Abstract
The phylogenetic relationships of 15 taxa from Hystrix and the related genera Leymus (NsXm), Elymus (StH), Pseudoroegneria (St), Hordeum (H), Psathyrostachys (Ns), and Thinopyrum (E) were examined by using the Giemsa C-banded karyotype. The Hy. patula C-banding pattern was similar to those of Elymus species, whereas C-banding patterns of the other Hystrix species were similar to those of Leymus species. The results suggest high genetic diversity within Hystrix, and support treating Hy. patula as E. hystrix L., and transferring Hy. coreana, Hy. duthiei ssp. duthiei and Hy. duthiei ssp. longearistata to the genus Leymus. On comparing C-banding patterns of Elymus species with their diploid ancestors (Pseudoroegneria and Hordeum), there are indications that certain chromosomal re-arrangements had previously occurred in the St and H genomes. Furthermore, a comparison of the C-banding patterns of the Hystrix and Leymus species with the potential diploid progenitors (Psathyrostachys and Thinopyrum) suggests that Hy. coreana and some Leymus species are closely related to the Ns genome of Psathyrostachys, whereas Hy. duthiei ssp. duthiei, Hy. duthiei ssp. longearistata and some of the Leymus species have a close relationship with the E genome. The results suggest a multiple origin of the polyploid genera Hystrix and Leymus.
Keywords: C-banding, Elymus, genome, Hystrix, Leymus, Triticeae
Introduction
Hystrix Moench is a small perennial genus of the tribe Triticeae (Poaceae). Moench (1794) established the genus Hystrix with Hy. patula Moench as the type-species through its distinctive morphological character of either lacking glumes entirely or, if present, of possessing long setaceous awn-shaped ones. Since then, 11 species have been included in Hystrix (Hitchcock, 1951; Bor, 1960; Tzvelev, 1976; Kuo, 1987; Osada, 1993). Baden et al. (1997) revised the genus and recognized six species, one of which is divided into three subspecies within Hystrix. All are tetraploids (2n = 4x = 28) except for Hy. californica, which is an octaploid (2n = 8x = 56). The natural distribution of Hystrix is disjunct with two species in North America (Hy. patula and Hy. californica), and the remainder in Central and Eastern Asia (Löve, 1984; Baden et al., 1997).
Although separated early as a genus in its own right, the recognition of Hystrix has been controversial ever since its establishment. Church (1967a, 1967b) reported that Hy. patula had a close affinity to species of the Elymus canadensis complex and treated Hy. patula as E. hystrix L. Consequently, Dewey (1982) and Löve (1984) recognized the genus Hystrix as a section of Elymus. However, Jensen and Wang (1997) reported that two species of Hystrix, Hy. coreana and Hy. californica, shared the genome of Leymus (NsXm), and so transferred Hy. coreana from Hystrix to Leymus. Based on the results of studies on meiotic pairing and genomic in situ hybridization (GISH), Zhang et al. (2006) reported that Hy. patula possesses the ElymusStH genome, whereas Hy. duthiei ssp. duthiei and Hy. duthiei ssp. longearistata contain the LeymusNsXm genome. However, based on results of GISH and southern genomic hybridization, Ellenskog-Staam et al. (2007) reported that Hy. coreana, Hy. longearistata and Hy. duthiei contained the Ns1Ns2 genomes, while Hy. patula contained the StH genomes, and Hy. komarovii most likely had a variant of the StH genomes. Although the varied genomic constitution of Hystrix species has been reported, morphological similarities have likewise occurred, such as the loosely tufted caespitose, relatively high culms, broadly lanceolate leaves and the obsolete, reduced, or setaceous glumes. Thus, there is every indication that genome relationships and genetic diversity among Hystrix species need to be further investigated.
