VEGFR2 expression has been observed in a subset of TNBC and correlates with shorter survival (102). Interestingly, VEGF also promotes breast cancer growth independently of its proangiogenic actions by an autocrine loop involving VEGFR1 and the VEGF coreceptor Neuropilin-1; EGFR activation acts synergistically with this VEGF autocrine loop by inducing VEGF, VEGFR, and Neuropilin-1 via a MAPK-dependent mechanism (103). Collectively, these results implicate the VEGF pathway in the etiology of TNBC and provide a strong rationale for targeting this pathway therapeutically.
Insights from microRNA (miRNA) expression analysis
miRNAs are noncoding RNAs that regulate gene expression predominately at the level of translation. After sequential processing, mature approximately 22-base miRNAs become part of silencing complexes that associate with the 3′-untranslated regions of target genes and inhibit translation and/or promote target RNA degradation (104). miRNA cancer signatures have been reported, as have miRNA signatures characteristic of different histological and molecular subtypes of breast cancer (105–107). The expression of the let-7 family of miRNAs, including let-7a, is commonly reduced in BLBC/TNBCs (105, 108). Let-7 miRNAs are also down-regulated in rare breast cancer cells with stem-like properties (breast cancer stem cells) and have been implicated in the self-renewal, tumor-initiating, and metastatic properties of breast cancer stem cells via their actions on several targets including H-Ras, HMGA2, and IL-6 (109, 110). These findings suggest a link between reduced expression of let-7 miRNAs and an aggressive cancer stem cell phenotype in BLBC. Moreover, miR-126, a suppressor of primary/metastatic tumor growth and cell proliferation, is down-regulated in a subset of BLBCs (105, 111), suggesting that reduced expression of miR-126 may promote tumor progression and metastasis in these breast carcinomas.
Integrating genomic and transcriptome analyses
Comparative genome hybridization (CGH) studies using bacterial artificial chromosome and higher resolution single nucleotide polymorphism and oligo arrays have revealed distinctive chromosomal aberrations in each molecular subtype of breast cancer. Copy number aberrations (CNAs) are distributed throughout the genome in BLBC resulting in a sawtooth pattern, which is similar to that seen in BRCA1-associated hereditary breast cancer (112). In contrast to luminal and HER2 tumors, regions of high-level amplification are rare in BLBC (112–114). Low-level copy number aberrations in BLBC often include gains at 1q, 6p, 8p, and 10p, with losses at 4p, 5q, 14q, and 15q (112–115). Cluster analyses of CGH data have identified three molecular classes of breast cancer based on CNAs (99); 64% of TNBCs fall into class I, characterized by frequent gains on chromosome 6p21-p23 and frequent losses on chromosome 15q14-q22. 6p21-p23 contains many cancer-relevant genes; specific probes for VEGFA and E2F3 were gained in approximately one third of TNBCs but only 10% or less of non-TNBCs. Furthermore, EGFR gain and PTEN loss were observed with similar frequencies in TNBC. These specific CNAs were accompanied by corresponding mRNA expression levels, i.e. high expression of VEGFA, E2F3, and EGFR and low expression of PTEN in TNBC. Another study integrating CGH and transcriptional profiles of TNBCs identified 40 genes that were both amplified and overexpressed, including FGFR2, BUB3, RAB20, NOTCH3, and PKN1 (116). FGFR2 is amplified in 4% of TNBC and encodes a receptor tyrosine kinase that confers resistance to apoptosis by activating the PI3-kinase/Akt/mTOR pathway (116, 117). TNBC cells with FGFR2 amplification are selectively sensitive to apoptosis induction by silencing FGFR2 or a pan-FGFR tyrosine kinase inhibitor, highlighting the functional relevance of this pathway in FGFR2-amplified TNBC cells (116). Taken together, these studies underscore the translational potential of integrating CGH and transcriptome platforms to illuminate molecular pathways deregulated in BLBC.
Translating Molecular Profiles into Targeted Therapies
Chemotherapy: a platinum lining?
Given the absence of validated molecular targets in TNBC, conventional chemotherapy (typically including a DNA-damaging anthracycline such as doxorubicin and a microtubule-stabilizing taxane) has been the only therapeutic option for women with these poor-prognosis tumors. Although the rates of pathological complete response (22–45%) for presurgery chemotherapy are higher for TNBC than luminal and normal-like tumors, the majority of women with TNBC have residual disease and are at high risk for relapse and death within the first 2–5 yr of diagnosis (7, 33, 34). Moreover, the nonspecific cytotoxicity of these agents results in significant dose-limiting side effects. Hence, the development of targeted therapies with improved therapeutic indices is of paramount importance (Fig. 1 and Table 2).
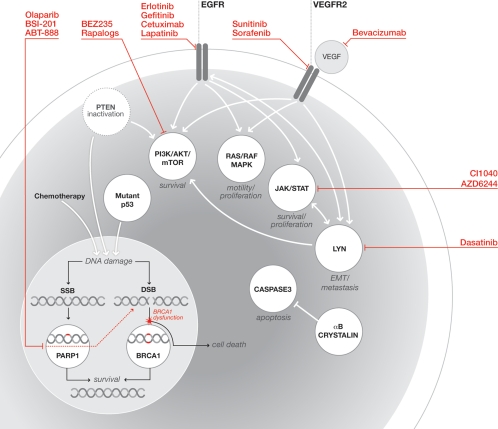
Fig. 1.
