Abstract
ZNF230 is a novel zinc finger gene cloned by our laboratory. In order to understand the potential functions of this gene in vertebrate development, we cloned the zebrafish orthologue of human ZNF230, named rnf141. The cDNA fragment of rnf141 was obtained by rapid amplification of cDNA ends (RACE). The open reading frame (ORF) encodes a polypeptide of 222 amino acids which shares 75.65% identity with the human ZNF230. RT-PCR analysis in zebrafish embryo and adult tissues revealed that rnf141 transcripts are maternally derived and that rnf141 mRNA has a broad distribution. Zygotic rnf141 message is strongly localized in the central nervous system, as shown by whole-mount in situ hybridization. Knockdown and over expression of rnf141 can induce abnormal phenotypes, including abnormal development of brain, as well as yolk sac and axis extendsion. Marker gene analysis showed that rnf141 may play a role in normal dorsoventral patterning of zebrafish embryos, suggesting that rnf141 may have a broad function during early development of vertebrates.
Keywords: rnf141, zebrafish (Danio rerio), development, zinc finger protein
Introduction
The zinc finger gene family, one of the largest gene families in mammals, is defined by a conserved cysteine and histidine rich domain essential for the binding of zinc ions (Freemont 1993; Klug and Schwabe, 1995). This gene family can be divided into several subfamilies, including ring finger, C2H2, glucocorticoid receptor, GATA1, GAL4, and LIM (Barlow et al., 1994; Borden and Freemont, 1996; Hammarstrom et al., 1996).
In accordance with their diverse structures, zinc finger proteins have been assigned multiple functions, including DNA recognition, transcriptional activation, RNA packaging, regulation of apoptosis, ubiquitination and many others (Coleman, 1992; Wolfe et al., 2000; Laity et al., 2001; Vazquez et al., 2007). More than 20 different zinc finger genes located on sex chromosomes or autosomes have been proposed to play a regulatory role in mammalian spermatogenesis (Noce et al., 1992; Pieler and Bellefroid, 1994; Yan et al., 2002).
The human ZNF230, which maps to the short arm of chromosome 11 (11p15), encodes a C3HC4-type zinc finger protein motif (ring finger motif), and, consistent with a role in premeiotic or postmeiotic sperm development, one of its transcripts has been identified in abundance in the testicular tissue of fertile men, but neither in fetus nor in azoospermic patients. This suggested that ZNF230 may be involved in spermatogenesis, and loss of its expression may lead to azoospermia (Zhang et al., 2001). But so far, no clear biological function and mechanism had been elucidated.
In order to analyse the function of this novel zinc finger gene during vertebrate development, we decided to identify a potential orthologue in the teleost fish Danio rerio. This animal has become widely used as a genetic model to uncover specific functions of unknown proteins (Dooley and Zon, 2000; Rubinstein, 2003). Being transparent early embryonicl stages, easy to manipulate and highly reproductive makes the zebrafish an ideal animal system for molecular studies (Moro et al., 2007).
We here report the cloning and characterization of an 816 bp cDNA sequence, named rnf141, which represents a candidate zebrafish orthologue of the human ZNF230 gene. By using whole-mount in situ hybridization, RT-PCR, gene knockdown and overexpression analysis, we further showed its spatiotemporal expression pattern during early developmental stages.
Materials and Methods
Zebrafish embryo maintenance
Zebrafish AB strain was provided by National Zebrafish Resources of China, and maintained under standard laboratory conditions at 28.5 °C (Westerfield, 1993). Embryonic stages were identified by morphological features (Kimmel et al., 1995), and embryos in developmental stages of interest were fixed in 4% paraformaldehyde.
RNA extraction and reverse transcription
Total RNAs was isolated from adult zebrafish tissues using RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. SuperScript TM Reverse Transcriptase (Invitrogen) was used for reverse transcription.
