Abstract
Retrotransposons (RTEs) are a principal component of most eukaryotic genomes, representing 50%-80% of some grass genomes. RTE sequences have been shown to be preferentially present in disease resistance gene clusters in plants. Arabidopsis thaliana has over 1,600 annotated RTE sequences and 56 of these appear to be expressed because of the exact expressed sequence tag (EST) matches and the presence of intact open reading frames. Of the 22 represented in the Affymetrix ATH1 array, AtCOPIA4 was found to be expressed at a higher level than all other RTEs across different developmental stages. Since AtCOPIA4 is located in the RPP5 gene cluster and is adjacent to RPP4 which confers resistance to the downy mildew oomycete Hyaloperonospora parasitica isolate EMWA1, we evaluated AtCOPIA4 mutants for resistance to this pathogen. T-DNA insertional and antisense knockout of AtCOPIA4 was found to reduce the resistance of wild type plants by 2-4 folds. Our results suggest that retrotransposon can be exapted to participate in plant defense response.
Keywords: Arabidopsis thaliana, retrotransposon, downy mildew resistance, knockout
Retrotransposons (RTEs) are a principal component of most eukaryotic genomes, representing more than 40% of the human genome (Kazazian, 2004; Lander et al., 2001) and 50%-80% of some grass genomes (Feschotte et al., 2002; SanMiguel and Bennetzen, 1998). Even in the compact genome of Arabidopsis thaliana, they account for 5.5% of the sequenced genome (Kazazian, 2004). Cellular functions of RTEs have been reported. They seem to play a role in the proliferation of cancer cells (Oricchio et al. 2007), globally interfere with the regulatory network of transcription factors p53 (Wang et al., 2007) and PSF (Song et al., 2005), and interact with the dynein light-chain which is a known component of the dynein microtubule motor (Havecker et al., 2005). In mammals, RTEs are more likely to be found in rapidly evolving gene clusters, such as those involved in defense and response to external signals, than in mRNAs of highly conserved genes involved in development, transcription, replication, cell structure and metabolism (Medstrand et al., 2005; van de Lagemaat et al., 2003). In plants, the pattern is similar. For example, Tos17 retrotransposon is preferably inserted into disease/defense-related and signal transduction (kinase) genes in the rice genome (Miyao et al., 2003). Furthermore, RTEs have been identified in disease resistance gene clusters in lettuces (Meyers et al., 1998; Michelmore and Meyers, 1998), rice (Song et al., 1995), barley (Marcel et al., 2007), the common bean (Vallejos et al., 2006), poplar (Lescot et al., 2004), and Arabidopsis (van der Biezen et al., 2002; Yi and Richards, 2007). Various RTEs have been shown to be induced by plant pathogens or elicitors in rice (Chen et al., 2007; Vergne et al., 2008), by Fusarium oxysporum in chickpea (Nimbalkara et al., 2006), and by fungal elicitors in tobacco (Pouteau et al., 1994; Melayah et al., 2001). In addition, RTE Tnt1A inserted in a tobacco resistance gene cluster has been shown to drive partial transcription of the neighboring disease resistance gene TNLL1 (Hernández-Pinzón et al., 2009).
RTE coding sequences are also known to form chimeric transcripts (Kashkush et al., 2003; Peaston et al., 2004) with non-RTE mRNA sequences and chimeric transcripts displaying a different expression pattern from that of the original transcripts (Peaston et al., 2004). Chimeric resistance and retrotransposon genes may function in disease resistance. For example, L10 is a Toll/Interleukin1 receptor-nucleotide binding site-leucine-rich repeat [TIR-NBS-LRR] class of resistance gene (Lawrence et al., 1995) and a chimera of the L10 TIR domain fused with a partial tobacco retrotransposon sequence at the 3' end has been reported. Expression of this chimera caused the same stunted phenotype produced by over-expressing full-length L6, and increased transcript abundance of a constitutive defense protein PR-1 (Frost et al., 2004). Similarly, Xa21D truncated at the 3' end with only the extracellular LRR domain by the retrotransposon Retrofit confers partial resistance to the bacterial pathogen Xanthomonas oryzae pv oryzae (Wang et al., 1998).