Schematic representation of key signal transduction pathways implicated in the pathogenesis of BLBC and targeted therapies. Commonly dysregulated pathways and their biological outcomes are depicted. Representative drug inhibitors are indicated in red. DSB, double-strand break; JAK, Janus family of tyrosine kinases; SSB, single-strand break; PI3K, phosphatidylinositol-3-kinase; STAT, signal transducer and activator of transcription.
Table 2.
A subset of basal-like markers associated with poor survival: candidate BLBC molecular targets
| Gene | Function | Expression pattern | Impact |
|---|---|---|---|
| αB-crystallin | Antiapoptotic small heat shock protein | High expression in 45% of BLBC | Hazard ratio 2.23 for reduced survival (67) |
| Cyclin E | G1-S phase cell cycle regulation | Preferentially expressed by BLBC | High expression associated with reduced survival (82, 83) |
| EGFR | Receptor tyrosine kinase | High expression in 39–54% of TNBC | Hazard ratio 1.98 for reduced survival (66) |
| LYN | Src family tyrosine kinase, EMT mediator | High expression in ∼50% of TNBC | Hazard ratio 2.29 for reduced survival (94) |
| PTEN | Inhibits PI3K/mTOR/ AKT; loss leads to chromosome instability | Expression lost in ∼1/3 of TNBC (99) | Hazard ratio 4.63 for reduced survival (not specific to BLBC) (144) |
| RB1 | Tumor suppressor | RB dysregulation and LOH common in BLBC | RB LOH gene signature predictive of pathological complete response and poor survival (79, 80) |
| VEGFA | Angiogenesis | High expression in 34% of TNBC (99) | VEGF 13-gene signature hazard ratio 1.54 for relapse-free survival (not specific to BLBC) (100) |
| VEGFR2 | Angiogenesis | Preferentially expressed by 22% of TNBC | Hazard ratio 2.6 for reduced survival (102) |
Based on the BRCA1 dysfunction phenotype of BLBC, one approach has been the exploration of platinum chemotherapy agents (carboplatin, cisplatin, and others) in these patients. Platinum agents produce DNA cross-links, which lead to DNA double-strand breaks, normally repaired by BRCA½-mediated high-fidelity homologous recombination repair mechanisms (47, 118). Consequently, BRCA½-deficient cells are highly sensitive to apoptosis induced by these agents (119). Cisplatin also promotes apoptosis in TNBC by disrupting a complex between the p53 family members ΔNp63 and TAp73 that is present selectively in TNBC with mutant TP53 (120). Specifically, cisplatin induces c-Abl-mediated phosphorylation of TAp73, which releases pro-apoptotic TAp73 from the inhibitory complex and triggers apoptosis. A recent small clinical study of 28 women with TNBC (including two BRCA1 mutation carriers) who were treated with presurgery cisplatin resulted in a 22% pathological complete response rate, similar to that observed with nonplatinum agents (121). Both women with BRCA1 mutations had a complete response, and breast tumors with low BRCA1 mRNA expression or TP53 mutations were associated with a favorable cisplatin response, suggesting that BCRA1 dysfunction may be linked to cisplatin response. Whether platinum agents will indeed improve survival in TNBC will have to await the outcome of several current clinical studies.
Synthetic lethality: a paradigm shift incancer therapy
One of the most exciting recent developments in translational cancer research is the concept of synthetic lethality (122). Two oncogenic pathways are in a synthetic lethal relationship if mutation of either oncogene is well tolerated, but mutation of both results in robust cell death. Synthetic lethal screens have been used to identify drugs or genes that induce cell death only in the presence of specific oncogenic alterations. For example, drug inhibitors of poly(ADP) ribose polymerase (PARP), an enzyme involved in DNA base-excision repair, prevent the repair of DNA single-strand breaks, which are converted to double-strand breaks at stalled DNA replication forks. These DNA double-strand breaks are normally repaired by BRCA½-mediated homologous recombination repair, and there are no untoward consequences for the cell. However, in the presence of BRCA1 or BRCA2 mutations, this repair mechanism is defective: cells accumulate DNA double-strand breaks and ultimately undergo apoptosis. Hence, PARP and BRCA½ are in a synthetic lethal relationship: PARP inhibitors potently induce cell death only in cancer cells with mutations in BRCA1 or BRCA2 (123, 124). Indeed, mutations in BRCA1 or BRCA2 confer 57- and 133-fold increase sensitivity to PARP inhibitors compared with cells with wild-type BRCA1 or BRCA2 (124). Preclinical studies of several PARP inhibitors have demonstrated impressive single-agent antitumor activity in multiple BRCA½-deficient tumor models and have shown robust synergy between PARP inhibitors and DNA-damaging agents, including platinum agents (123–125). In a small phase I clinical study, the oral PARP inhibitor olaparib (AZD2281) was well tolerated and induced partial and complete responses in some patients with BRCA1- or BRCA2-associated-cancer (12 of 19 patients had a clinical benefit), but the drug had no activity in nonmutation carriers (126). Preliminary analysis of a randomized phase II study in 86 patients with metastatic TNBC demonstrated that adding a PARP inhibitor (BSI-201) to chemotherapy (gemcitabine plus carboplatin) significantly improved progression-free (>2-fold increase) and overall survival compared with chemotherapy alone (127). There are currently 13 clinical trials listed in ClinicalTrials.gov to evaluate PARP inhibitors alone and in combination with platinum agents, which should provide invaluable information. As with all cancer therapies, de novo and/or acquired resistance to PARP inhibitors are likely to be encountered. Indeed, intragenic mutations in BRCA½, which restore expression of the wild-type protein, have been described as mechanisms of resistance to platinum agents and PARP inhibitors (128, 129). Nevertheless, PARP inhibitors have the potential to transform our therapeutic approach to hereditary and sporadic TNBC, and the pace of clinical translation in this area is likely to be unprecedentedly rapid given the dearth of existing options.
