Abstract
Several motile processes are responsible for the movement of proteins into and within the flagellar membrane, but little is known about the process by which specific proteins (either actin-associated or not) are targeted to protozoan flagellar membranes. Actin is a major cytoskeleton protein, while polymerization and depolymerization of parasite actin and actin-interacting proteins (AIPs) during both processes of motility and host cell entry might be key events for successful infection. For a better understanding the eukaryotic flagellar dynamics, we have surveyed genomes, transcriptomes and proteomes of pathogenic Leishmania spp. to identify pertinent genes/proteins and to build in silico models to properly address their putative roles in trypanosomatid virulence. In a search for AIPs involved in flagellar activities, we applied computational biology and proteomic tools to infer from the biological meaning of coronins and Arp2/3, two important elements in phagosome formation after parasite phagocytosis by macrophages. Results presented here provide the first report of Leishmania coronin and Arp2/3 as flagellar proteins that also might be involved in phagosome formation through actin polymerization within the flagellar environment. This is an issue worthy of further in vitro examination that remains now as a direct, positive bioinformatics-derived inference to be presented.
Keywords: actin-interacting proteins (AIPs), flagellar proteins, Leishmania, coronin and Arp2/3, phagosome
Introduction
Leishmaniaspp. is a trypanosomatid protozoan responsible for a complex group of diverse clinical forms of classically neglected diseases collectively known as leishmaniases (Peters and Killick-Kendrick, 1987). As an insinuating and persistent pathogen, Leishmania has a specialized organelle for motility, the flagellum, which is essential for parasite migration, invasion and persistence on host tissues. Several motile processes have been reported in the literature that could be responsible for the movement of proteins into and within the flagellar membrane (Kozminski et al., 1993). However, little is known about the process by which specific proteins (either actin-associated or not) are targeted to protozoan flagellar membranes, with few exceptions such as the work by Snapp and Landfear (1999) which targets a motif for a flagellar integral membrane protein in Leishmania enriettii. Actin is a major cytoskeleton protein, while polymerization and depolymerization of parasite actin and actin motor-associated proteins during both processes of motility and host cell entry might be key events for successful infection.
To gain a better understanding of flagellar dynamics in Leishmania, we have surveyed genomes, transcriptomes and proteomes of these pathogens to identify pertinent genes/proteins and to build in silico models to properly address their putative roles in trypanosomatid virulence. We have applied computational tools (hidden Markov models, Viterbi algorithm and comparative modeling) to infer biological meaning through detailed sequence-structural-functional analyses on actin-interacting proteins (AIPs) involved in flagellar activities of Leishmania spp. The actin system evolved to be fail-safe with multiple proteins sharing overlapping but novel roles, whereas these multifaceted roles are likely to provide the versatile scenario involving actin-interacting activities.
Proteins such as coronin and Arp2/3 complex have a common feature of actin-binding activity and might be involved in Leishmania intraflagellar pathways (Broadhead et al., 2006; Costa et al., 2007). The coronin family comprises two groups of evolutionary conserved WD-repeat proteins known to interact with Arp2/3 complex that help regulate the nucleation dynamics of actin filaments (Rybakin and Clemen, 2005). Invasion and differentiation processes in Leishmania directly involve phagocytosis and the formation of an intracellular membrane-bounded organelle called a phagosome, since Leishmania promastigotes must first evade complement-mediated lysis until they are engulfed by a mammalian host macrophage (Puentes et al., 1989; Olivier et al., 2005).
By phagocytosis we mean the process of ingestion per se. A phagosome is the endocytic compartment that contains a non-interfering particle, as opposed to a vacuole which describes a compartment containing a particle, such as a pathogen like Leishmania that diverts normal phagosome maturation. Phagosome maturation refers to the process of intracellular phagosome development after closure of the phagocytic cup (reviewed by Haas, 2007). Phagocytosis has evolved into a highly complex and regulated process in multicellular eukaryotes by which microbes and other particles are taken up into the phagosome (Griffiths and Mayorga, 2007). The precise mechanisms by which coronin and Arp2/3 contribute to phagocytosis are not known (Yan et al., 2005), but it is crucial to investigate elements that will shed more light on these mechanisms.
Although not clearly understood, it has been shown in vitro that, after the phagocytosis of L. donovani by macrophage-like cells J774, parasites are transiently located in phagosomes with poor fusogenic properties towards late endocytic compartments (Desjardins and Descoteaux, 1997). In contrast, after internalization, they are found in compartments that rapidly fuse with late endocytic organelles (Dermine et al., 2000). A possibility is that these distinctive features of the early phagosomes could be linked to different proteins expressed on the parasite surface, which may modify the fusion capacity of the phagosomal membrane (Desjardins and Descoteaux, 1997; Dermine et al., 2000). Furthermore, most of the Leishmania-containing phagosomes have been shown to accumulate F-actin, which is noted around these phagosomes (Holm et al., 2001).
We should recall, then, the early events following phagocytosis of Leishmania, pre-adapted to the encounter within intracellular conditions of mammals. As reported (Courret et al., 2002), young phagosomes containing Leishmania rapidly acquire a competence to fuse with late endosomes/lysosomes. If we take this version, AIPs have a greater chance of being key elements on the phagosome maturation than any of the many surface membrane proteins that have been widely investigated. For that reason, we have focused our studies on AIPs because they are rich in motif-binding activities, besides being excellent models for sequence and structural comparisons, predictions and examinations to be carried out with all kinds of restraints on spatial structure of the amino acid sequence(s) and ligands. Our coronin and Arp2/3 model restraints were derived from known related protein structures (comparative modeling) and from rules of secondary structure packing (combinatorial modeling). Our in silico examinations provide information about AIPs in the specific domain of phagosome function in Leishmania spp. infection. The role of the phagosome is to deliver particulate material to a hydrolytic environment that will lead to its degradation. The maturation process of the phagocytic compartment is linked intimately to both digestive housekeeping processes and innate sensing of molecules associated with infection (Vieira et al., 2002). Therefore, mechanisms and regulation behind this maturation process are strategic for both hosts and pathogens, whereas here we present and discuss results concerning Leishmania AIPs that are also flagellar proteins that might be targeted in phagosome formation.
Methods
Biological databases and bioinformatics tools
As previously (Oliveira et al., 2005; Gouveia et al., 2007; Vasconcelos et al., 2007, 2008), we have used publicly available datasets of individual/clusters of gene/protein data on Leishmania spp. (from GeneDB, a core part of Sanger Institute Pathogen Genomics) and from other available eukaryotic organisms (at NCBI and UniProt/Swiss-Prot/trEMBL knowledge DB), including sequences from the genome projects of C. reinhardtii in the U.S. Department of Energy (JGI) and all available data at The Chlamydomonas Flagellar Proteome Project. For databases (DB) searches, accession numbers/identifiers are those used in four main sources: NCBI, GeneDB, PDB and UniProt. As previously described (Oliveira et al., 2005; Gouveia et al., 2007; Vasconcelos et al., 2007, 2008), BLAST and its variants (Altschul et al., 1997) and MUSCLE (Edgar, 2004) were used for sequence similarity searches and comparisons/analysis through pairwise and multiple alignments. As previously detailed (Costa et al., 2007), for pattern recognition tasks of gene/protein predictions, motif finding and refinements for defining core domains of interest, HMM implementations such as HMMER (Eddy, 1998) and a structural prediction adaptation (Yoshizawa et al., 2006) were employed, with the additional improvement of the Viterbi algorithm (VA) (Forney, 1973; Black, 2004) for best possible alignments after recursion (Alexandrov and Gerstein, 2004). As previously (Costa et al., 2007), we assume that the HMMs generated the input sequence and are looking for the highest probability path. For sequence position i = 0, 1 ,…, L+1; for state l = 0, 1, …, n:
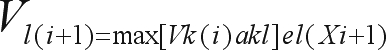 |
In-silico survey
As described in previous studies (Oliveira et al., 2005; Gouveia et al., 2007; Vasconcelos et al. 2008), we took alignments created with FASTA/BLAST as input and computed alignment tables, providing hierarchical and successive correlations between each of the two sets of sequences. FASTA files for amino-acid (aa) sequences of coding regions were downloaded from sources detailed above. All flagellar proteins and AIPs in Leishmania were collected from experimental papers and from the comprehensive GeneDB database. An illustration of the tools used is seen in Figure 1. Briefly, these sequences were used as queries against genomic datasets with PSI-BLAST (BLASTP2.2.10) (Altschul et al., 1997) to determine sequence similarity among all possible sequences, followed by multiple sequence alignments (MSA) performed with MUSCLE (Edgar, 2004) on target entries of main source DBs searched against various collections of protein motifs and families. Results of MSA were used as training datasets for HMM profiles (Eddy, 1998; Yoshizawa et al., 2006) and the SMART (Letunic et al., 2006) motif patterns of cluster motifs. The quality of trained HMM predictions and SMART pattern matching was examined. The HMM profiles for cluster motifs were trained, calibrated, and used for DB searches using HMMER package ver.2.3.1 with default parameters. Gene ontology (GO) terms were assigned, based on top matches to proteins with GO annotations from Swiss-Prot/trEMBL, AMIGO after GeneDB and TargetP access. Functional assignment of these genes/gene products was inferred using an RPS-BLAST search against conserved domain DBs (CDD) (Marchler-Bauer et al., 2005); information was taken into account about subcellular localization (Emanuelsson et al., 2000), sequence and structural features, domains/motifs conservation (von Mering et al., 2005; Letunic et al., 2006), and in vitro characterization (Avidor-Reiss et al., 2004; Tull et al., 2004). For three-dimensional (3D) modeling of AIPs we employed Modeller (Sali and Blundell, 1993; Marti-Renom et al., 2000) and ESyPred3D (Lambert et al., 2002). For visualization of 3D modeled structures, we employed the browser plug-in Chime. For addressing particular sites of structural-functional relationships of AIPs we used Cn3D (NCBI) and STING (Higa et al., 2004). Best models were evaluated by using PROCHECK (Laskowski et al., 1993) and a thorough analysis of consensus AIPs structures was carried out for model dissection, superposition and rms deviation (rmsd) calculations.