Here we show that knocking out the Arabidopsis retrotransposon AtCOPIA4 (At4g16870; Yi and Richards 2007) reduces resistance to the downy mildew pathogen Hyaloperonospora parasitica isolate EMWA1. AtCOPIA4 is represented in a single copy of the Arabidopsis genome based on BLAST search, and is located next to RPP4, separated only by its long terminal repeat (LTR; Figure 1). AtCOPIA4 protein contains the conserved domains of gag-integrase-reverse transcriptase. In silico EST analysis identified a chimeric cDNA consisting of the first exon of RPP4 which encodes the complete TIR domain upstream from the partial sequence of AtCOPIA4 (Figure 1), similar in configuration to the resistance gene domains truncated downstream by RTEs described above. Pathogenicity assays demonstrated that T-DNA insertional and antisense RNAi mutants were 2 to 4 times as likely to be infected by H. parasitica isolate EMWA1 to which Arabidopsis RPP4 (At4g16860), a TIR-NB-LRR class of disease resistance gene, confers host resistance (van der Biezen et al., 2002).
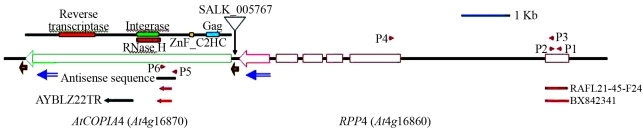
Figure 1.
The Arabidopsis genomic region of AtCOPIA4 (in green) and RPP4 (in red) based on Yi and Richards (2007) who have sequenced the full-length cDNA of genes in this region. Open boxes represent exons and lines between boxes represent introns in RPP4. AtCOPIA4 conserved domains are indicated above the gene. Location of T-DNA insertion is indicated for SALK_005767. Antisense sequence represented by a black line below was used for an AtCOPIA4 antisense construct. One cDNA match (AYBLZ22TR) to AtCOPIA4 is also shown. Chimeric cDNAs are drawn in red broken lines and arrows (RAFL21-45-F24 and BX842341). Two chimeric ESTs were also identified in GenBank: ES444452 and EL142415 (not shown). Affymetrix GeneChip probes for both genes are shown in blue arrows. Brown open arrows below the ends of AtCOPIA4 are the 130 bp long terminal repeats (LTRs; 9488607-9488478 and 9483894-9483755, respectively).
Potential AtCOPIA4 T-DNA insertional mutant SALK_005767 in the Col-0 background (Alonso et al., 2003) was obtained from the Arabidopsis Biological Resources Center at Ohio State University. To identify a homozygous insertion plant, two PCR reactions with primers LP+RP and LB+RP were set up using Ex Taq from Takarabio USA (Madison, WI). PCR was run with initial denaturing at 94 °C for 2 min and 35 cycles of 94 °C/30 s, 58 °C/30 s and 72 °C/2 min., followed by 72 °C for 5 min. A single PCR product from LB and RP primers was amplified and sequenced to determine the exact T-DNA insertion site in homozygous plants. Position of T-DNA insertion was thus determined and indicated in Figure 1. One heterozygous and one homozygous plant were identified and used in the study. Primers used for plant identification were:
LB: GCGTGGACCGCTTGCTGCAACT
LP: CTACTGATGTATTGTTGCCAGAGG
RP: ATCTCCGTAATAGAGGGAGTGTTG
Antisense RNAi plants were generated through the transformation of antisense sequence of AtCOPIA4 (see Figure 1), by using primers A1 (AACTAAAGACGAGCT CTATGAATG) and A2 (TCTAGATTAATGAAACAAT CCGAACAAG) which contain restriction sites for SacI and XbaI, respectively. The amplified PCR product was first cloned into a TA cloning vector pGEM T Easy (Promega, WI), and then into the binary vector pBI121 digested with SacI and XbaI. Arabidopsis Col-0 transformation followed a floral-dip protocol as described (Clough and Bent, 1998). T2 transgenic plants were used in the pathogenicity assay.