Antiangiogenics
Given the accumulating evidence of aberrant VEGF pathway activation in BLBC (and other neoplasms), antiangiogenic therapies targeting VEGF and its receptors have emerged as promising therapies for BLBC. Many small-molecule multikinase inhibitors, including sunitinib and sorafenib, have been developed as potential antiangiogenic agents (130). Sunitinib inhibits several receptor tyrosine kinases including VEGFR, platelet-derived growth factor receptor, c-KIT, RET, CSF-1R, and FMS-like tyrosine kinase 3. A phase II study of 64 women with metastatic breast cancer previously treated with an anthracycline and taxane found an 11% response rate to sunitinib as a single agent; three of 20 TNBCs had demonstrable response to treatment (131). In a later study of 22 patients with newly diagnosed locally advanced or metastatic breast cancer, the addition of sunitinib to paclitaxel was shown to produce an objective response in about one third of patients, including three of nine TNBCs (132). Other VEGFR multikinase inhibitors have not shown as much promise. A phase II trial of sorafenib, which has kinase specificity similar to sunitinib, failed to demonstrate any response in 23 metastatic breast cancer patients, 52% of whom were ER negative (133).
The anti-VEGF antibody bevacizumab has been shown to prolong disease-free survival of breast cancer patients, including TNBC patients also treated with paclitaxel by 4 months on average vs. treatment with paclitaxel alone; however, overall survival was not affected (134). When combined with the small-molecule reversible EGFR kinase inhibitor erlotinib, bevicizumab showed limited activity in metastatic breast cancer. In this study of 38 patients, 50% of the tumors were TNBC, and 10 of 19 demonstrated EGFR expression; however, response rates of the distinct tumor subtypes were not reported (135). Currently, 32 active trials are listed at ClinicalTrials.gov involving breast cancer and VEGF-based antiangiogenic agents. To date, the response to antiangiogenic agents has been disappointing in unselected TNBC patients. It remains to be seen whether the discovery of biomarkers to stratify patients who are likely to respond to these agents and/or identification of synergistic combination therapies will improve treatment response. An additional cautionary note is the recent observation in preclinical studies that antiangiogenic agents may paradoxically increase distant metastases (136).
Targeting EGFR and downstream kinases
Based on the frequent expression of EGFR in BLBC, small-molecule and antibody-based EGFR inhibitors are being explored as targeted therapies. In a small study of 41 previously untreated breast cancer patients, presurgery erolotinib reduced phospho-EGFR levels in most patients, but erolotinib inhibited cell proliferation, phospho-MAPK, and phospho-Akt levels only in ER-positive breast cancer (not in TNBC or HER2-positive tumors) (137). Phase I and II studies of another EGFR small-molecule inhibitor (gefitinib), a humanized anti-EGFR monoclonal antibody (cetuximab), and a dual EGFR/HER2 dual kinase small-molecule inhibitor (lapatinib), which is Food and Drug Administration (FDA) approved for relapsed HER2-positive breast cancer, have also failed to show efficacy of these agents as single agents or combined with chemotherapy in patients with largely pretreated metastatic TNBC (138–140). One potential explanation for the limited impact of EGFR-targeted therapies in TNBC to date is the lack of a biomarker (e.g. gene mutation or amplification) to identify potential responders; predictive biomarkers have been a cornerstone for the successful translation of targeted therapies (141). Moreover, constitutive activation of signaling pathways downstream of EGFR in TNBC, such as MEK/MAPK, PI3-kinase/Akt/mTOR, and Src family kinases (e.g. Lyn), may confer resistance to EGFR inhibitors. Indeed, preclinical studies suggest that TNBC cells may be particularly sensitive to MEK inhibitors by virtue of high expression of MAPK pathway genes (142). Intriguingly, MEK inhibition in TNBC cells led to activation of the PI3-kinase/Akt/mTOR pathway, whereas combined inhibition of the MEK and PI3-kinase/Akt/mTOR pathway resulted in synergistic cytotoxity or growth arrest. Similarly, BLBC cells highly express LYN, a Src family kinase, and are exquisitely sensitive to dasatinib, an inhibitor of Src and Abl kinases (94, 143). Collectively, these studies strongly suggest that identification of predictive biomarkers and targeting of multiple kinase pathways (e.g. MEK/MAPK and PI3-kinase/Akt/mTOR) is likely to be required for optimal therapeutic benefit in BLBC. Given the explosion of small-molecule inhibitors of these pathways, it seems likely that clinical data will be forthcoming shortly.
Concluding Comments
A decade has elapsed since the initial recognition of BLBC as a distinctive molecular subtype with a basal epithelial gene signature and an aggressive clinical course characterized by early relapses and poor survival. Major pathogenic insights include the apparent BRCA1 dysfunction phenotype of these sporadic tumors, resulting in widespread genomic instability and potentially profound vulnerability to PARP inhibitors due to synthetic lethality. Preclinical and clinical studies have also identified other signature molecular abnormalities in BLBC, including deregulated activation of αB-crystallin, EGFR and downstream kinases, and VEGF, resulting in an invasive apoptosis-resistant tumor phenotype. Clinical translation of these molecular insights is currently ongoing and will likely require careful patient selection based on the specific molecular targets of therapeutic agents and rationally designed combinatorial regimens to counteract treatment resistance. Although these obstacles are potentially daunting, early clinical success with some agents, particularly PARP inhibitors, suggests that targeted therapies for BLBC are within our grasp in the near future.