Proteomic analyses
Samples. Purified fractions of flagella from Leishmania amazonenesis, strain H21 (MHOM/BR/76/ MA-76), were kindly provided by Fiocruz (Fundação Oswaldo Cruz, Manguinhos, Rio de Janeiro). An amount of 250 μg of flagellar protein extract was normalized for use.
Two-dimensional (2D) gel electrophoresis. We performed proteomic analyses through 2D polyacrylamide gel electrophoresis (2D-PAGE), as previously described (Drummelsmith et al., 2003, Brobey et al., 2006). Briefly, proteins were treated with a lysis buffer with 7 M urea, 2 M thiourea, 2% NP-40, 2% DTT and 2% ampholytes (pH 3 to 10). In the first dimension, a protein separation by isoelectric point was performed applying IPG strips, soaked in the sample solution, at 300V for 5 min, 3,500 V for 5 h through Ettan IPGphor II Isoelectric Focusing System (GE Healthcare). To promote efficient protein transfer from the first to the second dimension, IPG strips were incubated in reducing buffer (1% DTT, 6M urea, 30% glycerol, 2% SDS, 50 mM Tris, pH 8.8) for 15 min, followed by 15 min incubation in alkylation buffer (2.5% iodoacetamide, 6 M urea, 30% glycerol, 2% SDS, 50 mM Tris, pH 8.8). The strips were immediately applied to the second dimension 12.5% polyacrylamide gel. Strips were, then, overlaid on freshly poured 10% Tricine-SDS gels (18 x 16 cm) and sealed with agarose solution (0.5% agarose, plus a few grains of bromophenol blue in a Tris-tricine cathode buffer). Protein standards of molecular mass range 14400 Da to 97000 Da were used (GE Healthcare). Gel electrophoresis was carried out at 250 V for 1 h, and then at 500 V until the dye front reached the bottom of the gel (around 8 h). Gels were silver-stained as previously described (Gromova and Celis, 2006).
After running and scanning the gels, the resulting protein gel was analyzed using ImageMaster 2-D Platinum 6.0® (Amersham Biosciences, Uppsala, Sweden). The spots detected automatically by the software were visually inspected. Spot filtering and editing were performed manually to remove artifacts and to correct for spots that did not split correctly or were not detected by the software's automatic spot detection process. Molecular mass (MW) and isoeletric point (pI) were predicted for the best spots.
Results
To identify putative flagellar proteins of Leishmania participating in the actin system, we have taken into account all actin-binding, interacting or related/regulated proteins as source data to investigate their possible involvement in assembly of F-actin (the principal driving force behind many forms of parasite locomotion, including those regarding or underlying internalization into the host cell). Our underlying hypothesis is that polymerization and depolymerization of parasite actin and actin motor-associated proteins, during both processes of motility and host cell entry, might be key events for successful infection, including the parasite survival within phagosomes. Recently (Vasconcelos et al., 2007, 2008) we reported an actin-polymerization Leishmania protein, profilin, and its partner, formin, as putative flagellar proteins due to their likely involvement in axonemal assembly/disassembly, therefore, flagellar dynamics/remodeling. In addition, we predicted other Leishmania AIPs, coronins and Arp2/3 complex as being flagellar proteins after detailed in silico structural alignments (Costa et al., 2007). Now, we focus on Leishmania coronin and Arp2/3 complex proteins as flagellar elements to phagosome formation in an attempt to directly associate the flagellum to the phagosomes, which are pivotal organelles in the ability of macrophages to perform several of their key functions, such as the handling of apoptotic cells, tissue remodeling, and restriction of the establishment and spread of intracellular pathogens (Méresse et al., 1999), such as Leishmania. Comparative bioinformatics analyses (performed after in silico genomic searches and in vitro proteomic screenings) have led to the identification of flagellar proteins that are involved not only in cytoskeleton activity, signaling, endocytosis, and lytic activity, but also putatively in the phagosome formation. With this study, we are able to confirm previous data on proteins already shown to be involved in actin systems, but not previously reported as Leishmania- or as flagellar-associated.
Flagellar coronin identification in Leishmania
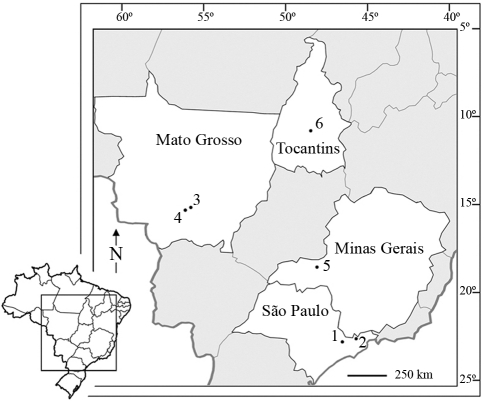
Coronins constitute an evolutionary conserved family of WD40-repeat (WDR) actin-binding proteins (reviewed by de Hostos, 1999; Rybakin and Clemen, 2005), originally described in the social amoeba Dictyosteliumdiscoideum (de Hostos et al., 1991). They can be divided, according to structure and function, into two subfamilies: one of shorter proteins (45-50kDa) implicated in, among other functions, nucleation of F-actin; and the other of longer, around 900 aa length, highly homologous coronins (POD-1 and Crn7) found in C. elegans, Drosophila and D. discoideum (Appleton et al., 2006; Rybakin et al., 2006). Until now, biochemical activities of coronins have been largely unknown, although one report (Humphries et al., 2002) identified how coronin and Arp2/3 complex interact in vitro. The comparative alignment of primary aa sequences of Leishmania coronins showed that they are highly conserved and have similar domain structures (Figure 2a), sharing ~45% aa identity with D. discoideum. Our models for the predicted 3D structure of Leishmania coronins, seen in Figure 2b, also reinforce their close proximity to PDB solved structures for coronin.
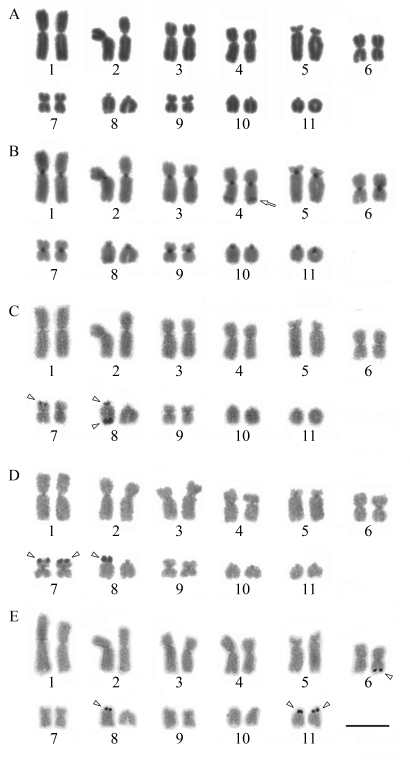
The proteomic screening performed on the flagellar fraction of L. amazonensis enabled us to anticipate evidence of coronin, among other proteins, as at least six putative spots, as shown in Figure 3 which provides a fairly good insight on Leishmania flagellar proteins (manuscript in preparation) that are also classical AIPs, such as coronin. Since coronin can be required for an early step in phagosome formation (Yan et al., 2005), which is consistent with its role in actin polymerization and accumulation around phagosomes formed during the ingestion of mycobacteria (Ferrari et al., 1999) and also in preventing phagosome maturation and mycobacterial killing (Ferrari et al., 1999), here we have considered this information as an important element for assigning a possible phagosome role for Leishmania coronin as well. We must recall that Leishmania has differential levels of survival inside phagosomes, as has been elegantly reported by Gueirard et al. (2007), whose work with L. donovani shows degraded parasites in spacious phagosomes, in contrast to morphologically intact parasites in tight compartments within neutrophils. These results suggest that the survival of parasites is linked to their ability to be targeted to tight/non-cidal and non-degradative compartments.