Karyotype analysis is considered to be an important method in genome analysis. Giemsa C-banded karyotyping stains constitutive heterochromatin, this resulting in unique banding patterns of individual chromosomes. This process thus provides more accurate evidence for identifying homologous chromosomes in karyologically similar species, thereby complementing studies of genome evolution among related species (Morris and Gill, 1987). Baden et al. (1997) undertook a pilot study on Giemsa C-banding patterns in Hy. patula, Hy. komarovii and Hy. coreana, and the results showed Hy. coreana as having large and conspicuous telomeric bands, different from the C-banding patterns of Hy. patula and Hy. komarovii. In this study, we investigated the genetic diversity among Hystrix species, as well as the phylogenetic relationship between Hystrix and its relatives (including related genera and their diploid ancestors) by using the Giemsa C-banded karyotype. The specific objectives were: (a) to report the Giemsa-C banded karyotypes of 15 perennial taxa in Triticeae representing nine genera; (b) to estimate genetic diversity among these perennial species; (c) to compare C-banding patterns among the diploid and tetraploid species; and (d) to explore the possible diploid ancestor of Hystrix species.
Materials and Methods
Plant material
A total of 15 taxa in Triticeae were used in this study, including four taxa of Hystrix (2n = 4x = 28), three species of Leymus (2n = 4x = 28, NsXm genome), two species of Elymus (2n = 4x = 28, StH genome), two species of Pseudoroegneria (2n = 2x = 14, St genome), two species of Psathyrostachys (2n = 2x = 14, Ns genome), Hordeum bogdanii (2n = 2x = 14, H genome), and Thinopyrum bessarabicum (2n = 2x = 14, Eb genome) (Table 1). All seed material was collected in the field by the authors of this paper or kindly provided by the American National Plant Germplasm System (Pullman, Washington, USA) and Dr. S. Sakamoto (Kyoto University, Japan). Voucher specimens were deposited in the Herbarium of the Triticeae Research Institute, Sichuan Agricultural University, China (SAUTI).
Table 1.
- Species of Hystrix and other closely related genera used in Giemsa-C banding analysis.
| Species | 2n | Genome | Accession n. | Origin |
| Pseudoroegneria libanotica (Hack.) D. R. Dewey | 14 | St | PI 228391 | Ardabil, Iran |
| Pseudoroegneria spicata (Pursh) Á. Löve | 14 | St | PI 232138 | Montana, United States |
| Hordeum bogdanii Wilensky | 14 | H | Y 1508 | Xinjiang, China |
| Psathyrostachys huashanica Keng ex Kuo | 14 | Ns | ZY 3157 | Shanxi, China |
| Psathyrostachys juncea (Fisch.) Nevski | 14 | Ns | PI 430871 | Former Soviet Union |
| Thinopyrum bessarabicum (Savul. & Rayss) Á. Löve | 14 | Eb | PI 531711 | Crimea, Ukraine |
| Elymus sibiricus L. | 28 | StH | ZY 3041 | Sichuan, China |
| Elymus canadensis L. | 28 | StH | PI 236805 | Canada |
| Leymus arenarius (L.) Hochst. | 28 | NsXm | PI 272126 | Alma-Ata, Kazakhstan |
| Leymus racemosus | 28 | NsXm | PI 478832 | Montana, United States |
| Leymus multicaulis (Kar. & Kir.) Tzvelev | 28 | NsXm | PI 440325 | Dzhambul, Kazakhstan |
| Hystrix patula Moench | 28 | StH | PI 372546 | Ottawa, Canada |
| Hystrix coreana (Honda) Ohwi | 28 | NsXm | W6 14259 | Vladivostack, Russian Federation |
| Hystrix duthiei (Stapf) Bor ssp. duthiei | 28 | NsXm | ZY 2004 | Sichuan, China |
| Hystrix duthiei ssp. longearistata (Hack.) Baden, Fred. & Seberg | 28 | NsXm | ZY 2005 | Tokyo, Japan |
Giemsa C-banding analysis
All seeds were germinated in Petri dishes on moistened filter paper at 22 °C. Root tips from the germinating seeds were pre-treated in ice-cold water for 24-28 h, fixed in ethanol: acetic acid (3:1, v/v) for 24 h at room temperature, and then stored in the refrigerator. Each root tip was squashed in a drop of 45% acetic acid.
The Giemsa C-banding technique followed the procedure of Gill et al. (1991). Metaphase cells with a complete chromosome complement were photographed, five cells being subsequently analyzed for each material. Idiograms so constructed were based on chromosome lengths, similarities in their morphology, banding patterns and relative arm-ratios. Chromosomes were arranged from the longest to the shortest and were designated with the Arabic numerals 1-7 in diploids and 1-14 in tetraploid species.