Acknowledgments
We thank Ruth Schmidt for her expert information design assistance. We regret that page limitations have precluded us from citing many relevant manuscripts.
Address all correspondence and requests for reprints to: Vincent L. Cryns, Department of Medicine, Lurie 4-113, Feinberg School of Medicine, Northwestern University, 303 East Superior Street, Chicago, Illinois 60611. E-mail: v-cryns@northwestern.edu.
This work was supported by National Institutes of Health grant T32DK007169 (to D.J.T.) and by grants from the Breast Cancer Research Foundation and the Lynn Sage Cancer Research Foundation (to V.L.C).
Disclosure Summary: V.L.C. has received honoraria for lectures related to this minireview.
Footnotes
- BLBC
- Basal-like breast cancer
- CGH
- comparative genome hybridization
- CNA
- copy number aberration
- EGFR
- epidermal growth factor receptor
- EMT
- epithelial-mesenchymal transition
- ER
- estrogen receptor
- HER2
- human epidermal growth factor receptor 2
- LOH
- loss of heterozygosity
- MEK
- MAPK kinase
- miRNA
- microRNA
- PARP
- poly(ADP) ribose polymerase
- PI3
- phosphatidylinositol-3
- PR
- progesterone receptor
- PTEN
- phosphatase and tensin analog
- TNBC
- triple-negative breast cancer
- VEGF
- vascular endothelial growth factor
- VEGFR
- VEGF receptor.
References
- 1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. 2009. Cancer statistics, 2009. CA Cancer J Clin 59:225–249 [DOI] [PubMed] [Google Scholar]
- 2. Horner MJ, Ries LAG, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinchcomb DG, Edwards BK, eds 2009. SEER cancer statistics review, 1975–2006, National Cancer Institute; Bethesda, MD [Google Scholar]
- 3. Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Børresen-Dale AL, Brown PO, Botstein D. 2000. Molecular portraits of human breast tumours. Nature 406:747–752 [DOI] [PubMed] [Google Scholar]
- 4. Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thørsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98:10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, Brown PO, Børresen-Dale AL, Botstein D. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100:8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM. 2006. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med 355:560–569 [DOI] [PubMed] [Google Scholar]
- 7. Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, Morandi P, Fan C, Rabiul I, Ross JS, Hortobagyi GN, Pusztai L. 2005. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 11:5678–5685 [DOI] [PubMed] [Google Scholar]
- 8. Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, Sledge GW, Carey LA. 2008. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res 14:8010–8018 [DOI] [PubMed] [Google Scholar]
- 9. Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. 2006. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med 12:537–544 [DOI] [PubMed] [Google Scholar]
- 10. Anders C, Carey LA. 2008. Understanding and treating triple-negative breast cancer. Oncology (Huntingt) 22:1233–1239; discussion 1239–1240:1243 [PMC free article] [PubMed] [Google Scholar]
- 11. Hudis CA. 2010. From the guest editor: triple-negative breast cancer. Cancer J 16:10–11 [DOI] [PubMed] [Google Scholar]
- 12. Turner NC, Reis-Filho JS. 2006. Basal-like breast cancer and the BRCA1 phenotype. Oncogene 25:5846–5853 [DOI] [PubMed] [Google Scholar]
- 13. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. 2009. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27:1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D. 2008. How basal are triple-negative breast cancers? Int J Cancer 123:236–240 [DOI] [PubMed] [Google Scholar]
- 15. Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. 2004. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10:5367–5374 [DOI] [PubMed] [Google Scholar]
- 16. Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC. 2006. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295:2492–2502 [DOI] [PubMed] [Google Scholar]
- 17. Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. 2008. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14:1368–1376 [DOI] [PubMed] [Google Scholar]
- 18. Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet JS, Akslen LA, Evans AJ, Blamey R, Reis-Filho JS, Foulkes WD, Ellis IO. 2009. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15:2302–2310 [DOI] [PubMed] [Google Scholar]
- 19. Perreard L, Fan C, Quackenbush JF, Mullins M, Gauthier NP, Nelson E, Mone M, Hansen H, Buys SS, Rasmussen K, Orrico AR, Dreher D, Walters R, Parker J, Hu Z, He X, Palazzo JP, Olopade OI, Szabo A, Perou CM, Bernard PS. 2006. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res 8:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM. 2006. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langerød A, Zhao H, Borgan Ø, Nesland JM, Bukholm IR, Ikdahl T, Kåresen R, Børresen-Dale AL, Jeffrey SS. 2007. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9:R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim MJ, Ro JY, Ahn SH, Kim HH, Kim SB, Gong G. 2006. Clinicopathologic significance of the basal-like subtype of breast cancer: a comparison with hormone receptor and Her2/neu-overexpressing phenotypes. Hum Pathol 37:1217–1226 [DOI] [PubMed] [Google Scholar]
- 23. Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, Hewitt SM, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Cartun R, Mandich D, Rymkiewicz G, Ligaj M, Lukaszek S, Kordek R, García-Closas M. 2007. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev 16:439–443 [DOI] [PubMed] [Google Scholar]
- 24. Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, Smith LV, Labbok MH, Geradts J, Bensen JT, Jackson S, Nyante S, Livasy C, Carey L, Earp HS, Perou CM. 2008. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat 109:123–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huo D, Ikpatt F, Khramtsov A, Dangou JM, Nanda R, Dignam J, Zhang B, Grushko T, Zhang C, Oluwasola O, Malaka D, Malami S, Odetunde A, Adeoye AO, Iyare F, Falusi A, Perou CM, Olopade OI. 2009. Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer. J Clin Oncol 27:4515–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. 2007. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California Cancer Registry. Cancer 109:1721–1728 [DOI] [PubMed] [Google Scholar]