It is of paramount interest to mention that a phagosome, when it normally matures into a phagolysosome, undergoes a transition that correlates with functional changes from an organelle with early endosomal characteristics to a compartment with lysosomal, degradative properties. If we consider that persistent accumulation of coronin around phagosomes containing pathogens is believed to prevent phagosome maturation and mycobacterial killing (Ferrari et al., 1999), it would be conceivable to determine how coronin, actively recruited by these pathogens, may be involved in the modulation of the actin cytoskeleton, thereby influencing intracellular trafficking and survival (Jayachandran et al., 2008). While broad investigations into this are ongoing, the few pieces of information available stem largely from studies on phagosomes containing pathogens such as Leishmania that force the maturation machinery to slow down but will not permanently inhibit phagolysosome formation, with growth dependent upon eventual phagolysosome formation (Haas, 2007).
To what extent these different killing pathways (delayed or not) are used in a given phagosome certainly depends on various cellular and environmental conditions (usually a plethora of macrophage receptors at variable proportions that may influence phagosome fate to variable degrees), including interactions with the flagellum apparatus. We must bear in mind that flagella are dynamic structures that exchange up to 20% of their polypeptides within 3 h without any changes in their overall length (Song and Dentler, 2001; Wiese et al., 2003). Because flagellar microtubules are constantly turning over, the phagosomal membrane remains tightly associated with the parasites but the compartments become shorter (Marshal and Rosembaum, 2001). This reduction in size correlates with the progressive loss of the flagellum, suggesting that the parasite remodeling is accompanied by the removal of the phagosomal membrane.
Proteins involved in actin cytoskeleton dynamics that were recently implicated in phagocytosis in another protozoan, Entamoeba histolytica, are actobindin, coactosin and formin-like proteins (Marion et al., 2005), members of the so-called “core machinery” involved in de novo polymerization of actin filaments, which includes several subunits of the Arp2/3 complex, CAP protein, profilin and ADF/cofilin protein family (Loisel et al., 1999; Marion et al., 2005). ADF/cofilin proteins are essential to activate the turnover of actin filaments in dynamic regions of the cell and have already been shown to promote the remodeling of the actin network beneath the phagocytic cup (Bamburg, 1999; Bierne et al., 2001) and they have also been the subject of our own bioinformatics analyses (Pacheco et al., 2007) and of a recent in vitro identification (Tammana et al., 2008) in Leishmania flagellum.
Here we have chosen to follow the hypothesis that the actin filaments beneath the phagocytic cup could be organized as a bundle-like structure, rather than a dendritic network, which is necessary to form a stiff actin network around the phagocytic cup thereby facilitating the contractile activity that closes the vacuole (Marion et al., 2005). It is tempting to recall the model in which Toxoplasma gondii-containing vacuoles possess deep invaginations supported by host microtubules that deliver host endosomes and lysosomes into the parasitophorous vacuole (Coppens et al., 2006), where the parasites feed on these endocytic organelles and not, as expected, on the Toxoplasma vacuole arising completely separate from the endocytic pathway (Sibley, 2003).
Leishmania Arp2/3 complex genes
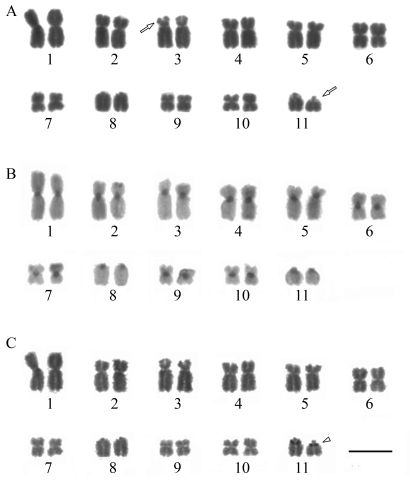
Arp2/3 complex has emerged as a central effector of actin assembly (Higgs and Pollard, 2001; Humphries et al., 2002). It is composed of seven evolutionarily conserved subunits: two actin-related proteins (Arp2 and Arp3) and five other subunits (yeast Arc40, Arc35, Arc18, Arc19, and Arc15). Since infection of mammalian hosts with Leishmania protozoa does depend on the ability of these parasites to replicate within macrophage phagolysosomes, it is noteworthy to link the function of Arps, such as the multiprotein Arp2/3 complex, and the phagosome formation, as a key step in parasite persistence within the host. Previously we have catalogued a list of Leishmania Arp2/3 complex genes comprising six putative sequences in L. braziliensis and L. infantum, and five in L. major(Costa et al., 2007), which are highly homologous with well-characterized Arp2/3 proteins in different organisms (as seen in Figure 4a). In Figure 4b we show a panel of four modeled Arp2/3 proteins in Leishmania where all of them possess the WDR, but no other known binding sites. WDR domains dominantly interfere with coronin function, acting as multimolecular scaffolding domains by bringing together interacting proteins on a single surface (Oku et al., 2003; Yan et al., 2005). They are likely to interfere with normal cell function by scavenging interacting proteins away from endogenous, full-length coronin.
If Arp2/3 complex is, in fact, one of those proteins that interact with coronin (Humphries et al., 2002; Yan et al., 2005), other portions (except WDR) of the protein are responsible for coronin recruitment to sites of actin remodeling, which would, then, link coronin and Arp2/3 to the sites of dynamic actin remodeling, with the contradiction that the WDR domain of coronin would act in a dominant-negative manner by preventing Arp2/3 accumulation. Conflicting interpretations (Humphries et al., 2002; Yan et al., 2005) pose a likely, unforeseen alternative: that coronin might play both positive (stimulating) and negative (inhibiting) roles in actin polymerization, depending on whether Arp2/3 functions prevent branching of F-actin or not.
Since our flagellar proteomic analyses on L. amazonensis did demonstrate clear support for the presence of at least four Arp2/3 proteins, as shown in Figure 3, we put forward an interesting point of view regarding the possibility that different sources of Arp2/3 proteins, whether flagellar or from the cytoskeleton, might be responsible in part for such paradox on activities of the coroninArp2/3 interaction.
Some Leishmania flagellar AIPs are homologous to phagosome proteins
To find out which AIPs in Leishmania were homologous to known phagosome proteins, as characterized in other studies (Desjardins et al., 1994; Garin et al., 2001; Marion et al., 2005; Stuart et al., 2007; Haas, 2007), we have searched lists of reported in vitro phagosome assays that gave us a panel ranging from 140-617 proteins (Garin et al., 2001; Gotthardt et al., 2006; Stuart et al., 2007) that are studied due to their direct involvement in phagosome formation. Therefore, we have compared their data to that found in Leishmania genomes, proteomes and particularly in our L. amazonensis flagellar proteome. We then collected a selected list of Leishmania proteins, including coronin and Arp2/3, ascribed as highly homologous to mammalian phagosome proteins (Table 1). We built a chart to emphasize these comparisons (Figure 5), which is a modified view of the full virtual phagosome (Garin et al., 2001) to cope with our predictive analysis of parasite flagellar proteins as homologous to mammalian phagosome counterparts. Therefore, in this study, we attempt to make a small contribution to further comprehensive analysis of Leishmania-containing phagosomes from which most of proteins have yet to be identified.
Table 1.