Cloning of the zebrafish rnf141 gene
Primers for 5'-RACE (366L 5'-ACGAGACGC CTCTACCATTCCATCC-3') and the other three pairs of primers (70U 5'-TCTCCATTTGGAGCCAAGATGGGC C-3' and 763L 5'- TAGATTTTTAAGGTCTGTGTGGG TG-3'; 338U 5'-AGGAGGATGGAATGGTAGAGGCGT C-3' and 619L 5'- GGCTCTGGCCGCTCCACTTGTCA AT-3'; and 432U 5'- TAGTTCAAATGTGGCGGCAG AGGGA-3' and 816L 5'- ATATATAGGTGGTCTTTT ATGGGGA-3') were designed to obtain the complete coding sequence, based on the potential orthologue of ZNF230 in zebrafish. This orthologue sequence was acquired by PSI-BLAST alignment. 5'-RACE experiments were performed using SMART RACE cDNA Amplification Kit (Clontech) (Frohman et al., 1988). cDNAs were reverse-transcribed from total RNAs of zebrafish tissue. The PCR products, including 5'- RACE products, were ligated into the pGEM-T Easy Vector (Promega), cloned and sequenced bidirectionally. Sequences obtained by 5'-RACE and the other three fragments were assembled, and the contig was queried to the zebrafish genome database to determine its chromosomal location, and analyse its genomic structure. The deduced amino acid sequence was searched against InterPro Database for possible functional domains.
Multi-tissue RT-PCR
To reveal the tissue distribution and expression of zebrafish rnf141 gene, total RNA was extracted from embryos of various developmental stages (Kimmel et al., 1995) and several tissues of adult zebrafish. The gene-specific primers 488U 5'-GGATGGGCAGGGTAAAAC AGTTGA- 3'(forward) and 683L 5'-GCATCCGACATG ACCCAGGATTCATTAG-3' (reverse) were designed to amplify a 196 bp fragment of rnf141. Amplification was performed in 30 cycles as follows: 30 s denaturation at 94 °C, 30 s primer annealing at 62 °C and 1 min extension at 72 °C. The PCR products were electrophoresed on 1% agarose gel in 1 x TAE buffer and ethidium bromide stained.
Primer sequences used for amplifying 470 bp ß-actin were 5'-TGTGGCCCTTGACTTTGAGCAG-3' (forward) and 5'-TAGAAGCATTTGCGGTGGACGA-3'(reverse), according to Kaslin et al., (2004),. In negative controls, ddH2O was used instead of cDNA template. The gene-specific primers were selected from two exons separated by an intronic sequence to identify possible amplicons from contaminating genomic DNA. All synthetic oligonuleotides were purchased from Invitrogen Corporation (CA, USA).
rnf141 gene knockdown and overexpression experiments
rnf141 morpholino antisense oligonucleotide (rnf141-MO, 5'-CCAGAAAGCTGCTGGCCCATCTTG G-3') was used to target rnf141 mRNA, and 5'mispaired control morpholino (5'-CCAcAAAcCTcCTGcCCCATgT TGG-3') served as a control. Both were designed by using Gene-tools (Philomath, OR). The coding region of rnf141 was ligated into vector pcDNA3 (Invitrogen) and linearized by appropriate restriction enzymes for mRNA was synthesis by using mMESSAGE mMACHINE® Kit (Ambion). rnf141-MO and control-MO were injected into 1-2 cell zebrafish embryos by using a Model PLI-90 Pico-Injector.
Whole-mount in situ hybridization
rnf141 sense and antisense RNA probes were labeled with digoxigenin-11-UTP and synthesized by using DIG RNA Labeling Kit (SP6/T7) (Roche). The template for the probe was the entire 668 bp long cDNA. Whole-mount in situ hybridizations was performed as described by Westerfield (1995). Images were captured using an Olympus digital camera.