- 27. Brown M, Tsodikov A, Bauer KR, Parise CA, Caggiano V. 2008. The role of human epidermal growth factor receptor 2 in the survival of women with estrogen and progesterone receptor-negative, invasive breast cancer: the California Cancer Registry, 1999–2004. Cancer 112:737–747 [DOI] [PubMed] [Google Scholar]
- 28. Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D. 2006. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol 24:5652–5657 [DOI] [PubMed] [Google Scholar]
- 29. Lund MJ, Trivers KF, Porter PL, Coates RJ, Leyland-Jones B, Brawley OW, Flagg EW, O'Regan RM, Gabram SG, Eley JW. 2009. Race and triple negative threats to breast cancer survival: a population-based study in Atlanta, Georgia. Breast Cancer Res Treat 113:357–370 [DOI] [PubMed] [Google Scholar]
- 30. Maiti B, Kundranda MN, Spiro TP, Daw HA. 2010. The association of metabolic syndrome with triple-negative breast cancer. Breast Cancer Res Treat 121:479–483 [DOI] [PubMed] [Google Scholar]
- 31. Dolle JM, Daling JR, White E, Brinton LA, Doody DR, Porter PL, Malone KE. 2009. Risk factors for triple-negative breast cancer in women under the age of 45 years. Cancer Epidemiol Biomarkers Prev 18:1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma H, Wang Y, Sullivan-Halley J, Weiss L, Marchbanks PA, Spirtas R, Ursin G, Burkman RT, Simon MS, Malone KE, Strom BL, McDonald JA, Press MF, Bernstein L. 2010. Use of four biomarkers to evaluate the risk of breast cancer subtypes in the women's contraceptive and reproductive experiences study. Cancer Res 70:575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM. 2007. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 13:2329–2334 [DOI] [PubMed] [Google Scholar]
- 34. Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L. 2008. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26:1275–1281 [DOI] [PubMed] [Google Scholar]
- 35. Early Breast Cancer Trialists' Collaborative Group 2005. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365:1687–1717 [DOI] [PubMed] [Google Scholar]
- 36. Berry DA, Cirrincione C, Henderson IC, Citron ML, Budman DR, Goldstein LJ, Martino S, Perez EA, Muss HB, Norton L, Hudis C, Winer EP. 2006. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA 295:1658–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Calza S, Hall P, Auer G, Bjöhle J, Klaar S, Kronenwett U, Liu ET, Miller L, Ploner A, Smeds J, Bergh J, Pawitan Y. 2006. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res 8:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. 2007. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13:4429–4434 [DOI] [PubMed] [Google Scholar]
- 39. Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. 2008. Subtypes of breast cancer show preferential site of relapse. Cancer Res 68:3108–3114 [DOI] [PubMed] [Google Scholar]
- 40. Luck AA, Evans AJ, Green AR, Rakha EA, Paish C, Ellis IO. 2008. The influence of basal phenotype on the metastatic pattern of breast cancer. Clin Oncol (R Coll Radiol) 20:40–45 [DOI] [PubMed] [Google Scholar]
- 41. Lin NU, Claus E, Sohl J, Razzak AR, Arnaout A, Winer EP. 2008. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: high incidence of central nervous system metastases. Cancer 113:2638–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dawood S, Broglio K, Esteva FJ, Yang W, Kau SW, Islam R, Albarracin C, Yu TK, Green M, Hortobagyi GN, Gonzalez-Angulo AM. 2009. Survival among women with triple receptor-negative breast cancer and brain metastases. Ann Oncol 20:621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heitz F, Harter P, Lueck HJ, Fissler-Eckhoff A, Lorenz-Salehi F, Scheil-Bertram S, Traut A, du Bois A. 2009. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer 45:2792–2798 [DOI] [PubMed] [Google Scholar]
- 44. Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massagué J. 2009. Genes that mediate breast cancer metastasis to the brain. Nature 459:1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. 2003. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3:537–549 [DOI] [PubMed] [Google Scholar]
- 46. Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, Kreike B, Zhang Y, Wang Y, Ishwaran H, Foekens JA, van de Vijver M, Massagué J. 2007. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci USA 104:6740–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Venkitaraman AR. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171–182 [DOI] [PubMed] [Google Scholar]
- 48. Wooster R, Weber BL. 2003. Breast and ovarian cancer. N Engl J Med 348:2339–2347 [DOI] [PubMed] [Google Scholar]
- 49. Turner N, Tutt A, Ashworth A. 2004. Hallmarks of ‘BRCAness' in sporadic cancers. Nat Rev Cancer 4:814–819 [DOI] [PubMed] [Google Scholar]
- 50. Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. 2002. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol 20:2310–2318 [DOI] [PubMed] [Google Scholar]
- 51. Consortium BCL. 1997. Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 and sporadic cases. Lancet 349:1505–1510 [PubMed] [Google Scholar]
- 52. Crook T, Brooks LA, Crossland S, Osin P, Barker KT, Waller J, Philp E, Smith PD, Yulug I, Peto J, Parker G, Allday MJ, Crompton MR, Gusterson BA. 1998. p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene 17:1681–1689 [DOI] [PubMed] [Google Scholar]
- 53. Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, Rasmussen TP, Klimke A, Marrese C, Marahrens Y, Deng CX, Feunteun J, Livingston DM. 2002. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell 111:393–405 [DOI] [PubMed] [Google Scholar]
- 54. Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. 2006. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9:121–132 [DOI] [PubMed] [Google Scholar]