List of selected Leishmania actin-interacting proteins (AIPs) that are seemingly homologous to the phagosome proteins, as described by Garin et al. (2001). The proteins are presented with respective sequence ID (as on GeneDB), length (in number of amino acids), predicted molecular weight and isoelectric point (pI).
| Protein | Organism | Accession number | MW (kDa) | pI | Amino acids |
| Prohibitin | L.braziliensis | LbrM34.0070/LbrM16.1230 | 32.2/30.2 | 10.0/8.3 | 292/268 |
| L. infantum | LinJ16.1700/ LinJ35.0170 | 30.2/32.3 | 8.3/9.9 | 268/292 | |
| L. major | LmjF35.0070/ LmjF16.1610 | 32.3/30.2 | 9.9/8.3 | 292/268 | |
| RAB2 | L.braziliensis | LbrM30.1540 | 25.6 | 8.3 | 235 |
| L. infantum | LinJ30.2050 | 25.6 | 7.3 | 235 | |
| L. major | LmjF30.1710 | 25.6 | 7.3 | 235 | |
| RAB7 | L.braziliensis | LbrM18.0760/ LbrM10.0880 | 24.0/24.6 | 5.3/5.0 | 223/221 |
| L. infantum | LinJ10.1520 | 24.9 | 5.4 | 222 | |
| L. major | LmjF10.1170/ LmjF18.0890 | 24.7/24.1 | 5.7/5.1 | 221/223 | |
| RAB11B | L.braziliensis | LbrM32.1770 | 24.8 | 6.6 | 219 |
| L. infantum | LinJ32.2250 | 25.2 | 7.1 | 224 | |
| L. major | LmjF32.1840 | 25.2 | 7.1 | 224 | |
| Stomatin | L.braziliensis | LbrM05.0940 | 40.2 | 8.6 | 358 |
| L. infantum | LinJ05.1050 | 39.6 | 7.8 | 357 | |
| L. major | LmjF05.1040 | 39.7 | 8.3 | 357 | |
| Ubiquitin C | L.braziliensis | LM24.42/ LbrM25.0230 | 34.5/25.1 | 4.7/5.3 | 307/233 |
| LbrM32.0660 | 26.2 | 4.5 | 233 | ||
| L. infantum | LM24.42/ LinJ32.0850 | 34.3/26.3 | 4.9/4.3 | 307/234 | |
| LinJ25.0190 | 25.1 | 4.7 | 233 | ||
| L. major | LM24.42 / LmjF32.0700 | 34.4/26.4 | 4.8/4.4 | 307/234 | |
| Ubiquitin C | L.major | LmjF25.0190 | 25.1 | 4.9 | 233 |
| 14-3-3 | L.braziliensis | LbrM35_V2.3430/LbrM11_V2.0040 | 29.6/29.0 | 4.6/4.9 | 258/253 |
| L.infantum | LinJ36_V3.3360/ LinJ11_V3.0350 | 29.7/29.2 | 4.6/4.9 | 258/253 | |
| L.major | LmjF36.3210/ LmjF11.0350 | 29.6/29.1 | 4.6/5.0 | 258/253 | |
| ARP3 | L.braziliensis | LbrM15_V2.1360 | 38.1 | 5.4 | 348 |
| L.infantum | LinJ15_V3.1410 | 43.7 | 6.1 | 405 | |
| L.major | LmjF15.1360 | 43.7 | 5.9 | 405 | |
| Ash | L.infantum | LinJ30_V3.3620 | 18.6 | 5.7 | 162 |
| L.major | LmjF30.3560 | 18.5 | 6.1 | 162 | |
| L.braziliensis | LbrM30_V2.3590 | 18.5 | 6.9 | 162 | |
| Calreticulin | L.infantum | LinJ31_V3.2670 | 45.0 | 4.4 | 400 |
| L.major | LmjF31.2600 | 45.0 | 4.4 | 400 | |
| L.braziliensis | LbrM31_V2.2940 | 48.4 | 4.1 | 422 | |
| Cathepsin L | L.major | LmjF08.1030/ LmjF08.1040 | 48.0/37.7 | 7.6/7.1 | 443/348 |
| LmjF08.1010/ LmjF08.1020 | 37.9/37.7 | 7.3/6.7 | 348/348 | ||
| LmjF08.1050/ LmjF08.1060 | 37.8/48.0 | 6.7/7.6 | 348/443 | ||
| LmjF08.1070/ LmjF08.1080 | 37.8/48.0 | 6.7/7.6 | 348/443 | ||
| L.infantum | LinJ08_V3.0960/ LinJ08_V3.0950 | 47.9/41.3 | 7.2/7.5 | 443/381 | |
| L.braziliensis | LbrM08_V2.0810/ LbrM08_V2.0820 | 47.9/47.9 | 6.0/7.1 | 441/441 | |
| LbrM08_V2.0830 | 47.9 | 6.7 | 441 | ||
| Coronin | L.infantum | LinJ23_V3.1400 | 56.2 | 7.2 | 510 |
| L.major | LmjF23.1165 | 56.6 | 6.6 | 510 | |
| L.braziliensis | LbrM23_V2.1260 | 57.0 | 6.9 | 510 | |
| Cytochrome P450 | L.braziliensis | LbrM20_V2.2920/LbrM34_V2.2490 | 61.7/91.3 | 7.8/8.0 | 546/832 |
| LbrM20_V2.2230/ LbrM27_V2.0100 | 68.4/67.2 | 6.2/8.9 | 624/592 | ||
| LbrM30_V2.3580 | 58.0 | 7.3 | 509 | ||
| L.infantum | LinJ35_V3.2600/ LinJ34_V3.3110 | 91.1/61.3 | 8.0/6.7 | 832/546 | |
| LinJ34_V3.3610/ LinJ34_V3.2500 | 22.8/67.9 | 10.1/5.6 | 197/624 | ||
| LinJ27_V3.0090 | 66.7 | 8.6 | 592 | ||
| L.major | LmjF27.0090/ LmjF35.2560 | 68.5/90.9 | 8.1/7.9 | 606/831 | |
| LmjF34.3330/ LmjF30.3550 | 59.9/58.0 | 7.6/7.1 | 533/508 | ||
| LmjF34.2670 | 68.0 | 6.1 | 624 | ||
| HSP-60 | L.braziliensis | LbrM32_V2.2030/ LbrM30_V2.2790 | 64.3/58.2 | 6.0/5.4 | 594/539 |
| LbrM35_V2.2240/ LbrM35_V2.2250 | 60.2/59.5 | 5.3/5.1 | 564/562 | ||
| L. infantum | LinJ32_V3.1940/ LinJ36_V3.2130 | 64.3/60.5 | 6.4/5.1 | 594/566 | |
| LinJ36_V3.2140/ LinJ30_V3.2830 | 59.3/58.1 | 5.1/5.2 | 562/538 | ||
| L. major | LmjF32.1850/ LmjF36.2020 | 64.3/60.1 | 6.3/5.2 | 594/565 | |
| LmjF36.2030/ LmjF30.2820 | 59.3/58.0 | 5.1/5.3 | 562/538 | ||
| Prohibitin | L.braziliensis | LbrM34.0070/LbrM16.1230 | 32.2/30.2 | 10.0/8.3 | 292/268 |
| L. infantum | LinJ16.1700/ LinJ35.0170 | 30.2/32.3 | 8.3/9.9 | 268/292 | |
| L. major | LmjF35.0070/ LmjF16.1610 | 32.3/30.2 | 9.9/8.3 | 292/268 | |
| RAB2 | L.braziliensis | LbrM30.1540 | 25.6 | 8.3 | 235 |
| L. infantum | LinJ30.2050 | 25.6 | 7.3 | 235 | |
| L. major | LmjF30.1710 | 25.6 | 7.3 | 235 | |
| RAB7 | L.braziliensis | LbrM18.0760/ LbrM10.0880 | 24.0/24.6 | 5.3/5.0 | 223/221 |
| L. infantum | LinJ10.1520 | 24.9 | 5.4 | 222 | |
| L. major | LmjF10.1170/ LmjF18.0890 | 24.7/24.1 | 5.7/5.1 | 221/223 | |
| RAB11B | L.braziliensis | LbrM32.1770 | 24.8 | 6.6 | 219 |
| L. infantum | LinJ32.2250 | 25.2 | 7.1 | 224 | |
| L. major | LmjF32.1840 | 25.2 | 7.1 | 224 | |
| Stomatin | L.braziliensis | LbrM05.0940 | 40.2 | 8.6 | 358 |
| L. infantum | LinJ05.1050 | 39.6 | 7.8 | 357 | |
| L. major | LmjF05.1040 | 39.7 | 8.3 | 357 | |
| Ubiquitin C | L.braziliensis | LM24.42/ LbrM25.0230 | 34.5/25.1 | 4.7/5.3 | 307/233 |
| LbrM32.0660 | 26.2 | 4.5 | 233 | ||
| L. infantum | LM24.42/ LinJ32.0850 | 34.3/26.3 | 4.9/4.3 | 307/234 | |
| LinJ25.0190 | 25.1 | 4.7 | 233 | ||
| L. major | LM24.42 / LmjF32.0700 | 34.4/26.4 | 4.8/4.4 | 307/234 | |
| Ubiquitin C | L.major | LmjF25.0190 | 25.1 | 4.9 | 233 |
| 14-3-3 | L.braziliensis | LbrM35_V2.3430/LbrM11_V2.0040 | 29.6/29.0 | 4.6/4.9 | 258/253 |
| L.infantum | LinJ36_V3.3360/ LinJ11_V3.0350 | 29.7/29.2 | 4.6/4.9 | 258/253 | |
| L.major | LmjF36.3210/ LmjF11.0350 | 29.6/29.1 | 4.6/5.0 | 258/253 | |
| ARP3 | L.braziliensis | LbrM15_V2.1360 | 38.1 | 5.4 | 348 |
| L.infantum | LinJ15_V3.1410 | 43.7 | 6.1 | 405 | |
| L.major | LmjF15.1360 | 43.7 | 5.9 | 405 | |
| Ash | L.infantum | LinJ30_V3.3620 | 18.6 | 5.7 | 162 |
| L.major | LmjF30.3560 | 18.5 | 6.1 | 162 | |
| L.braziliensis | LbrM30_V2.3590 | 18.5 | 6.9 | 162 | |
| Calreticulin | L.infantum | LinJ31_V3.2670 | 45.0 | 4.4 | 400 |
| L.major | LmjF31.2600 | 45.0 | 4.4 | 400 | |
| L.braziliensis | LbrM31_V2.2940 | 48.4 | 4.1 | 422 | |
| Cathepsin L | L.major | LmjF08.1030/ LmjF08.1040 | 48.0/37.7 | 7.6/7.1 | 443/348 |
| LmjF08.1010/ LmjF08.1020 | 37.9/37.7 | 7.3/6.7 | 348/348 | ||
| LmjF08.1050/ LmjF08.1060 | 37.8/48.0 | 6.7/7.6 | 348/443 | ||
| LmjF08.1070/ LmjF08.1080 | 37.8/48.0 | 6.7/7.6 | 348/443 | ||