Results and Discussion
Zebrafish rnf141 has a C3HC4 zinc finger domain
We cloned zebrafish rnf141 based on the amino acid sequence of human ZNF230 and mouse znf230 by PSI-BLAST alignment and RT-PCR including RACE. As a result, four fragments were obtained that formed a816 bp cDNA contig. This was submitted to GenBank (accession number: AY621088). Previous studies have shown that human ZNF230 has two transcripts (Zhang et al., 2001). In mouse, there are three transcripts of znf230 (Qiu et al. 2003). In contrast, in our analyses on zebrafish we found only one transcript of this gene. Furthermore, our result was confirmed by another mRNA sequence record (GenBank accession numbe BC071534) that encodes the same protein submitted by Strausberg et al. Also, the sequence of our 3'-RACE fragment is identical to this sequence record.
The predicted open reading frame from 88 to 756 is 668 bp in length and encodes a polypeptide of 222 amino acid residues with a C3HC4 zinc finger domain from 146 to 187 amino acid residues (Figure 1). Proteins with such a structure generally are nuclear transcriptional factors with the motifs being involved in both protein-DNA and protein-protein interactions. We found that zebrafish rnf141 protein shares 75.65% and 75.22% identity in amino acid sequence with the human and mouse homologues, respectively. Furthermore, aligning amino acid sequence of zebrafish rnf141 with ZNF230 of other vertebrate species indicated that this gene domain is highly conserved (date not shown), suggesting functional similarity and conservation.
Figure 1.
Nucleotide and predicted amino acid sequence of rnf141 gene. The coding region (nucleotides 88-756) is in uppercase letters. The translation initiation codon is underlined. The stop codon at the 3'-end of the sequence is underlined and shaded. The deduced amino acid sequence (222 amino acids) is also shown below the nucleotide sequence. The predicted Ring-finger motif is shaded.
rnf141 is expressed in the CNS, primarily during early embryogenesis
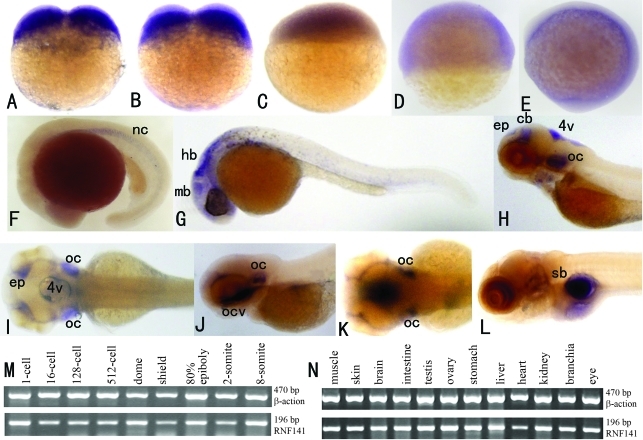
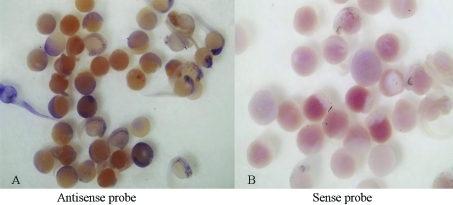
To analyse the spatiotemporal expression of rnf141 during early embryonic development, whole-mount in situ hybridizations were performed on two-cell stage to five-day-old embryos using an antisense probe. As a result, rnf141 transcripts were already detected at the two-cell stage (Figure 2A), thus suggesting a maternal origin of the transcript. From the sphere stage (4 hpf) to the tail bud stage (10 hpf) (Figure 2C-E), the rnf141 transcripts have a broad distribution. However, at the 5-somite stage (11.6 hpf), a characteristic pattern was displayed with marked staining in the notochord (Figure 2F), and at the Prim-5 stage of the pharyngula period, this pattern was displayed in the midbrain and hindbrain (Figure 2G). Following the long-pec stage (48 hpf), restricted signal localization was evident in the otic capsule, 4th ventricle, epiphysis and cerebellum (Figure 2H-I). When embryos reached the protruding-mouth stage (72 hpf), obvious signals were detected in the oral cavity and otic capsule (Figure 2J-K). In 5 dpf (120 hpf) embryos, an extensive rnf141 expression was visible in the gut, with a restricted localization in the swim bladder (Figure 2L). To assess the specificity of the antisense probe, a sense probe was used in a parallel control experiment at all stages. With this sense probe no staining was detected in any embryo (Figure 3).