- 55. Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, Eddington K, McClure M, Frye C, Weaver-Feldhaus J, Ding W, Gholami Z, Soederkvist P, Terry L, Jhanwar S, Berchuck A, Iglehart JD, Marks J, Ballinger DG, Barrett JC, Skolnick MH, Kamb A, Wiseman R. 1994. BRCA1 mutations in primary breast and ovarian carcinomas. Science 266:120–122 [DOI] [PubMed] [Google Scholar]
- 56. Catteau A, Harris WH, Xu CF, Solomon E. 1999. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 18:1957–1965 [DOI] [PubMed] [Google Scholar]
- 57. Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, Gabrielson E, Schutte M, Baylin SB, Herman JG. 2000. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 92:564–569 [DOI] [PubMed] [Google Scholar]
- 58. Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC, Ashworth A, Tutt AN. 2007. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26:2126–2132 [DOI] [PubMed] [Google Scholar]
- 59. Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, Wong-Staal F. 2001. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA 98:130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14:927–939 [PMC free article] [PubMed] [Google Scholar]
- 61. Wang W. 2007. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 8:735–748 [DOI] [PubMed] [Google Scholar]
- 62. Vousden KH, Lane DP. 2007. p53 in health and disease. Nat Rev Mol Cell Biol 8:275–283 [DOI] [PubMed] [Google Scholar]
- 63. Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. 1999. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet 22:37–43 [DOI] [PubMed] [Google Scholar]
- 64. Hynes NE, Lane HA. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 5:341–354 [DOI] [PubMed] [Google Scholar]
- 65. Rimawi MF, Shetty PB, Weiss HL, Schiff R, Osborne CK, Chamness GC, Elledge RM. 2010. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer 116:1234–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kamradt MC, Chen F, Cryns VL. 2001. The small heat shock protein αB-crystallin negatively regulates cytochrome c- and caspase-8-dependent activation of caspase-3 by inhibiting its autoproteolytic maturation. J Biol Chem 276:16059–16063 [DOI] [PubMed] [Google Scholar]
- 67. Moyano JV, Evans JR, Chen F, Lu M, Werner ME, Yehiely F, Diaz LK, Turbin D, Karaca G, Wiley E, Nielsen TO, Perou CM, Cryns VL. 2006. AlphaB-crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J Clin Invest 116:261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ivanov O, Chen F, Wiley EL, Keswani A, Diaz LK, Memmel HC, Rademaker A, Gradishar WJ, Morrow M, Khan SA, Cryns VL. 2008. αB-Crystallin is a novel predictor of resistance to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat 111:411–417 [DOI] [PubMed] [Google Scholar]
- 69. Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmström PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. 2005. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 65:2554–2559 [DOI] [PubMed] [Google Scholar]
- 70. Saal LH, Gruvberger-Saal SK, Persson C, Lövgren K, Jumppanen M, Staaf J, Jönsson G, Pires MM, Maurer M, Holm K, Koujak S, Subramaniyam S, Vallon-Christersson J, Olsson H, Su T, Memeo L, Ludwig T, Ethier SP, Krogh M, Szabolcs M, Murty VV, Isola J, Hibshoosh H, Parsons R, Borg A. 2008. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet 40:102–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Marty B, Maire V, Gravier E, Rigaill G, Vincent-Salomon A, Kappler M, Lebigot I, Djelti F, Tourdès A, Gestraud P, Hupé P, Barillot E, Cruzalegui F, Tucker GC, Stern MH, Thiery JP, Hickman JA, Dubois T. 2008. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res 10:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. 2007. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128:157–170 [DOI] [PubMed] [Google Scholar]
- 73. Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, et al. 2010. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464:999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294:173–177 [DOI] [PubMed] [Google Scholar]
- 75. Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. 2008. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 321:1499–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. 2006. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol 19:264–271 [DOI] [PubMed] [Google Scholar]
- 77. Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD. 2007. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 12:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sherr CJ, McCormick F. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103–112 [DOI] [PubMed] [Google Scholar]
- 79. Hu X, Stern HM, Ge L, O'Brien C, Haydu L, Honchell CD, Haverty PM, Peters BA, Wu TD, Amler LC, Chant J, Stokoe D, Lackner MR, Cavet G. 2009. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res 7:511–522 [DOI] [PubMed] [Google Scholar]
- 80. Herschkowitz JI, He X, Fan C, Perou CM. 2008. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res 10:R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Agarwal R, Gonzalez-Angulo AM, Myhre S, Carey M, Lee JS, Overgaard J, Alsner J, Stemke-Hale K, Lluch A, Neve RM, Kuo WL, Sorlie T, Sahin A, Valero V, Keyomarsi K, Gray JW, Borresen-Dale AL, Mills GB, Hennessy BT. 2009. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res 15:3654–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Keyomarsi K, Tucker SL, Buchholz TA, Callister M, Ding Y, Hortobagyi GN, Bedrosian I, Knickerbocker C, Toyofuku W, Lowe M, Herliczek TW, Bacus SS. 2002. Cyclin E and survival in patients with breast cancer. N Engl J Med 347:1566–1575 [DOI] [PubMed] [Google Scholar]
- 83. Voduc D, Nielsen TO, Cheang MC, Foulkes WD. 2008. The combination of high cyclin E and Skp2 expression in breast cancer is associated with a poor prognosis and the basal phenotype. Hum Pathol 39:1431–1437 [DOI] [PubMed] [Google Scholar]
- 84. Thiery JP. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2:442–454 [DOI] [PubMed] [Google Scholar]