Results
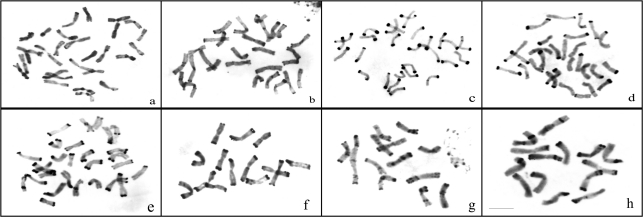
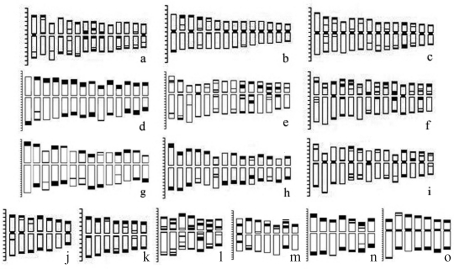
The Giemsa-C banded metaphase chromosomes in representative species of Hystrix and relatives are shown in Figure 1. The C-banded karyotypes and ideograms of all the taxa in Hystrix and related genera are shown in Figures 2 and 3, respectively.
C-banding in Hystrix species
There were small telomeric bands in all of the 14 chromosomes in Hy. patula most of which with minor intercalary bands (Figure 1a). Large centromeric bands were also present in both arms of chromosomes 7, 8 and 9 (Figures 2a, 3a).
Hy. duthiei ssp. duthiei and Hy. duthiei ssp. longearistata revealed similar basic C-banding patterns. All the 14 chromosomes presented minor to small terminal and centromeric C-bands, except for chromosome 2 of Hy. duthiei ssp. duthiei, where centromeric bands were absent (Figures 2b, 2c). In Hy. duthiei ssp. longearistata, nine of the fourteen chromosomes contained minor interstitial bands, whereas these were present in only four of the 14 chromosomes in Hy. duthiei ssp. duthiei (Figures 3b, 3c).
Large terminal bands in all the 14 chromosomes were characteristic of the C-banding pattern in Hy. coreana. Except for a minor interstitial band in chromosome 2, no centromeric or interstitial bands were encountered in this species (Figures 1c, 3d).
C-banding in Elymus (StH), Pseudoroegneria (St) and Hordeum (H) species
Giemsa-C banded karyotypes in two Elymus species containing the StH genome were comprised of terminal, interstitial and a few centromeric bands. In E. sibiricus, small to medium terminal C-bands were observed in one or both arms of all the chromosomes, besides rather large interstitial bands in chromosomes 9-13 (Figures2e, 3e). In E. canadensis, all the chromosomes presented terminal C-bands in one or both arms. Furthermore, distinct bands were located near the centromere in both arms of chromosome 10, besides two pairs of chromosomes containing centromeric bands (Figures 2f, 3f). The banding pattern of E. canadensis is similar to that of Hy. patula.
In Pseudoroegneriaspicata (St) and Pse. libanotica (St), the C-banding patterns were rather similar, being characterized by large terminal bands in both arms or only in the short arm of all the seven chromosomes. Centromeric bands were found in chromosomes 1, 2 and 7 in Pse. spicata (Figures 2j, 3j), whereas in Pse. libanotica these were observed in chromosomes 1, 4, 6 and 7 (Figures 2k, 3k).
In H. bogdanii (H), small to medium interstitial bands were present in one or both arms of all the chromosomes, all of which showed terminal bands (Figures 2l, 3l).
C-banding in Leymus (NsXm), Psathyrostachys (Ns) and Thinopyrum (E) species
Although distinct terminal bands were present in the chromosomes of L. arenarius and L. racemosus, they were rather faint in those of L. multicaulis. All the 14 chromosomes of L. arenarius and L. racemosus, with the exception of chromosome 10 of L. arenarius, presented large terminal or interstitial C-bands and the absence of centromeric bands (Figures 2g, 3g; 2h, 3h). Small to medium terminal, interstitial and centromeric bands were found in the chromosomes of L. multicaulis (Figures 2i, 3i).