Figure 2.
Expression analysis of rnf141 in early embryos and adult zebrafish tissues. rnf141 mRNA was initially detected at the 2-cell stage (A) and 4-cell stage (B), the signal become weaker at the sphere stage (C), shield stage (D) and bud stage (E). At the 5-somite stage (11.6 hpf), a characteristic pattern was displayed with marked staining in the notochord (F). At the 5-prim stage (24 hpf), a strong signal was detected in the head, particularly in the midbrain, hindbrain and the otic capsule (G). At the long-pec stage (48 hpf) the signal localization became restricted to the otic capsule, the 4th ventricle, as well as the epiphysis and tegmentum (H-lateral view from left; I-dorsal view). An even more restricted expressions was detected in the oral cavity and otic capsule when embryos reached the mouth-protruding stage (72 hpf) (J- lateral view from left; K-dorsal view). An extensive expression in the gut and a restricted localization in swim bladder was found in 5 dpf embryo (L). Expression analysis of rnf141 detected by RT-PCR in different developmental stage embryos (M) and adult zebrafish tissues (N). Abbreviations: mb, midbrain; hb, hindbrain; ep, epiphysis; cb, cerebellum; 4v, 4th ventricle; oc, otic capsule; ocv, oral cavity; sb, swim bladder.
Figure 3.
Whole-mount in situ hybridization with rnf141 antisense and sense probe. Hybridizations were performed on two-cell stage to five-day-old embryos using rnf141 antisense probe (A) and sense probe (B).
The consistency of hybridization experiments was confirmed by RT-PCR expression analysis performed on cDNAs from whole zebrafish embryos at various early developmental stages (Figure 2M).
Since the 1 kb transcript of human ZNF230 is only expressed in fertile male testis, whereas another 4.4 kb transcript was detected in many tissues; include heart, brain, skeletal muscle, kidney and pancreas (Zhang et al., 2001), we further addressed the question as to whether zebrafish rnf141 maintains its ubiquitous spatial expression in adult stages. As shown in Figure 2N, RT-PCR based analysis demonstrated that almost all analysed tissues of adult fish do display a high content of rnf141 transcripts.
In conclusion, these results of whole-mount in situ hybridization and RT-PCR analyses performed both on zebrafish embryos and adult tissues provide evidence that rnf141 may have multiple functions. The detection of its transcripts in the CNS of early embryos, especially restricted in the notochord at the 5-somite of the segmentation period, suggests a function for rnf141 in zebrafish development. Further analysis is ongoing in order to improve knowledge on the role of rnf141.