- 85. Yang J, Weinberg RA. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14:818–829 [DOI] [PubMed] [Google Scholar]
- 86. Chen J, Imanaka N, Chen J, Griffin JD. 2010. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer 102:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sarrió D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. 2008. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 68:989–997 [DOI] [PubMed] [Google Scholar]
- 88. Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, Thompson EW. 2008. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis 25:629–642 [DOI] [PubMed] [Google Scholar]
- 89. Storci G, Sansone P, Trere D, Tavolari S, Taffurelli M, Ceccarelli C, Guarnieri T, Paterini P, Pariali M, Montanaro L, Santini D, Chieco P, Bonafé M. 2008. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol 214:25–37 [DOI] [PubMed] [Google Scholar]
- 90. DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C. 2009. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res 69:5364–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL, Weinberg RA. 2007. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA 104:10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lo HW, Hsu SC, Xia W, Cao XY, Shih JY, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung MC. 2007. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res 67: 9066–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kedrin D, Wyckoff J, Boimel PJ, Coniglio SJ, Hynes NE, Arteaga CL, Segall JE. 2009. ERBB1 and ERBB2 have distinct functions in tumor cell invasion and intravasation. Clin Cancer Res 15:3733–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Choi YL, Bocanegra M, Kwon MJ, Shin YK, Nam SJ, Yang JH, Kao J, Godwin AK, Pollack JR. 2010. LYN is a mediator of epithelial-mesenchymal transition and a target of dasatinib in breast cancer. Cancer Res 70:2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, Liu W, Stivers D, Baggerly K, Carey M, Lluch A, Monteagudo C, He X, Weigman V, Fan C, Palazzo J, Hortobagyi GN, Nolden LK, Wang NJ, Valero V, Gray JW, Perou CM, Mills GB. 2009. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69:4116–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kerbel RS. 2008. Tumor angiogenesis. N Engl J Med 358:2039–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Linderholm BK, Hellborg H, Johansson U, Elmberger G, Skoog L, Lehtiö J, Lewensohn R. 2009. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann Oncol 20:1639–1646 [DOI] [PubMed] [Google Scholar]
- 99. Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L. 2009. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 15:441–451 [DOI] [PubMed] [Google Scholar]
- 100. Hu Z, Fan C, Livasy C, He X, Oh DS, Ewend MG, Carey LA, Subramanian S, West R, Ikpatt F, Olopade OI, van de Rijn M, Perou CM. 2009. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Linderholm BK, Lindahl T, Holmberg L, Klaar S, Lennerstrand J, Henriksson R, Bergh J. 2001. The expression of vascular endothelial growth factor correlates with mutant p53 and poor prognosis in human breast cancer. Cancer Res 61:2256–2260 [PubMed] [Google Scholar]
- 102. Rydén L, Jirström K, Haglund M, Stål O, Fernö M. 2010. Epidermal growth factor receptor and vascular endothelial growth factor receptor 2 are specific biomarkers in triple-negative breast cancer. Results from a controlled randomized trial with long-term follow-up. Breast Cancer Res Treat 120:491–498 [DOI] [PubMed] [Google Scholar]
- 103. Lichtenberger BM, Tan PK, Niederleithner H, Ferrara N, Petzelbauer P, Sibilia M. 2010. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell 140:268–279 [DOI] [PubMed] [Google Scholar]
- 104. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 105. Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavaré S, Caldas C, Miska EA. 2007. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 8:R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838 [DOI] [PubMed] [Google Scholar]
- 107. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070 [DOI] [PubMed] [Google Scholar]
- 108. Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. 2007. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67:11612–11620 [DOI] [PubMed] [Google Scholar]
- 109. Iliopoulos D, Hirsch HA, Struhl K. 2009. An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139:693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. 2007. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131:1109–1123 [DOI] [PubMed] [Google Scholar]
- 111. Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. 2008. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451:147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Natrajan R, Lambros MB, Rodríguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchió C, Vatcheva R, Rayter S, Mahler-Araujo B, Fulford LG, Hungermann D, Mackay A, Grigoriadis A, Fenwick K, Tamber N, Hardisson D, Tutt A, Palacios J, Lord CJ, Buerger H, Ashworth A, Reis-Filho JS. 2009. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res 15:2711–2722 [DOI] [PubMed] [Google Scholar]
- 113. Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Børresen-Dale AL, Pollack JR. 2006. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 45:1033–1040 [DOI] [PubMed] [Google Scholar]
- 114. Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10:529–541 [DOI] [PubMed] [Google Scholar]
- 115. Adélaïde J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, Schoenmakers E, Viens P, Jacquemier J, Birnbaum D, Bertucci F, Chaffanet M. 2007. Integrated profiling of basal and luminal breast cancers. Cancer Res 67:11565–11575 [DOI] [PubMed] [Google Scholar]
- 116. Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, Ashworth A, van de Vijver MJ, Reis-Filho JS. 2010. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 29:2013–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Turner N, Grose R. 2010. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10:116–129 [DOI] [PubMed] [Google Scholar]