The C-banding patterns of the two Psathyrostachys (Ns) species were characterized by diagnostic terminal or interstitial bands in all the seven chromosomes (Figures 2m, n; 3m, n). One satellited chromosome (chromosome 6) and one chromosome with centromeric C-bands (chromosome 1) were observed in Psa. juncea (Figures 2m, 3m). Large terminal C-bands were observed in one or both arms of all the chromosomes in Psa. huashanica, although there were no centromeric bands (Figures 2n, 3n).
Figure 1.
Giemsa-C banded metaphase chromosomes in representative species of Hystrix and relatives. a.Hystrix patula. b.Hy. duthiei ssp. longearistata. c.Hy. coreana. d.Leymus arenarius.e.L. racemosus.f.Pseudoroegneria libanotica. g.Hordeum bogdanii. h.Psathyrostachys juncea. Bar = 10 μm.
Distinct terminal and centromeric C-bands were noted in one or both arms of the seven Th. bessarabicum (Eb) chromosomes. No interstitial bands were observed (Figures 2o, 3o).
Discussion
Relationships among Hystrix, Elymus and Leymus
Cytological and molecular studies showed that species of Hystrix differed as to genomic constitution (Jensen and Wang, 1997; Mason-Gamer et al., 2002; Zhang and Zhou, 2006; Zhang et al., 2006; Ellenskog-Staam et al., 2007; Fan et al., 2007). Hy. patula, the type species of Hystrix, shared the StH genome of Elymus, whereas Hy. coreana, Hy. duthiei ssp. duthiei, Hy. duthiei ssp. longearistata and Hy. californica contained the NsXm genome of Leymus. In this study, the Giemsa C-banding patterns of four taxa of Hystrix were different. Furthermore, the C-banding patterns of Hy. patula were similar to those of E. canadensis and E. sibiricus. Darkly stained centromeric bands were observed in all the three species, although these were absent in the remaining Hystrix species. The results were consistent with those of chromosome pairing and GISH, hence suggesting a close relationship between Hy. patula and the Elymus species and a distant one between Hy. patula and the other species of Hystrix.
C-banding patterns of Hy. duthiei ssp. duthiei and Hy. duthiei ssp. longearistata were characterized by minor terminal and centromeric bands in almost all of the 14 chromosomes, thus displaying a certain degree of similarity with those of L. multicaulis. Nevertheless, there were differences in the number of minor interstitial bands. Hy. duthiei ssp. longearistata revealed 23 terminal bands, whereas Hy. duthiei ssp. duthiei only 16. Zhou et al. (1999) reported a certain morphological divergence and sterility barrier between the two taxa due to a difference in distribution and habitat. From previous cytological and molecular studies on our part, it was shown that the NsXm genomes of Hy. duthiei ssp. duthiei and Hy. duthiei ssp. longearistata were the same as those of the genus Leymus (Zhang et al., 2006, 2008). In this study, the C-banding patterns of the two taxa were similar to those of L. multicaulis, although less centromeric and more interstitial bands were found in the latter. This indicated that Hy. duthiei ssp. duthiei and Hy. duthiei ssp. longearistata were closely related to L. multicaulis, which is congruent with results from cytological and molecular studies.
Figure 2.
C-banded karyotypes in 15 taxa of Hystrix, Elymus, Leymus,Pseudoroegneria, Hordeum, Psathyrostachys and Thinopyrum. a.Hystrix patula. b. Hy. duthiei ssp. duthiei. c.Hy.duthiei ssp. longearistata. d.Hy. coreana. e.Elymus sibiricus. f.E. canadensis. C-banded karyotypes in 15 taxa of Hystrix, Elymus, Leymus,Pseudoroegneria, Hordeum, Psathyrostachys and Thinopyrum. g.Leymus arenarius. h.L. racemosus. i.L. multicaulis. j.Pseudoroegneria spicata. k.Pse. libanotica. l.Hordeum bogdanii. m.Psathyrostachys juncea. n.Psa. huashanica. o.Thinopyrum bessarabicum.
Jensen and Wang (1997) reported that Hy. coreana contained the NsXm of Leymus and so transferred the species to this genus. In this study, Hy. coreana revealed distinct terminal bands in all the 14 chromosomes, which similar to the banding patterns of L. arenarius and L. racemosus, and consistent with cytological and molecular studies.