For RT-PCR analysis, total RNA was isolated from two week-old Hyaloperonospora parasitica EMWA1 infected seedlings using TRIzol (Invitrogen, CA), and then treated with DNase I (Ambion, TX) according to manufacturer's protocol. RT-PCR was performed using the Verso 1-Step RT-PCR kit (Thermo Scientific/Fisher, PA). PCR was run for 15 min at 50 °C, 15 min at 95 °C, followed by 25 cycles of 95 °C/30 s, 58 °C/30 s, 72 °C/2 min, and the final extension of 5 min at 72 °C. All RT-PCR primers were tested for their target specificity using Seqviewer (www.arabidopsis.org). All the primers used showed desired specificity:
P1: GTAGATGTTCGCAAAACGTTCCTC
P2: AATCACCATTTGTTCCCCTTTCTT
P3: TTAAGAGCAAGACCTTGAGATGGC
P4: GAGGACAAACCAGAGGATCAGAAA
P5: TGTTGCTCCAAGGGAGAACTAAAG
P6: ATGAAACAATCCGAACAAGCAAGT
UBQ1: GATCTTTGCCGGAAAACAATTGGAGGATGGT
UBQ2: CGACTTGTCATTAGAAAGAAAGAGATAACAGG
To conduct pathogenicity assay, seeds were planted in soils (Metromix 360, SunGro, Canada) saturated with water and stratified at 4 °C for 48 h. Pathogenicity assays followed those described previously (Holub et al., 1994; Yoshioka et al., 2006). Briefly, 10 to 14 day-old conidiophores of H. parasitica isolate EMWA1 (kindly provided by Daniel Klessig) were collected from susceptible live plants of Nd-0 and re-suspended in cold, sterile water. The spores were vortexed for 30 s for release from the sporangia. Spore concentration was adjusted to 104-106 per mL, and 1-2 μL of the spore suspension was dropped onto each cotyledon of 6 to 7 day-old plants (10 to 20 plants for each line in each replicate). The inoculated plants were covered with plastic wrap and incubated at 16 °C with 10 hour-photoperiods. At 10 to 14 days after inoculation, the number of conidiophores on each cotyledon leaf, number of cotyledon leaves with conidiophores and the total number of plants, were recorded using a dissection microscope. The experiment was replicated three times with similar trends. Both resistant (Col-0) and susceptible (Nd-0; Holub et al., 1994) lines were used as pathogenicity assay controls, although only Col-0 data are shown in Figure 2 and Figure 3.
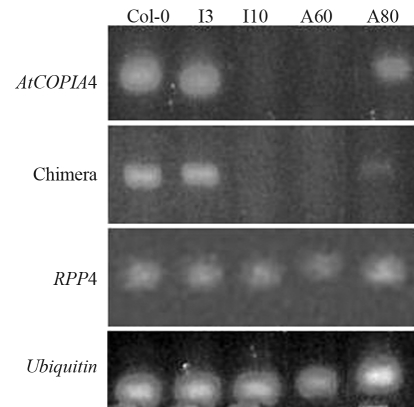
Figure 2.
RT-PCR of Arabidopsis T-DNA insertion and antisense mutants in AtCOPIA4. Primers used for AtCOPIA4 are P5 and P6, P1 and P6 for the chimeric transcript as shown in Figure 1 and P3 and P4 for RPP4. Total RNA from seedlings was used. Lines used are: Col-0-Columbia wild type; I3, I10-heterozygous and homozygous T-DNA insertion lines, respectively; A60, and A80 are antisense lines. Chimera indicates RPP4- AtCOPIA4 chimeric mRNA.