| L.infantum | LinJ08_V3.0960/ LinJ08_V3.0950 | 47.9/41.3 | 7.2/7.5 | 443/381 | |
| L.braziliensis | LbrM08_V2.0810/ LbrM08_V2.0820 | 47.9/47.9 | 6.0/7.1 | 441/441 | |
| LbrM08_V2.0830 | 47.9 | 6.7 | 441 | ||
| Coronin | L.infantum | LinJ23_V3.1400 | 56.2 | 7.2 | 510 |
| L.major | LmjF23.1165 | 56.6 | 6.6 | 510 | |
| L.braziliensis | LbrM23_V2.1260 | 57.0 | 6.9 | 510 | |
| Cytochrome P450 | L.braziliensis | LbrM20_V2.2920/LbrM34_V2.2490 | 61.7/91.3 | 7.8/8.0 | 546/832 |
| LbrM20_V2.2230/ LbrM27_V2.0100 | 68.4/67.2 | 6.2/8.9 | 624/592 | ||
| LbrM30_V2.3580 | 58.0 | 7.3 | 509 | ||
| L.infantum | LinJ35_V3.2600/ LinJ34_V3.3110 | 91.1/61.3 | 8.0/6.7 | 832/546 | |
| LinJ34_V3.3610/ LinJ34_V3.2500 | 22.8/67.9 | 10.1/5.6 | 197/624 | ||
| LinJ27_V3.0090 | 66.7 | 8.6 | 592 | ||
| L.major | LmjF27.0090/ LmjF35.2560 | 68.5/90.9 | 8.1/7.9 | 606/831 | |
| LmjF34.3330/ LmjF30.3550 | 59.9/58.0 | 7.6/7.1 | 533/508 | ||
| LmjF34.2670 | 68.0 | 6.1 | 624 | ||
| HSP-60 | L.braziliensis | LbrM32_V2.2030/ LbrM30_V2.2790 | 64.3/58.2 | 6.0/5.4 | 594/539 |
| LbrM35_V2.2240/ LbrM35_V2.2250 | 60.2/59.5 | 5.3/5.1 | 564/562 | ||
| L. infantum | LinJ32_V3.1940/ LinJ36_V3.2130 | 64.3/60.5 | 6.4/5.1 | 594/566 | |
| LinJ36_V3.2140/ LinJ30_V3.2830 | 59.3/58.1 | 5.1/5.2 | 562/538 | ||
| L. major | LmjF32.1850/ LmjF36.2020 | 64.3/60.1 | 6.3/5.2 | 594/565 | |
| LmjF36.2030/ LmjF30.2820 | 59.3/58.0 | 5.1/5.3 | 562/538 |
*Predicted Molecular Weight and Isoeletric Point (pI).
14-3-3 proteins are a family of conserved regulatory molecules expressed in all eukaryotic cells. 14-3-3 proteins have the ability to bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors, being considered evolved members of the Tetratricopeptide Repeat (TPR) superfamily.
*Predicted features and general scheme are the same as at GeneDB, including accession number / ID (Lmj = L. major; Lin = L. infantum; Lbr = L.braziliensis), according to the manuscript text.
Many groups are currently looking into these various aspects of phagocyte biology and progress is steady, although no comprehensive phagosome proteome or lipidome has been published for any pathogen-containing vacuole (Haas, 2007). There is an amazing diversity of compartments that intracellular pathogens inhabit, and they appear to use a stunning number of different factors to establish these compartments. The real challenge remains in understanding how phagocytes and their phagosomes co-operate in pathological situations to phagosome biogenesis, as well as how to turn a phagosome from a hospitable into a killing environment for Leishmania.
Figure 1.
Bioinformatics methods employed in this work, shown as a briefly simplified scheme of tools to illustrate a stepwise process in biological information technology.
Figure 2.
Sequence-structural alignments of four Leishmania coronins. a), b), c) and d): 3D models of L. major (LmjF23.1165/CAC44941.1), L. infantum (LinJ23.1360), L. donovani (AAY56362.1) and L. braziliensis (LbrM23.1230), respectively, illustrating the putative actin-binding region (N-terminus). Left-panel: schematic diagram of corresponding regions of other coronin family proteins. The first 30 amino acids are shown to illustrate residues conserved across members of the coronin family (Trypanosoma brucei, Cryptosporidium parvum, Toxoplasma gondii, Plasmodium falciparum, Mus musculus and Bos taurus).
Figure 3.
3D visualization of Arp2/3 complex modeling, including possible binding site to a coronin WDR domain at central (blue) region and multiple alignment of the amino acid sequences of Arp2/3 complex proteins from different organisms. A) PDB solved structure of Bos taurus Arp2/3 complex (1K8K) used as a template for building comparative models of Leishmania Arps with sequence from B) L. braziliensis (LbrM02.0360); C) L. infantum (LinJ02.0520) and D) L. major (XP_822258.1). *N-terminus; **C-terminus. E) Multiple alignment of the amino acid sequences of Arp2/3 complex proteins. Accession numbers are: T. cruzi (XP_810627.1), T. brucei (XP_951567.1), D. rerio (NP_991100.1), H. sapiens (NP_005709.1), S. cerevisiae (NP_012912.1), L. infantum (LinJ02.0520), L. major (XP_822258.1) and L. braziliensis (LbrM02.0360). Residues identical or similar are shaded dark or light gray, respectively. Dashes indicate gaps introduced for optimal alignment.
Figure 4.
Two dimensional gel electrophoresis of Leishmania amazonensis flagellar fraction. Proteomic analysis after visualization by silver staining. Imaging covering narrow range IPG strips of pH 3-10 - observed in the gel top. The standard of molecular mass range used is 14400 Da to 97000 Da; -1 is a software configuration parameter. Six (06) putative spots for coronin are circled and indicated by a red arrow; four (04) Arp2/3 putative spots are circled and pointed by a yellow arrow. The molecular weight (in KDa) and the corresponding isoeletric point of each protein are shown in white.
Figure 5.
Schematic illustration of Leishmania spp. proteins that are seemingly homologous (by sequence alignments) to the proteins of the virtual phagosome depicted on the article by Garin et al., 2001. Selected proteins were used in the present study in an attempt to suggest their potential interaction with phagosomes. Assigned localization to the lumen, the membrane, or the cytoplasmic aspect of the phagosome was the same indicated by Garin et al. (2001).
Discussion
Phagocytosis is initiated by recognition of a pathogen by a host cell (usually macrophages and neutrophils) receptors that trigger its engulfment into the phagosome. Following their attachment to the macrophage, Leishmania promastigotes are internalized to the relatively benign environment of the endosome, where they begin to differentiate into amastigotes. Unlike amastigotes, promastigotes are vulnerable to degradation by the acidic and hydrolytic environment of the phagolysosome (Olivier et al., 2005). They must therefore retard endosome maturation and phagosome-endosome fusion, a process that has been observed by the absence or delayed arrival of late endosomal markers such as rab7 and LAMP-1 (Scianimanico et al., 1999) and is related to the accumulation of F-actin (Holm et al., 2001). The mechanism is not completely understood (Olivier et al., 2005), but it has been shown that the delay in phagolysosome maturation provides a window during which promastigotes can differentiate into the more resistant amastigotes.