Relationships between tetraploids and their diploid ancestors
From studies on chromosome pairing, there are indications that the St and H genome in Elymus originated from Pseudoroegneria and Hordeum, respectively (Dewey, 1967, 1971). In this study, C-banding diversity was observed among Elymus (including Hy. patula), Pseudoroegneria and Hordeum. Distinct terminal C-bands were observed, for example, in Pseudoroegneria (St), these being absent in tetraploid Elymus (StH) species. Similar results were found in E. trachycaulus (2n = 4x = 28, StH) and Pse. spicata (St) (Morris and Gill, 1987). These results suggested the occurrence of chromosomal re-arrangement between the St and H genomes in polyploidization events during the speciation process.
Previous cytological and molecular studies showed that species of Leymus have either JN, or Ns1Ns2, or NsXm genomes (Zhang and Dvorak, 1991; Wang et al., 1994; Sun et al., 1995; Anamthawat-Jónsson, 2005). The J (E) genome is from Thinopyrum and the Ns from Psathyrostachys. From this study it was indicated that two Psathyrostachys (Ns) species, besides L. arenarius (NsXm), L. racemosus (NsXm) and Hy. coreana (NsXm), had large terminal bands. However, the C-banding patterns of Th. bessarabicum (Eb), L. multicaulis (NsXm), Hy. duthiei ssp. duthiei (NsXm) and Hy. duthiei ssp. longearistata (NsXm) were characterized by centromeric and terminal bands. From the results it could be inferred that Hy. coreana and some species of Leymus were closely related to the Ns genome of Psathyrostachys, whereas for Hy. duthiei ssp. duthiei, Hy. duthiei ssp. longearistata and a part of Leymus species this was so with the E genome. The present data are consistent with previous cytological and molecular data, thereby suggesting large genetic diversity within the genera Hystrix and Leymus, and the multiple-origin of the polyploid genera Hystrix and Leymus (Sun et al., 1995; Anamthawat-Jónsson and Bödvarsdóttir, 2001; Yang et al., 2006; Zhang and Zhou, 2006).
The use of C-banding in the phylogeny of Triticeae
Giemsa C-binding has been widely used in chromosome identification, genetic mapping and studies on genome evolution of Triticeae species, ever since it was first reported (e.g., Morris and Gill, 1987; Gill et al., 1991; Linde-Laursen and Baden, 1994). The basic C-banding patterns of Pse. spicata, H. bogdanii, Psa. juncea, Psa. huashanica, L. racemosus, L. multicaulis, Hy. coreana, and Hy. patula, as exposed in the present study, are consistent with the findings from previous studies on C-banding (Linde-Laursen and von Bothmer, 1986; Morris and Gill, 1987; Wei et al., 1995; Baden et al., 1997; Wang et al., 1999; Ge et al., 2004). These findings imply that the C-banding technique is relatively stable and repeatable. The C-banding analysis undertaken in this study revealed genetic diversity and phylogenetic relationships among species from nine genera in Triticeae, consistent with available data on chromosome pairing and molecular evidence. Thus, the Giemsa C-banding technique can be used as a supplementary method for analyzing the genomic constitution of wild species, especially the Triticeae.
Figure 3.
Ideograms in 15 taxa of Hystrix, Elymus, Leymus,Pseudoroegneria, Hordeum, Psathyrostachys and Thinopyrum. a.Hystrix patula. b.Hy. duthiei ssp. duthiei. c.Hy.duthiei ssp. longearistata. d.Hy. coreana. e.Elymus sibiricus. f.E. canadensis. g.Leymus arenarius. h.L. racemosus. i.L. multicaulis. j.Pseudoroegneria spicata. k.Pse. libanotica. l.Hordeum bogdanii. m.Psathyrostachys juncea. n.Psa. huashanica. o.Thinopyrum bessarabicum.