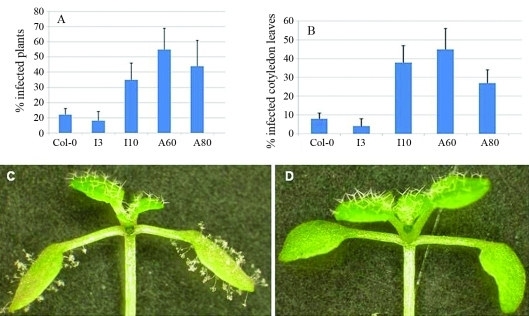
Figure 3.
Percentage of infected Arabidopsis wild type and mutant plants 10 days after inoculation of Hyaloperonospora parasitica EMWA1. (A) Percent of infected plants. (B) Percent of infected cotyledon leaves. (C) and (D) show an infected I10 and an uninfected wild type plant, respectively. Lines tested are: Col-0-Columbia wild type; I3, I10-heterozygous and homozygous T-DNA insertion lines, respectively; A60, and A80 are antisense lines.
To identify Arabidopsis retrotransposons that had acquired cellular functions, we searched the genome sequences of about 1,600 annotated retrotransposon genes curated in VirtualPlants (virtualplant.bio.nyu.edu; www.virtualplant.org) for matches to ESTs. Among these, 56 had exact matches to EST sequences and intact open reading frames. Twenty two of the genes were represented in the Affymetrix Arabidopsis ATH1 GeneChip and expression of those was searched in over 3,000 GeneChips in the Genevestigator database (Zimmermann et al., 2004). AtCOPIA4 was selected because it represents a typical retrotransposon which encodes gag, integrase and reverse transcriptase proteins (Feschotte et al., 2002), and it is the most highly expressed retrotransposon throughout the development stages, although generally their expression level is low due to regulation by the host. AtCOPIA4 transcript level was found to be highest in developing leaves and flowers (Table 1). This expression pattern was also confirmed by the Massively Parallel Signature Sequencing (MPSS) mRNA signature data (Nakano et al., 2006). Genes in this region have been shown to be co-expressed, probably due to local chromatin structural changes (Yi and Richards, 2007; Zhan et al., 2006). Cluster analysis in Genevestigator also revealed that AtCOPIA4, RPP4 and At4g16880, which are three adjacent genes on chromosome 4 based on the latest genome annotation release (TIGR/AGI V8) and Yi and Richards (2007), were coexpressed under salt, cold, heat, wound, oxidative, and genotoxic conditions (data not shown). Correlation of AtCOPIA4 expression is 0.59 with At4g16880 and 0.55 with RPP4, as calculated in the ATTED-II database of Arabidopsis microarray data (Obayashi et al., 2007). This correlation between AtCOPIA4 and RPP4 is noticeable in Table 1 as well.
Table 1.
Average signal intensity of selected retrotransposon genes in different developmental stages in Arabidopsisa.
| Germinated seed | Seedling | Young rosette | Developed rosette | Bolting | Young flower | Developed flower | Flowers and siliques | Siliques | |
| AtCOPIA4 (At4g16870) | 310 | 619 | 646 | 741 | 503 | 681 | 699 | 450 | 460 |
| RPP4 (At4g16860) | 80 | 432 | 1,053 | 978 | 380 | 1,291 | 771 | 332 | 536 |
| At3g21020 | 295 | 166 | 154 | 170 | 314 | 216 | 268 | 205 | 280 |
| At2g15510 | 125 | 145 | 134 | 147 | 112 | 193 | 195 | 124 | 201 |
| At2g17490 | 23 | 20 | 17 | 18 | 43 | 19 | 27 | 14 | 20 |
| ACT2 (At3g18780) | 14,828 | 18,867 | 15,468 | 16,580 | 13,033 | 14,243 | 13,732 | 16,403 | 5,333 |
| Total arraysb | 169 | 944 | 419 | 173 | 150 | 277 | 619 | 121 | 57 |
aOnly four of the 22 RTEs are presented in the table. The RTEs are randomly selected (except AtCOPIA4) to show that AtCOPIA4 has the highest transcript abundance. RPP4 is included as a comparison for its expression pattern with that of AtCOPIA4. Actin2 (ACT2) is included as a control. bTotal number of arrays (GeneChips) used to obtain the averaged signal for each stage. Data are gathered from the Genevestigator database (https://www.genevestigator.ethz.ch).