Nascent phagosomes have a composition that resembles that of the plasmalemma and are unable to digest the vacuolar contents. This capability is acquired by remodeling of the phagosomal membrane and contents through a complex series of fusion events with compartments of the endocytic and secretory pathways (Desjardins and Descoteaux, 1997). The components delivered to maturing phagosomes include acid hydrolases, as well as vacuolar-type ATPases that are responsible for acidification of the phagosomal lumen. This maturation process culminates in the formation of the phagolysosome, a highly acidic organelle (pH < 5.5) where degradation occurs.
Here we report the association of two classical AIPs (coronins and Arp2/3) to the flagellar compartment in Leishmania, certainly a reasonable indication to link flagellar proteins to a possible F-actin binding in this protozoan. In vitro, purified coronin is said to bind specifically to F-actin, to bundle actin filaments, and to weakly promote actin assembly (David et al., 1998; Fukui et al., 1999), whereas, in Acanthamoeba, coronin was localized in the cell's periphery (on the leading edge) consistent with that of actin located around the phagocytic cups (Baldo et al., 2005). Understanding the molecular mechanisms of phagosome maturation is critical because a number of pathogens, particularly Leishmania, survive inside host cells through subversion of this process (Rosenberger and Finlay, 2003). Keeping these facts in mind, together with our own results, we, then, believe that flagellar AIPs, such as coronin and Arp2/3/, might play a role in these subversion mechanisms that maintain the parasite viable within the phagosome.
Some reports have shown that Leismania can persist in mammalian neutrophils for up to 48 h, a period that largely exceeds the normal life span of neutrophils, indicating that the phagocytosis/infectious process might alter the ability of neutrophils to initiate their programmed cell death, increasing the lifetime of neutrophils by delaying apoptosis (Aga et al., 2002; Gueirard et al., 2007). It seems that Leishmania may have co-evolved with their mammalian hosts (Table 1 and Figure 5) to take advantage of phagosomes to establish a privileged niche for the transient parasitism of leukocytes and their subsequent invasion of macrophages, since it has been shown that the delay in phagolysosome maturation provides a window during which promastigotes can differentiate into the more resistant amastigotes (Olivier et al., 2005).
One could expect that phagosomes containing a given type of particle that entered cells simultaneously via the same receptor would behave the same, at least in a single cell. Surprisingly, however, phagosomes formed via the same receptors are found in different chemical states even within the same macrophage (Griffiths, 2004), a notion that is very useful to help our speculations on Leishmania AIPs rich content and at the same time homologous to a few phagosome proteins. Each phagosome is an individual entity whose behavior depends on a finite number of stable equilibrium states in its membrane signaling networks (Griffiths, 2004), which would, thus, underline a diversity of available elements to disturb effective phagosome formation.
As stated by Rasmusson and Descoteaux (2004), L. donovani promastigotes inhibit phagolysosome biogenesis in a lipophosphoglycan (LPG)-dependent manner, which correlates with an accumulation of periphagosomal F-actin, forming a physical barrier that prevents L. donovani promastigote-containing phagosomes from interacting with endocytic vacuoles. Such inhibition of phagosome maturation may constitute a strategy to provide an environment propitious to promastigote-to-amastigote differentiation. Interestingly, some molecules, known to be involved in apoptotic signaling, are thought to accumulate in phagosome membrane microdomains, suggesting that this location might be important to their specific function, such as LPG in a disruptive process in the phagosome (Gueirard et al., 2007), which could imply an involvement in the ability of Leishmania to inhibit neutrophil apoptosis and, thus, the posterior macrophage activity that also depends on actin.
Assuming that the driving force for host cell entry involves polymerization of parasite actin and its AIPs, plus the recognized subcellular localization of functional coronins and Arps shown in the phagosome (Bricheux et al., 2000; Asano et al., 2001; Baldo et al., 2005), then we can propose a possible viable role for flagellar coronin and Arp2/3 within phagosome. As Love et al. (1998) have shown, periphagosomal actin is rapidly lost when parasites are internalized. These authors have mentioned a higher percentage of L. major promastigotes surrounded by actin after experimentally adding parasites to macrophages. These findings suggest a clear binding to F-actin and implicit evidence of intense actin-interacting activity during phagosome formation after host cell entry (promastigote phagocytosis).
It seems quite obvious that intracellular parasites living within the harsh environment of phagocytes have developed strategies that allow them to adapt quickly, escape from first-line defense systems, and inhibit several functions of their host cells (Olivier et al., 2005). One of these strategies might be explained through the complex interactions of the actin system (Sturgill-Koszycki et al., 1996; Tardieux et al., 1998; Ullrich et al., 1999; Vieira et al., 2002; Yan et al., 2005). Coronins were first identified as AIPs, but, although they are crucial to the dynamics of actin filaments and cell movement, their actin-binding sites have been difficult to pin down, which reinforces the significance of computational biology and proteomic predictions that focus on coronin direct ligands, such as Arp2/3. Moreover, establishing relationships of subcellular/organellar localizations, such as those we now report for coronin and Arp2/3 on the flagellum of L. amazonensis, will help to clarify unforeseen roles that parasite AIPs might play on phagocytosis and phagosome formation through direct actin polymerization within the flagellar dynamic environment. This is an issue worthy of further in vitro examination that remains now as a direct, positive bioinformatics-derived inference to be presented. Research into phagosome biogenesis has flourished in recent years (Haas, 2007) and it will surely lead to a better understanding of Leishmania pathogenesis and mechanisms of immune invasion that involve the flagellar apparatus.
Acknowledgments
We wish to thank Fundação Oswaldo Cruz (FIOCRUZ, Brazil) for providing biological samples and GeneDB (The Wellcome Trust Sanger Institute, Pathogen Sequencing Unit) for full access to bioinformatics tools and databases on Leishmania genomes. This work is supported in part by CNPq and FUNCAP through individual grants to DMO and graduate fellowships to MCD, MPC, ACLP and SCS. MCD is a PhD student from RENORBIO (Rede Nordeste de Biotenologia) and this research is part of her thesis work.