Acknowledgments
We wish to thank Mr. Huan-Lin Shu (Triticeae Research Institute, Sichuan Agricultural University, China) for his valuable suggestions on C-banding analysis. We are grateful to Dr. S. Sakamoto (Kyoto University, Japan) and the American National Plant Germplasm System (Pullman, Washington, USA) for providing a part of the seed-material used in this study. This study was support by grants from the Program for Changjiang Scholars and the Innovative Research Team in University (PCSIRT) China (No. IRT 0453), the National Natural Science Foundation of China (No. 30670150) and the Science and Technology Bureau and the Education Bureau of Sichuan Province, China.
Footnotes
Associate Editor: Marcelo Guerra
References
- Anamthawat-Jónsson K. The Leymus Ns-genome. In: Holubec V., Barkworth M., von Bothmer R., editors. Proceedings of the 5th International Triticeae Symposium; Prague: 2005. pp. 13–20. [Google Scholar]
- Anamthawat-Jónsson K., Bödvarsdóttir S.K. Genomic and genetic relationships among species of Leymus (Poaceae, Triticeae) inferred from 18S-26S ribosomal genes. Am J Bot. 2001;88:553–559. [PubMed] [Google Scholar]
- Baden C., Frederiksen S., Seberg O. A taxonomic revision of the genus Hystrix (Triticeae, Poaceae) Nord J Bot. 1997;17:449–467. [Google Scholar]
- Bor N.L. The Grasses of Burma, Ceylon, India and Pakistan. New York: Pergamon Press; 1960. [Google Scholar]
- Church G.L. Taxonomic and genetic relationships of eastern North American species of Elymus with setaceous glumes. Rhodora. 1967a;69:121–162. [Google Scholar]
- Church G.L. Pine Hills Elymus. Rhodora. 1967b;69:330–351. [Google Scholar]
- Dewey D.R. Synthetic hybrids of Agropyron scribneri x Elymus juncea. Bull Torrey Bot Club. 1967;94:388–395. [Google Scholar]
- Dewey D.R. Synthetic hybrids of Hordeum bogdanii with Elymus canadensis and Sitanion hystrix. Am J Bot. 1971;58:902–908. [Google Scholar]
- Dewey D.R. Estes JR, Tyrl RJ and Brunken JN Grasses and Grasslands: Systematics and Ecology. Norman: University of Oklahoma Press; 1982. Genomic and phylogenetic relationships among North American perennial Triticeae; pp. 51–88. [Google Scholar]
- Ellenskog-Staam P., von Bothmer R., Anamthawat-Jónsson K., Salomom B. Genome analysis of species in the genus Hystrix (Triticeae, Poaceae) Plant Syst Evol. 2007;265:241–249. [Google Scholar]
- Fan X., Zhang H.Q., Sha L.N., Zhang L., Yang R.W., Ding C.B., Zhou Y.H. Phylogenetic analysis among Hystrix, Leymus and its affinitive genera (Poaceae, Triticeae) based on the sequences of a gene encoding plastid acetyl-CoA carboxylase. Plant Sci. 2007;172:701–707. [Google Scholar]
- Ge R.C., Zhao B.C., Chen Y.Z., Huang Z.J., Zhao M.L. Study on C-banding and karyotype of Leymus multicaulis. Grassland of China. 2004;26:72–74. [Abstract in English] [Google Scholar]
- Gill B.S., Friebe B., Endo T.R. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum) Genome. 1991;34:830–839. [Google Scholar]
- Hitchcock A.S. Manual of the Grasses of the United States. USDA Miscellaneous Publication 200. New York: Dover Press; 1951. pp. 230–280. [Google Scholar]
- Jensen K.B., Wang R.R.-.C. Cytological and molecular evidence for transferring Elymus coreanus from the genus Elymus to Leymus and molecular evidence for Elymus californicus (Poaceae, Triticeae) Int J Plant Sci. 1997;158:872–877. [Google Scholar]
- Kuo P.C. Pooideae Flora Reipublicae Popularis Sinicae 9 (3) Beijing: Science Press; 1987. pp. 34–37. [Google Scholar]
- Linde-Laursen I., Baden C. Giemsa C-banded karyotypes of two cytotypes (2x, 4x) of Psathyrostachys lanuginosa (Poaceae; Triticeae) Hereditas. 1994;120:11–120. [Google Scholar]