To elucidate the function of AtCOPIA4, a homozygous T-DNA insertion mutant was identified from SALK_005767 and antisense RNAi mutants were generated, as described above. Sequencing analysis indicated that T-DNA was inserted 70 bp before the start codon of AtCOPIA4 and 117 bp after the stop codon of RPP4 in SALK_005767 (Figure 1) in the LTR. RT-PCR analysis of the mutant seedlings indicates that the AtCOPIA4 transcript was undetectable in the homozygous T-DNA insertional mutant (I10) but present in the heterozygote (I3; Figure 2), indicating that transcription of AtCOPIA4 had been knocked out in the T-DNA insertion mutant. Among the two antisense mutants tested (A60, and A80), AtCOPIA4 transcript levels were undetectable in A60 and significantly reduced in A80 (Figure 2). In the mutants with no or reduced AtCOPIA4 transcript, the level of the AtCOPIA4-RPP4 chimeric transcript was also either not apparent or was at a reduced level (Figure 2). However, the abundance of RPP4 transcript was not affected in these lines, when compared to Col-0 and based on RT-PCR analysis, using primers P3 and P4, as shown in Figure 1 (Figure 2).
No noticeable morphological difference was observed between the mutants and Col-0. However, because AtCOPIA4 is located in the cluster of RPP5 class of resistance genes (van der Biezen et al., 2002; Yi and Richards, 2007), right next to RPP4 and in silico EST analysis had revealed a chimeric AtCOPIA4-RPP4 mRNA (Figure 1), we sought to evaluate the mutants for resistance to H. parasitica isolate EMWA1. Pathogenicity assays showed that on average, homozygous insertional and antisense mutants were 2 to 4 times as likely to be infected by the isolate based on percentage of infected plants, whereas heterozygous insertional mutants were as resistant to the isolate as Col-0 (Figure 3). Notwithstanding, the number of conidiophores was not significantly higher in the mutants examined 10 days after inoculation, when compared to wild type Col-0. Overall, the number of conidiophores ranges from 2 to 5 per cotyledon leaf on average for all lines and replicates and the highest number of conidiophores was 15 found in the mutants.
How AtCOPIA4 functions in the disease resistance is not clear. AtCOPIA4 may contribute to resistance to H. parasitica isolate EMWA1 either through the chimeric transcript (TIR-RTE) or through other mechanisms, since knockout undermines resistance conferred by RPP4. Even so, RPP4 transcript level was not noticeably different among the five lines tested (Figure 2). It has previously been shown that L10 TIR-RTE chimeric expression increases PR-1 transcription (Frost et al. 2004) and that a chimeric Xa21D-Retrofit confers partial resistance to Xanthomonas oryzae pv oryzae (Wang et al.. 1998). Retrofit is 41% identical and 57% similar to AtCOPIA4, based on a comparison of the whole protein sequence, thus making Retrofit the most homologous RTE from another species to AtCOPIA4. The coding region of Retrofit contains all the domains as in AtCOPIA4 and the truncated Xa21D encodes the LRR domain (Song et al.. 1997; Wang et al.. 1998). Both LRR and TIR domains affect resistance gene specificity in plants (Ellis et al.. 1999; Luck et al.. 2000). Apparently, expression of these domains alone could have an impact on disease resistance. While the chimera of L10 TIR-RTE and Xa21D-Retrofit are caused by RTE insertion in the DNA sequence, the RPP4 TIR-AtCOPIA4 fusion is due to the fact that the two genes are adjacent and chimeras formed at the RNA level (Figure 1). Therefore, it will be of interest to see whether increasing RPP4-AtCOPIA4 chimera expression would boost resistance as conferred by RPP4 because the level of the chimerical transcript was much lower in the mutants tested (Figure 2). AtCOPIA4 expression is driven by the 130 bp LTRs flanking the coding region. Future studies should focus on how the chimeric transcript is generated with the AtCOPIA4 sequence downstream from the RPP4 TIR domain, in contrast to what has been reported in other cases.