- 118. Isakoff SJ. 2010. Triple-negative breast cancer: role of specific chemotherapy agents. Cancer J 16:53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG, Harkin DP. 2003. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res 63:6221–6228 [PubMed] [Google Scholar]
- 120. Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. 2007. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest 117:1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE. 2010. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 28:1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ashworth A. 2008. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol 26:3785–3790 [DOI] [PubMed] [Google Scholar]
- 123. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913–917 [DOI] [PubMed] [Google Scholar]
- 124. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917–921 [DOI] [PubMed] [Google Scholar]
- 125. Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, Ghoreishi-Haack NS, Grimm DR, Guan R, Han EK, Holley-Shanks RR, Hristov B, Idler KB, Jarvis K, Johnson EF, Kleinberg LR, Klinghofer V, Lasko LM, Liu X, Marsh KC, et al. 2007. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res 13:2728–2737 [DOI] [PubMed] [Google Scholar]
- 126. Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361:123–134 [DOI] [PubMed] [Google Scholar]
- 127. O'Shaughnessy J, Osborne C, Pippen J, Yoffe M, Patt D, Monaghan G, Rocha C, Ossovskaya V, Sherman B, Bradley C. 2009. Efficacy of BSI-201, a poly(ADP-ribose) poymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized phase II trial. J Clin Oncol 27(Suppl):18s [Google Scholar]
- 128. Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. 2008. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res 68:2581–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. 2008. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451:1111–1115 [DOI] [PubMed] [Google Scholar]
- 130. Ivy SP, Wick JY, Kaufman BM. 2009. An overview of small-molecule inhibitors of VEGFR signaling. Nat Rev Clin Oncol 6:569–579 [DOI] [PubMed] [Google Scholar]
- 131. Burstein HJ, Elias AD, Rugo HS, Cobleigh MA, Wolff AC, Eisenberg PD, Lehman M, Adams BJ, Bello CL, DePrimo SE, Baum CM, Miller KD. 2008. Phase II study of sunitinib malate, an oral multitargeted tyrosine kinase inhibitor, in patients with metastatic breast cancer previously treated with an anthracycline and a taxane. J Clin Oncol 26:1810–1816 [DOI] [PubMed] [Google Scholar]
- 132. Kozloff M, Chuang E, Starr A, Gowland PA, Cataruozolo PE, Collier M, Verkh L, Huang X, Kern KA, Miller K. 2010. An exploratory study of sunitinib plus paclitaxel as first-line treatment for patients with advanced breast cancer. Ann Oncol 21:1436–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Moreno-Aspitia A, Morton RF, Hillman DW, Lingle WL, Rowland Jr KM, Wiesenfeld M, Flynn PJ, Fitch TR, Perez EA. 2009. Phase II trial of sorafenib in patients with metastatic breast cancer previously exposed to anthracyclines or taxanes: North Central Cancer Treatment Group and Mayo Clinic Trial N0336. J Clin Oncol 27:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, Shenkier T, Cella D, Davidson NE. 2007. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med 357:2666–2676 [DOI] [PubMed] [Google Scholar]
- 135. Dickler MN, Rugo HS, Eberle CA, Brogi E, Caravelli JF, Panageas KS, Boyd J, Yeh B, Lake DE, Dang CT, Gilewski TA, Bromberg JF, Seidman AD, D'Andrea GM, Moasser MM, Melisko M, Park JW, Dancey J, Norton L, Hudis CA. 2008. A phase II trial of erlotinib in combination with bevacizumab in patients with metastatic breast cancer. Clin Cancer Res 14:7878–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. 2009. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell 15:232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Guix M, Granja Nde M, Meszoely I, Adkins TB, Wieman BM, Frierson KE, Sanchez V, Sanders ME, Grau AM, Mayer IA, Pestano G, Shyr Y, Muthuswamy S, Calvo B, Krontiras H, Krop IE, Kelley MC, Arteaga CL. 2008. Short preoperative treatment with erlotinib inhibits tumor cell proliferation in hormone receptor-positive breast cancers. J Clin Oncol 26:897–906 [DOI] [PubMed] [Google Scholar]
- 138. Green MD, Francis PA, Gebski V, Harvey V, Karapetis C, Chan A, Snyder R, Fong A, Basser R, Forbes JF. 2009. Gefitinib treatment in hormone-resistant and hormone receptor-negative advanced breast cancer. Ann Oncol 20:1813–1817 [DOI] [PubMed] [Google Scholar]
- 139. Modi S, D'Andrea G, Norton L, Yao TJ, Caravelli J, Rosen PP, Hudis C, Seidman AD. 2006. A phase I study of cetuximab/paclitaxel in patients with advanced-stage breast cancer. Clin Breast Cancer 7:270–277 [DOI] [PubMed] [Google Scholar]
- 140. Finn RS, Press MF, Dering J, Arbushites M, Koehler M, Oliva C, Williams LS, Di Leo A. 2009. Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J Clin Oncol 27:3908–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. McDermott U, Settleman J. 2009. Personalized cancer therapy with selective kinase inhibitors: an emerging paradigm in medical oncology. J Clin Oncol 27:5650–5659 [DOI] [PubMed] [Google Scholar]
- 142. Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM, Guan Y, Hu Z, Knight Z, Feiler HS, Gascard P, Parvin B, Spellman PT, Shokat KM, Wyrobek AJ, Bissell MJ, McCormick F, Kuo WL, Mills GB, Gray JW, Korn WM. 2009. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res 69:565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Huang F, Reeves K, Han X, Fairchild C, Platero S, Wong TW, Lee F, Shaw P, Clark E. 2007. Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res 67:2226–2238 [DOI] [PubMed] [Google Scholar]
- 144. Capodanno A, Camerini A, Orlandini C, Baldini E, Resta ML, Bevilacqua G, Collecchi P. 2009. Dysregulated PI3K/Akt/PTEN pathway is a marker of a short disease-free survival in node-negative breast carcinoma. Hum Pathol 40:1408–1417 [DOI] [PubMed] [Google Scholar]