- Linde-Laursen I., von Bothmer R. Comparison of the karyotypes of Psathyrostachys juncea and P. huashanica studied by banding techniques. Plant Syst Evol. 1986;151:203–213. [Google Scholar]
- Löve A. Conspectus of the Triticeae. Feddes Repert. 1984;95:425–521. [Google Scholar]
- Mason-Gamer R.J., Orme N.L., Anderson C.M. Phylogenetic analysis of North American Elymus and the monogenomic Triticeae using three chloroplast DNA data sets. Genome. 2002;45:991–1002. doi: 10.1139/g02-065. [DOI] [PubMed] [Google Scholar]
- Moench C. Methodus Plantas Horti Botanici et Agri Marburgensis a Staminum Situ Describendi. Marburg: Marburgi Cattorum; 1794. [Google Scholar]
- Morris K.L.D., Gill B.S. Genomic affinities of individual chromosomes based on C- and N-banding analyses of tetraploid Elymus species and their diploid progenitor species. Genome. 1987;29:247–252. [Google Scholar]
- Osada T. Illustrated Grasses of Japan. Enlarged edition. Tokyo: Heibonsha Ltd Press; 1993. [Google Scholar]
- Sun G.L., Wu B.H., Liu F. Cytogenetic and genomic relationships of Thinopyrum elongatum with two Psathyrostachys species and with Leymus secalinus. Plant Syst Evol. 1995;197:225–231. [Google Scholar]
- Tzvelev N.N. Poaceae, URSS. Leningrad: Nauka; 1976. [Google Scholar]
- Wang R.R.-.C., von Bothmer R., Dvorák J., Fedak G., Linde-Laursen I., Muramatsu M. Genome symbols in the Triticeae. In: Wang R.R.-.C., Jensen K.B., Jaussi C., editors. Proceedings of the 2nd International Triticeae Symposium; Utah: Logan; 1994. pp. 29–34. [Google Scholar]
- Wang S.L., Qi L.L., Chen P.D., Liu D.J. Study on chromosome C-banding in Leymus racemosus and its related species. Acta Bot Sin. 1999;41:258–262. [Abstract in English] [Google Scholar]
- Wei J.Z., Campbell F.W., Wang R.R.-.C. Standard Giemsa C-banded karyptype of Russian wild rye (Psathyrostachysjuncea) and its use in identification of a deletion-translocation heterozygote. Genome. 1995;38:1262–1270. doi: 10.1139/g95-166. [DOI] [PubMed] [Google Scholar]
- Yang R.W., Zhou Y.H., Zhang Y., Zheng Y.L., Ding C.B. The genetic diversity among Leymus species based on random amplified microsatellite polymorphism. Genet Resour Crop Evol. 2006;53:139–144. [Google Scholar]
- Zhang H.B., Dvorák J. The genome origin of tetraploid species of Leymus (Poaceae, Triticeae) inferred from variation in repeated nucleotide sequences. Am J Bot. 1991;78:871–884. [Google Scholar]
- Zhang H.Q., Yang R.W., Dou Q.W., Tsujimoto H., Zhou Y.H. Genome constitution of Hystrix patula, Hystrix duthiei ssp. duthiei and Hystrix duthiei ssp. longearistata (Poaceae, Triticeae) revealed by meiotic pairing behaviour and genomic in situ hybridization. Chromosome Res. 2006;14:595–604. doi: 10.1007/s10577-006-1057-2. [DOI] [PubMed] [Google Scholar]
- Zhang H.Q., Zhou Y.H. Meiotic pairing behaviour reveals differences in genomic constitution between Hystrixpatula and other species of genus Hystrix Moench (Poaceae, Triticeae) Plant Syst Evol. 2006;258:129–136. [Google Scholar]
- Zhang H.Q., Fan X., Sha L.N., Zhang C., Yang R.W., Zhou Y.H. Phylogeny of Hystrix and related genera (Poaceae, Triticeae) based on nuclear rDNA ITS sequences. Plant Biology. 2008;10:635–642. doi: 10.1111/j.1438-8677.2008.00065.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y.H., Yang J.L., Yen C., Zheng Y.L. Biosystematic studies on Hystrix longearistata from Japan and Hystrix duthiei from China (Poaceae, Triticeae) Acta Phytotaxon Sin. 1999;37:386–393. [Abstract in English] [Google Scholar]