rnf141 may play a part in normal dorsoventral patterning of zebrafish embryos
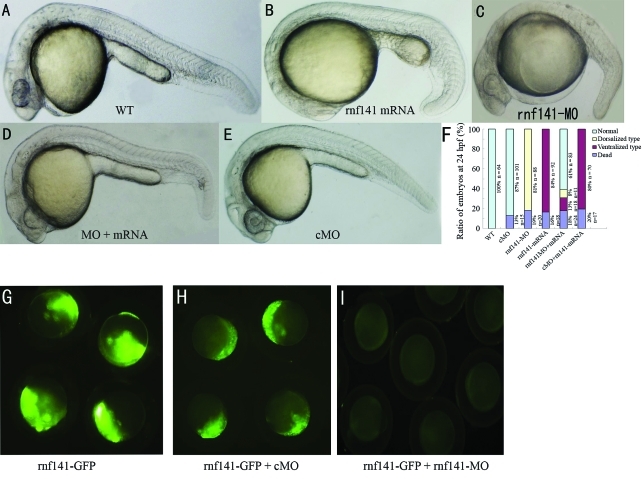
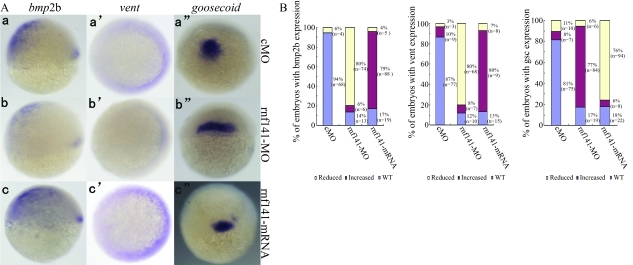
To further study the potential function of rnf141, we first injected zebrafish embryos with synthetic rnf141 mRNA. Injection of 200 pg rnf141 mRNA caused 84% (n = 92) of the embryos to show phenotypes that are characteristic of embryonic ventralization at 24 hpf (Figure 4B). The expression of the shield-specific gene goosecoid was decreased at the shield stage (Figure 5Ac”). In contrast, the ventral markers bmp2b and vent expanded dorsally during gastrulation (Figure 5Ac-c'). The ratios of embryos with altered marker gene expression are summarized in Figure 5B.
Figure 4.
Knockdown and overexpression analysis of rnf141. All images are lateral views of live embryos at 24 hpf, anterior is to the left. (A) Wild-type embryo. (B) Injection with 200 pg rnf141 mRNA resulted in enlargement of the yolk sac, in addition an extension and broadening of caudal ventral fin. (C) Injection with 12 ng rnf141-MO led to caudal ventral fin loss of and yolk sac extension. (D) Coinjection with 100 pg rnf141 mRNA and 12 ng rnf141-MO resulted in a normal morphology. (E) Injection with 15 ng of a control morpholino, which differed from rnf141-MO in five mismatched nucleotides, did not show visible developmental defects. (F) The ratios of embryos showing different phenotype in experiments represented in A-E. Data were averaged from three independent experiments and expressed as means and standard deviations. The numbers of analysed embryos are indicated beneath each bar. (G-I) Live embryos at the shield stage, (G) embryos injected with 100 pg prnf141-GFP DNA. (H) Embryos injected with 100 pg prnf141-GFP DNA and 12 ng rnf141-5mis-MO. (I) embryos injected with 100 pg prnf141-GFP DNA and 12 ng rnf141-MO.
Figure 5.
Expression patterns of marker genes in injected embryos at shield stage. (A) Expression patterns of bmp2b , vent, goosecoid in embryos injected with 15 ng rnf141-5mis-MO, 12 ng rnf141-MO, or 200 pg rnf141 mRNA, respectively. For bmp2b expression, the embryo is shown in lateral view with dorsal pointed towards right; the embryo showing vent expression is depicted in animal pole view with dorsal oriented towards left; and the embryo withgoosecoid expression is shown in a dorsal view with the animal pole pointed towards the top. (B) Statistical data for each marker gene of shown in A.
To investigate the role of endogenous rnf141, a morpholino antisense oligonucleotide (rnf141-MO) was injected into one-cell embryos. As a result, 81% (n = 88) of the embryos injected with 12 ng rnf141-MO exhibited dorsalized phenotypes at 24 hpf: complete loss of the yolk sac extension and partial loss of the caudal ventral fin (Figure 4C). The effects of rnf141 knockdown on the expression of the marker genes bmp2b, vent and goosecoid (Figure 5Ab-b”) tended to be opposite to those of rnf141 overexpression. In contrast, injection with 15 ng of control morpholino, which differs from rnf141-MO in five mismatched nucleotides, did not cause developmental defects (Figure 4E).