We have shown here that knockout of an RTE compromises plant resistance to the downy mildew pathogen H. parasitica EMWA1. RTEs have been shown to play a role in defense response in other eukaryotes as well. In mammals, degraded reverse transcribed RTEs can trigger defense response from the immune system (Stetson et al., 2008). Our evidence suggests that RTEs also function in defense response in plants.
Acknowledgments
We thank Ralph A. Dean of North Carolina State University, Daniel F. Voytas of the University of Minnesota, Daniel F. Klessig and Hong-Gu Kang of Boyce Thompson Institute and an anonymous reviewer for suggestions to improve the manuscript. Hyaloperonospora parasitica isolate EMWA1 was kindly provided by Daniel F. Klessig. The project was initiated at Penn State Erie. Funding for the project was provided by Penn State Erie-The Behrend College and the University of Louisiana at Lafayette.
Footnotes
Associate Editor: Márcio de Castro Silva Filho
References
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., et al. Genomewide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Chen F., Li Q., He Z. Proteomic analysis of rice plasma membrane-associated proteins in response to chitooligosaccharide elicitors. J Integr Plant Biol. 2007;49:863–870. [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Ellis J.G., Lawrence G.J., Luck J.E., Dodds P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Jiang N., Wessler S.R. Plant transposable elements: Where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Frost D., Way H., Howles P., Luck J., Manners J., Hardham A., Finnegan J., Ellis J. Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol Plant Microbe Interact. 2004;17:224–232. doi: 10.1094/MPMI.2004.17.2.224. [DOI] [PubMed] [Google Scholar]
- Havecker E.R., Gao X., Voytas D.F. The Sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain 8. Plant Physiol. 2005;139:857–868. doi: 10.1104/pp.105.065680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Pinzón I., de Jesús E., Santiago N., Casacuberta J.M. The frequent transcriptional readthrough of the tobacco Tnt1 retrotransposon and its possible implications for the control of resistance genes. J Mol Evol. 2009;68:269–278. doi: 10.1007/s00239-009-9204-y. [DOI] [PubMed] [Google Scholar]
- Holub E.B., Beynon J.L., Crute I.R. Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol Plant-Microbe Interact. 1994;2:223–239. [Google Scholar]
- Kashkush K., Feldman M., Levy A.A. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lawrence G.J., Finnegan E.J., Ayliffe M.A., Ellis J.G. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell. 1995;7:1105–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Rombauts S., Zhang J., Aubourg S., Mathé C., Jansson S., Rouzé P., Boerjan W. Annotation of a 95-kb Populus deltoides genomic sequence reveals a disease resistance gene cluster and novel class I and class II transposable elements. Theor Appl Genet. 2004;109:10–22. doi: 10.1007/s00122-004-1621-0. [DOI] [PubMed] [Google Scholar]
- Marcel T.C., Aghnoum R., Durand J., Varshney R.K., Niks R.E. Dissection of the barley 2L1.0 region carrying the ‘Laevigatum‘ quantitative resistance gene to leaf rust using near-isogenic lines (NIL) and subNIL. Mol Plant Microbe Interact. 2007;20:1604–1615. doi: 10.1094/MPMI-20-12-1604. [DOI] [PubMed] [Google Scholar]
- Medstrand P., van de Lagemaat L.N., Dunn C.A., Landry J.R., Svenback D., Mager D.L. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet. Genome Res. 2005;110:342–352. doi: 10.1159/000084966. [DOI] [PubMed] [Google Scholar]