References
- Aga E., Katschinski D.M., van Zandbergen G., Laufs H., Hansen B., Muller K., Solbach W., Laskay T. Inhibition of the spontaneous apoptosis of neutrophil granulocytes by the intracellular parasite Leishmania major. J Immunol. 2002;169:898–905. doi: 10.4049/jimmunol.169.2.898. [DOI] [PubMed] [Google Scholar]
- Alexandrov V., Gerstein M. Using 3D Hidden Markov Models that explicitly represent spatial coordinates to model and compare protein structures. BMC Bioinform. 2004;5:2. doi: 10.1186/1471-2105-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton B.A., Wu P., Wiesmann C. The crystal structure of murine coronin-1: A regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14:87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Asano S., Mishima M., Nishida E. Coronin forms a stable dimer through its C-terminal coiled coil region: An implicated role in its localization to cell periphery. Genes Cells. 2001;6:225–235. doi: 10.1046/j.1365-2443.2001.00416.x. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A.M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C.S. Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Baldo E.T., Moon E.K., Kong H.H., Chung D.I. Acanthamoeba healyi: Molecular cloning and characterization of a coronin homologue an actin-related protein. Exp Parasitol. 2005;110:114–122. doi: 10.1016/j.exppara.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Bamburg J.R. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bierne H., Gouin E., Roux P., Caroni P., Yin H.L., Cossart P. A role for cifinin and LIM kinase in Listeria-induced phagocytosis. J Cell Biol. 2001;155:101–112. doi: 10.1083/jcb.200104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricheux G., Coffe G., Bayle D., Brugerolle G. Characterization, cloning and immunolocalization of a coronin homologue in Trichomonas vaginalis. Eur J Cell Biol. 2000;79:413–422. doi: 10.1078/0171-9335-00065. [DOI] [PubMed] [Google Scholar]
- Broadhead R., Dawe H.R., Farr H., Griffiths S., Hart S.R., Portman N., Shaw M.K., Ginger M.L., Gaskell S.J., McKean P.G., et al. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- Brobey R.K.B., Mei F.C., Cheng X., Soong L. Comparative two-dimensional gel electrophoresis maps for promastigotes of Leishmania amazonensis and Leishmania major. Braz J Infect Dis. 2006;10:1–6. doi: 10.1590/s1413-86702006000100001. [DOI] [PubMed] [Google Scholar]
- Coppens I., Dunn J.D., Romano J.D., Pypaert M., Zhang H., Boothroyd J.C., Joiner K.A. Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell. 2006;125:261–274. doi: 10.1016/j.cell.2006.01.056. [DOI] [PubMed] [Google Scholar]
- Costa M.P., Lucas H.R., Maia A.R.S., Pacheco A.C.L., Pinheiro D.P., Kamimura M.T., Araújo-Filho R., Oliveira D.M. Flagellar proteins prediction after sequence-structure alignments of Coronin and Arp2/3 complex in Leishmania spp. Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine - Workshop on Computational Structural Bioinformatics; Piscitaway: IEEE Press; 2007. pp. 80–88. [Google Scholar]
- Courret N., Fréhel C., Gouhier N., Pouchelet M., Prina E., Roux P., Antoine J.C. Biogenesis of Leishmania-harbouring parasitophorous vacuoles following phagocytosis of the metacyclic promastigote or amastigote stages of the parasites. J Cell Sci. 2002;115:2303–2316. doi: 10.1242/jcs.115.11.2303. [DOI] [PubMed] [Google Scholar]
- David V., Gouin E., Troys M.V., Grogan A., Segal A.W., Ampe C., Cossart P. Identification of cofilin, coronin, Rac and capZ in actin tails using a Listeria affinity approach. J Cell Sci. 1998;111:2877–2884. doi: 10.1242/jcs.111.19.2877. [DOI] [PubMed] [Google Scholar]
- de Hostos E.L. The coronin family of actin-associated proteins. Trends Cell Biol. 1999;9:345–350. doi: 10.1016/s0962-8924(99)01620-7. [DOI] [PubMed] [Google Scholar]
- de Hostos E.L., Bradtke B., Lottspeich F., Guggenheim R., Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections has sequence similarities to G protein beta subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermine J.F., Scianimanico S., Privé C., Descoteaux A., Desjardins M. Leishmania promastigotes require lipophosphoglycan to actively modulate the fusion properties of phagosomes at an early step of phagocytosis. Cell Microbiol. 2000;2:115–126. doi: 10.1046/j.1462-5822.2000.00037.x. [DOI] [PubMed] [Google Scholar]
- Desjardins M., Descoteaux A.J. Inhibition of phagolysosomal biogenesis by the Leishmania lipophosphoglycan. Exp Med. 1997;185:2061–2068. doi: 10.1084/jem.185.12.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins M., Huber L.A., Parton R.G., Griffiths G. Biogenesis of phagosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummelsmith J., Brochu V., Girard I., Messier N., Ouellette M. Proteome mapping of the protozoan parasite Leishmania and application to the study of drug targets and resistance mechanisms. Mol Cell Proteomics. 2003;2:146–155. doi: 10.1074/mcp.M200085-MCP200. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O.H., Nielsen S., Brunak Heijne G.V. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Ferrari G., Langen H., Naito M., Pieters J. A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell. 1999;97:435–447. doi: 10.1016/s0092-8674(00)80754-0. [DOI] [PubMed] [Google Scholar]
- Forney D.G., Jr The Viterbi algorithm. Proceedings of the IEEE 61. 1973;3:268–278. [Google Scholar]
- Fukui Y., Engler S., Inoue S., de Hostos E.L. Architectural dynamics and gene replacement of coronin suggest its role in cytokinesis. Cell Motil Cytoskel. 1999;42:204–217. doi: 10.1002/(SICI)1097-0169(1999)42:3<204::AID-CM4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Garin J., Diez R., Kieffer S., Dermine J.-.F., Duclos S., Gagnon E., Sadoul R., Rondeau C., Desjardins M. The phagosome proteome: Insight into phagosome functions. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt D., Blancheteau V., Bosserhoff A., Ruppert T., Delorenzi M., Soldati T. Proteomic fingerprinting of phagosome maturation and evidence for the role of a G alpha during uptake. Mol Cell Proteomics. 2006;5:2228–2243. doi: 10.1074/mcp.M600113-MCP200. [DOI] [PubMed] [Google Scholar]
- Gouveia J.J., Vasconcelos E.J.R., Pacheco A.C.L., Araújo-Filho R., Maia A.R., Kamimura M.T., Costa M.P., Viana D.A., Costa R.B., Maggioni R., et al. Intraflagellar transport (IFT) complex in Leishmania spp: In silico genome-wide screening and annotation of gene function. Genet Mol Res. 2007;6:675–689. [PubMed] [Google Scholar]
- Gueirard P., Laplante A., Rondeau C., Milon G., Desjardins M. Trafficking of Leishmania donovani promastigotes in non-lytic compartments in neutrophils enables the subsequent transfer of parasites to macrophages. Cell Microbiol. 2007;10:100–111. doi: 10.1111/j.1462-5822.2007.01018.x. [DOI] [PubMed] [Google Scholar]
- Griffiths G. On phagosome individuality and membrane signalling networks. Trends Cell Biol. 2004;14:343–351. doi: 10.1016/j.tcb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Griffiths G., Mayorga L. Phagosome proteomes open the way to a better understanding of phagosome function. Genome Biology. 2007;8:207. doi: 10.1186/gb-2007-8-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova I., Celis J.E. Protein detection in gels by silver staining: A procedure compatible with mass-spectrometry. In: Celis J.E., Carter N., Hunter T., Simons K., Small J.V., Shotton D., editors. Cell Biology: A Laboratory Handbook. 3rd edition. v. 4. San Diego: Elsevier. Academic Press; 2006. pp. 421–429. [Google Scholar]
- Haas A. The Phagosome: Compartment with a license to kill. Traffic. 2007;8:311–330. doi: 10.1111/j.1600-0854.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- Higa R.H., Togawa R.C., Montagner A.J., Palandrani J.C., Okimoto I.K., Kuser P.R., Yamagishi M.E., Mancini A.L., Neshich G. STING Millennium Suite: Integrated software for extensive analyses of 3d structures of proteins and their complexes. BMC Bioinform. 2004;5:107–111. doi: 10.1186/1471-2105-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs H.N., Pollard T.D. Regulation of actin filament network formation through ARP2/3 complex: Activation by a diverse array of proteins. Annu Rev Biochem. 2001;70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- Holm A., Tejle K., Magnusson K.E., Descoteaux A., Rasmusson B. Leishmania donovani lipophosphoglycan causes periphagosomal actin accumulation: Correlation with impaired translocation of PKCalpha and defective phagosome maturation. Cell Microbiol. 2001;3:439–447. doi: 10.1046/j.1462-5822.2001.00127.x. [DOI] [PubMed] [Google Scholar]