To test the efficiency of the morpholinos, fertilized eggs were injected with 12 ng rnf141-MO in combination with 100 pg of prnf141-GFP DNA, an expression construct containing the full coding region of rnf141 cDNA fused in-frame to a GFP coding sequence. At this dose of rnf141-MO, the injected embryos almost lacked green fluorescence from the GFP fusion protein (Figure 5I), while the same dose of rnf141-cMO injected embryos retained visible fluorescence (Figure 5H), suggesting that rnf141-MO could effectively block translation of rnf141 mRNA.
To test the specificity of rnf141-MO, a 5 mis-pair rnf141 mRNA corresponding to 5 mis-pair control morpholino was synthesized for rescuing the phenotype mediated with rnf141-MO. These results showed that the rnf141-MO-induced dorsalization could be neutralized by coinjection with a smaller amount of 5 mis-pair rnf141 (Figure 4D), suggesting that rnf141-MO specifically targets rnf141.
In conclusion, knockdown of rnf141 by using special morpholino-induced abnormal outcomes, including inordinate development of the CNS with an atrophic hindbrain, thin and crooked notochord, as well as disappearance of yolk sac extension, abnormality of axis, and partial loss of the caudal ventral fin. These embryos are characteristic of weakly dorsalized phenotypes, reminiscent of mini fin (mfn) and lost-a-fin (laf) mutant embryos, which were first described by Mullins et al. (1996) and were subsequently found to be caused by inefficient BMP signaling (Bauer et al., 2001; Connors et al., 1999; Mintzer et al., 2001). Overexpressing this gene by injection of rnf141 mRNA may caused embryos to show ventralized phenotypes. We also noted that rnf141-MO-induced dorsalization could be neutralized by coinjection of a smaller amount of rnf141 mRNA, suggesting that rnf141-MO specifically targets rnf141 mRNA. The expression of ventral markers (bmp2b and vent) and of a dorsal marker (goosecoid) were impacted by altered expression of rnf141, thus suggesting that zebrafish rnf141 may participate in normal dorsoventral embryonic patterning. Further research is needed to better understand the respective biological pathway(s) and improve the knowledge on the function of rnf141.
Acknowledgments
This research is supported by the National Natural Science Foundation of China (30770812, 90408025 and 30500186) and National High-Tech Research and Development Program of China (2008AA02Z102).
Footnotes
Associate Editor: André Luiz Paranhos Perondini
References
- Barlow P.N., Luisi B., Milner A., Elliott M., Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- Bauer H., Lele Z., Rauch G.J., Geisler R., Hammerschmidt M. The type I serine/threonine kinase receptor Alk8/Lost-a-fin is required for Bmp2b/7 signal transduction during dorsoventral patterning of the zebrafish embryo. Development. 2001;128:849–858. doi: 10.1242/dev.128.6.849. [DOI] [PubMed] [Google Scholar]
- Borden K.L., Freemont P.S. The RING finger domain: A recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- Coleman J.E. Zinc proteins: Enzymes, storage proteins, transcription factors, and replication proteins. Annu Rev Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- Connors S.A., Trout J., Ekker M., Mullins M.C. The role of tolloid/mini fin in dorsoventral pattern formation of the zebrafish embryo. Development. 1999;126:3119–3130. doi: 10.1242/dev.126.14.3119. [DOI] [PubMed] [Google Scholar]
- Dooley K., Zon L.I. Zebrafish: A model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Freemont P.S. The RING finger. A novel protein sequence motif related to the zinc finger. Ann NY Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- Frohman M.A., Dush M.K., Martin G.R. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarstrom A., Berndt K.D., Sillard R., Adermann K., Otting G. Solution structure of a naturally-occurring zinc-peptide complex demonstrates that the N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit. Biochemistry. 1996;35:12723–12732. doi: 10.1021/bi961149j. [DOI] [PubMed] [Google Scholar]