- Melayah D., Bonnivard E., Chalhoub B., Audeon C., Grandbastien M.A. The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J. 2001;28:159–168. doi: 10.1046/j.1365-313x.2001.01141.x. [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Chin D.B., Shen K.A., Sivaramakrishnan S., Lavelle D.O., Zhang Z., Michelmore R.W. The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell. 1998;10:1817–1832. doi: 10.1105/tpc.10.11.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R.W., Meyers B.C. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Miyao A., Tanaka K., Murata K., Sawaki H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Nobuta K., Vemaraju K., Tej S.S., Skogen J.W., Meyers B.C. Plant MPSS databases: Signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimbalkar S.B., Harsulkara A.M., Giri A.P., Sainani M.N., Franceschi V., Gupta V.S. Differentially expressed gene transcripts in roots of resistant and susceptible chickpea plant (Cicer arietinum L. ) upon Fusarium oxysporum infection. Physiol Mol Plant Pathol. 2006;68:176–188. [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oricchio E., Sciamanna I., Beraldi R., Tolstonog G.V., Schumann G.G., Spadafora C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26:4226–4233. doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- Peaston A.E., Evsikov A.V., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Pouteau S., Grandbastien M.A., Boccara M. Microbial elicitors of plant defense responses activate transcription of a retrotransposon. Plant J. 1994;5:535–542. [Google Scholar]
- SanMiguel P., Bennetzen J.L. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot. 1998;82:37–44. [Google Scholar]
- Song W.Y., Wang G.L., Chen L.L., Kim H.S., Pi L.Y., Holsten T., Gardner J., Wang B., Zhai W.X., Zhu L.H., et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Song W.Y., Pi L.Y., Wang G.L., Gardner J., Holsten T., Ronald P.C. Evolution of the rice Xa21 disease resistance gene family. Plant Cell. 1997;9:1279–1287. doi: 10.1105/tpc.9.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Sun Y., Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci USA. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos C.E., Astua-Monge G., Jones V., Plyler T.R., Sakiyama N.S., Mackenzie S.A. Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics. 2006;172:1229–1242. doi: 10.1534/genetics.105.050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat L.N., Landry J.R., Mager D.L., Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- van der Biezen E.A., Freddie C.T., Kahn K., Parker J.E., Jones J.D. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 2002;29:439–451. doi: 10.1046/j.0960-7412.2001.01229.x. [DOI] [PubMed] [Google Scholar]
- Vergne E., Ballini E., Droc G., Tharreau D., Nottéghem J.L., Morel J.B. RCHIPELAGO: A dedicated resource for exploiting past, present, and future genomic data on disease resistance regulation in rice. Mol Plant Microbe Interact. 2008;21:869–878. doi: 10.1094/MPMI-21-7-0869. [DOI] [PubMed] [Google Scholar]
- Wang G.L., Ruan D.L., Song W.Y., Sideris S., Chen L., Pi L.Y., Zhang S., Zhang Z., Fauquet C., Gaut B.S., et al. Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell. 1998;10:765–779. doi: 10.1105/tpc.10.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Zeng J., Lowe C.B., Sellers R.G., Salama S.R., Yang M., Burgess S.M., Brachmann R.K., Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H.G., Kachroo P., Masmoudi K., Berkowitz G., Klessig D.F. The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell. 2006;18:747–747. doi: 10.1105/tpc.105.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S., Horrocks J., Lukens L.N. Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J. 2006;45:347–357. doi: 10.1111/j.1365-313X.2005.02619.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]