- Humphries C.L., Balcer H.I., DAgostino J.L., Winsor B., Drubin D.G., Barnes G., Andrews B.J., Goode B.L. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayachandran R., Gatfield J., Massner J., Albrecht I., Zanolari B., Pieters J. RNA interference in J774 macrophages reveals a role for coronin 1 in mycobacterial trafficking but not in actin-dependent processes. Mol Biol Cell. 2008;19:1241–1251. doi: 10.1091/mbc.E07-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C., Leonard N., De Bolle X., Depiereux E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Letunic L., Copley R.C., Pils B., Pinkert S., Schultz J., Bork P. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel T.P., Boujemaa R., Pantaloni D., Carlier M.-.F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Love D.C., Kane M.M., Mosser D.M. Leishmania amazonensis: The phagocytosis of amastigotes by macrophages. Exp Parasitol. 1998;88:161–171. doi: 10.1006/expr.1998.4232. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., et al. CDD: A conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion S., Laurent C., Guillén N. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: A proteomic approach. Cell Microbiol. 2005;7:1504–1518. doi: 10.1111/j.1462-5822.2005.00573.x. [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Rosenbaum J.L. Intraflagellar transport balances continuous turnover of outer doublet microtubules: Implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom M.A., Stuart A., Fiser A., Sánchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Méresse S., Steele-Mortimer O., Moreno E., Desjardins M., Finlay B.B., Gorvel J.P. Controlling the maturation of pathogen-containing vacuoles: A matter of life or death. Nat Cell Biol. 1999;1:E183–E188. doi: 10.1038/15620. [DOI] [PubMed] [Google Scholar]
- Oku T., Itoh S.M., Okano M., Suzuki A., Suzuki K., Nakajin S., Tsuji T., Nauseef W.M., Toyoshima S. Two regions responsible for the actin binding of p57, a mammalian coronin family actin-binding protein. Biol Pharm Bull. 2003;26:409–416. doi: 10.1248/bpb.26.409. [DOI] [PubMed] [Google Scholar]
- Oliveira D.M., Gouveia J.J.S., Diniz N.B., Pacheco A.C., Vasconcelos E.J., Diniz M.C., Viana D.A., Ferreira T.D., Albuquerque M.C., Fortier D.C., et al. Pathogenomics analysis of Leishmania spp: Flagellar gene families of putative virulence factors. OMICS. 2005;9:171–191. doi: 10.1089/omi.2005.9.173. [DOI] [PubMed] [Google Scholar]
- Olivier M., Gregory D.J., Forget G. Subversion mechanisms by which Leishmania parasites can escape the host immune response: A signaling point of view. Clin Microbiol Rev. 2005;18:293–305. doi: 10.1128/CMR.18.2.293-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco A.C.L., Araujo F.F., Kamimura M.T., Medeiros S.R., Viana D.A., Oliveira F.C.E., Araújo-Filho R., Costa M.P., Oliveira D.M. Following the Viterbi path to deduce flagellar actin-interacting proteins of Leishmania spp.: Report on Cofilins and Twinfilins. Proceedings of Computational Models For Life Sciences; International Symposium on Computational Models of Life Sciences; Melville: American Institute of Physics; 2007. pp. 315–324. AIP Proceedings of International Symposium on Computational Models of Life Sciences. [Google Scholar]
- Peters W., Killick-Kendrick R. The Leishmaniases in Biology and Medicine v. II: Clinical Aspects and Control. In: Peters W., Killick-Kendrick R., editors. London: Academic Press; 1987. pp. 551–941. [Google Scholar]
- Puentes S.M., Dwyer D.M., Bates P.A., Joiner K.A. Binding and release of C3 from Leishmania donovani promastigotes during incubation in normal human serum. J Immunol. 1989;143:3743–3749. [PubMed] [Google Scholar]
- Rasmusson B., Descoteaux A. Contribution of electron and confocal microscopy in the study of Leishmania-macrophage interactions. Microsc Microanal. 2004;10:656–661. doi: 10.1017/S1431927604040851. [DOI] [PubMed] [Google Scholar]
- Rosenberger C.M., Finlay B.B. Phagocyte sabotage: Disruption of macrophage signalling by bacterial pathogens. Nat Rev Mol Cell Biol. 2003;4:385–396. doi: 10.1038/nrm1104. [DOI] [PubMed] [Google Scholar]
- Rybakin V., Clemen C.S. Coronin proteins as multifunctional regulators of the cytoskeleton and membrane trafficking. BioEssays. 2005;27:625–632. doi: 10.1002/bies.20235. [DOI] [PubMed] [Google Scholar]
- Rybakin V., Gounko N.V., Späte K., Höning S., Majoul I.V., Duden R., Noegel A.A. Crn7 interacts with AP-1 and is required for the maintenance of Golgi morphology and protein export from the Golgi. Biol Chem. 2006;281:31070–31078. doi: 10.1074/jbc.M604680200. [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L.J. Comparative protein modelling by satisfaction of spatial restraints. Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Scianimanico S., Desrosiers M., Dermine J.F., Meresse S., Descoteaux A., Desjardins M. Impaired recruitment of the small GTPase rab7 correlates with the inhibition of phagosome maturation by Leishmania donovani promastigotes. Cell Microbiol. 1999;1:19–32. doi: 10.1046/j.1462-5822.1999.00002.x. [DOI] [PubMed] [Google Scholar]
- Sibley L.D. Toxoplasma gondii: Perfecting an intracellular life style. Traffic. 2003;4:581–586. doi: 10.1034/j.1600-0854.2003.00117.x. [DOI] [PubMed] [Google Scholar]
- Snapp E.L., Landfear S.M.J. Characterization of a targeting motif for a flagellar membrane protein in Leishmania enriettii. Biol Chem. 1999;274:29543–29548. doi: 10.1074/jbc.274.41.29543. [DOI] [PubMed] [Google Scholar]
- Song L., Dentler W.L. Flagellar protein dynamics in Chlamydomonas. J Biol Chem. 2001;276:29754–29763. doi: 10.1074/jbc.M103184200. [DOI] [PubMed] [Google Scholar]
- Stuart L.M., Boulais J., Charriere G.M., Hennessy E.J., Brunet S., Jutras I., Goyette G., Rondeau C., Letarte S., Huang H., et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S., Schaible U.E., Russell D.G. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 1996;15:6960–6696. [PMC free article] [PubMed] [Google Scholar]
- Tammana T.V., Sahasrabuddhe A.A., Mitra K., Bajpai V.K., Gupta C.M. Actin-depolymerizing factor, ADF/cofilin, is essentially required in assembly of Leishmania flagellum. Mol Microbiol. 2008;70:837–852. doi: 10.1111/j.1365-2958.2008.06448.x. [DOI] [PubMed] [Google Scholar]
- Tardieux I., Liu X., Poupel O., Parzy D., Dehoux P., Langsley G. A Plasmodium falciparum novel gene encoding a coronin-like protein which associates with actin filaments. FEBS Lett. 1998;441:251–256. doi: 10.1016/s0014-5793(98)01557-9. [DOI] [PubMed] [Google Scholar]
- Tull D., Vince J.E., Callaghan J.M., Naderer T., Spurck T., McFadden G.I., Currie G., Ferguson K., Bacic A., McConville M.J. SMP-1 a member of a new family of small myristoylated proteins in kinetoplastid parasites is targeted to the flagellum membrane in Leishmania. Mol Biol Cell. 2004;15:4775–4786. doi: 10.1091/mbc.E04-06-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich H.J., Beatty W.L., Russell D.G. Direct delivery of procathepsin D to phagosomes: Implications for phagosome biogenesis and parasitism by Mycobacterium. Eur J Cell Biol. 1999;78:739–748. doi: 10.1016/S0171-9335(99)80042-9. [DOI] [PubMed] [Google Scholar]
- Vasconcelos E.J.R., Pacheco A.C.L., Gouveia J.J.S., Araujo F.F., Diniz M.C., Kamimura M.T., Costa M.P., Maggioni R., Araujo-Filho R., Costa R.B., et al. Profilins, formins and katanins as flagellar proteins of Leishmania spp: A genome-based multi-step bioinformatics-driven description. In: Bourbakis N.G., editor. Conference Proceedings IEEE 7th International Symposium on Bioinformatics & Bioengineering-2007; Boston: IEEE Press; 2007. pp. 880–887. [Google Scholar]
- Vasconcelos E.J.R., Pacheco A.C.L., Gouveia J.J.S., Araujo F.F., Diniz M.C., Kamimura M.T., Costa M.P., Araujo-Filho R., Oliveira D.M. Actin-interacting proteins in flagellated pathogenic Leishmania spp. : A genome-based bioinformatics report on profilins, formins and katanins. Int J Funct Informat Personal Med. 2008;1:234–252. [Google Scholar]
- Vieira O.V., Botelho R.J., Grinstein S. Phagosome maturation: Aging gracefully. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: Known and predicted protein-protein associations integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese M., Kuhn D., Grünfelder C.G. Protein kinase involved in flagellar-length control. Eukaryot Cell. 2003;2:769–777. doi: 10.1128/EC.2.4.769-777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Collins R.F., Grinstein S., Trimble W.S. Coronin-1 function is required for phagosome formation. Mol Biol Cell. 2005;16:3077–3087. doi: 10.1091/mbc.E04-11-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa A.C., Kawashima S., Okuda S., Fujita M., Itoh M., Moriya Y., Hattori M., Kanehisa M. Extracting sequence motifs and the phylogenetic features of SNARE-dependent membrane traffic. Traffic. 2006;7:1104–1118. doi: 10.1111/j.1600-0854.2006.00451.x. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Black P.E. Viterbi algorithm. In: Black Paul E., editor. Dictionary of Algorithms and Data Structures [online] U.S. National Institute of Standards and Technology; 2004. [28 June 2009]. Available from: http://www.itl.nist.gov/div897/sqg/dads/HTML/viterbiAlgorithm.html. [Google Scholar]
- GeneDB, a core part of The Wellcome Trust Sanger Institute - Pathogen Sequencing Unit. [accessed02May 2009]. Available from: http://www.genedb.org.
- The Chlamydomonas Flagellar Proteome Project. [accessed28June 2009]. Available from: http://labs.umassmed.edu/chlamyfp/index.php.
- HMMER. package ver.2.3.1. [accessed17January 2009]. Available from: http://www.hmmer.wustl.edu.
- Swiss-Prot/trEMBL. [accessed17January 2009]. Available from: http://www.expasy.org/sprot.
- TargetP. [accessed17January 2009]. Available from: http://www.cbs.dtu.dk/services/TargetP.
- Chime browser plug-in. [accessed17January 2009]. Available from: http://www.mdl.com.