- Kaslin J., Nystedt J.M., Ostergard M., Peitsaro N., Panula P. The orexin/hypocretin system in zebrafish is connected to the aminergic and cholinergic systems. J Neurosci. 2004;24:2678–2689. doi: 10.1523/JNEUROSCI.4908-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klug A., Schwabe J.W. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- Laity J.H., Lee B.M., Wright P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Mintzer K.A., Lee M.A., Runke G., Trout J., Whitman M., Mullins M.C. Lost-a-fin encodes a type I BMP receptor, Alk8, acting maternally and zygotically in dorsoventral pattern formation. Development. 2001;128:859–869. doi: 10.1242/dev.128.6.859. [DOI] [PubMed] [Google Scholar]
- Moro E., Maran C., Slongo M.L., Argenton F., Toppo S., Onisto M. Zebrafish spata2 is expressed at early developmental stages. Int J Dev Biol. 2007;51:241–246. doi: 10.1387/ijdb.062220em. [DOI] [PubMed] [Google Scholar]
- Mullins M.C., Hammerschmidt M., Kane D.A., Odenthal J., Brand M., van Eeden F.J., Furutani-Seiki M., Granato M., Haffter P., Heisenberg C.P., et al. Genes establishing dorsoventral pattern formation in the zebrafish embryo: The ventral specifying genes. Development. 1996;123:81–93. doi: 10.1242/dev.123.1.81. [DOI] [PubMed] [Google Scholar]
- Noce T., Fujiwara Y., Sezaki M., Fujimoto H., Higashinakagawa T. Expression of a mouse zinc finger protein gene in both spermatocytes and oocytes during meiosis. Dev Biol. 1992;153:356–367. doi: 10.1016/0012-1606(92)90120-6. [DOI] [PubMed] [Google Scholar]
- Pieler T., Bellefroid E. Perspectives on zinc finger protein function and evolution - An update. Mol Biol Rep. 1994;20:1–8. doi: 10.1007/BF00999848. [DOI] [PubMed] [Google Scholar]
- Qiu W., Zhang S., Xiao C., Xu W., Ma Y., Liu Y., Wu Q. Molecular cloning and characterization of a mouse spermatogenesis-related ring finger gene znf230. Biochem Biophys Res Commun. 2003;306:347–353. doi: 10.1016/s0006-291x(03)00970-7. [DOI] [PubMed] [Google Scholar]
- Rubinstein A.L. Zebrafish: From disease modeling to drug discovery. Curr Opin Drug Discov Dev. 2003;6:218–223. [PubMed] [Google Scholar]
- Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez O., Vazquez M.E., Blanco J.B., Castedo L., Mascarenas J.L. Specific DNA recognition by a synthetic, monomeric Cys2His2 zinc-finger peptide conjugated to a minor-groove binder. Angew Chem Int Ed Engl. 2007;46:6886–6890. doi: 10.1002/anie.200702345. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish. Oregon: University of Oregon Press; 1993. pp. 8–48.The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish. Oregon: Eugene Press; 1995. pp. 200–204.The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish [Google Scholar]
- Wolfe S.A., Nekludova L., Pabo C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Yan W., Burns K.H., Ma L., Matzuk M.M. Identification of Zfp393, a germ cell-specific gene encoding a novel zinc finger protein. Mech Dev. 2002;118:233–239. doi: 10.1016/s0925-4773(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Zhang S., Qiu W., Wu H., Zhang G., Huang M., Xiao C., Yang J., Kamp C., Huang X., Huellen K., et al. The shorter zinc finger protein ZNF230 gene message is transcribed in fertile male testes and may be related to human spermatogenesis. Biochem J. 2001;359:721–727. doi: 10.1042/0264-6021:3590721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- Zebrafish Genome Resources. Available from: http://www.ncbi.nlm.nih.gov/projects/genome/guide/zebrafish/(U.S. National Library of Medicine, Maryland MD, 2005)
- InterPro Database. Available from: http://www.ebi.ac.uk/interpro/ (European Bioinformatics Institute, 2006)