Abstract
In this study, we detected new sequence variations in LAMA2 and SGCG genes in 5 ethnic populations, and analysed their effect on enhancer composition and mRNA structure. PCR amplification and DNA sequencing were performed and followed by bioinformatics analyses using ESEfinder as well as MFOLD software. We found 3 novel sequence variations in the LAMA2 (c.3174+22_23insAT and c.6085 +12delA) and SGCG (c. * 102A/C) genes. These variations were present in 210 tested healthy controls from Tunisian, Moroccan, Algerian, Lebanese and French populations suggesting that they represent novel polymorphisms within LAMA2 and SGCG genes sequences. ESEfinder showed that the c. * 102A/C substitution created a new exon splicing enhancer in the 3'UTR of SGCG genes, whereas the c.6085 +12delA deletion was situated in the base pairing region between LAMA2 mRNA and the U1snRNA spliceosomal components. The RNA structure analyses showed that both variations modulated RNA secondary structure. Our results are suggestive of correlations between mRNA folding and the recruitment of spliceosomal components mediating splicing, including SR proteins. The contribution of common sequence variations to mRNA structural and functional diversity will contribute to a better study of gene expression.
Keywords: muscular dystrophies, exon splicing enhancer, RNA fold, SR proteins
Introduction
Whole-genome sequencing of many organisms is producing enormous amounts of data, useful in elucidating the transcriptional regulatory mechanisms of genes. Moreover, comparative sequence analysis of non-coding elements has helped to find new regulatory elements within many genes. New motifs have been discovered from evolutionarily conserved regions (Wasserman et al., 2000), a list of co-regulated genes (Roth et al., 1998), or from a list of functionally related genes (Elkon et al., 2003).
Pre-messenger RNA (mRNA) splicing requires the accurate recognition of splice sites by cellular RNA processing machinery. In addition to sequences that comprise the branch point and the 3' and 5' splice sites, cellular splicing machinery relies on additional information in the form of exonic and intronic splicing enhancer and silencer sequences (Zhang et al., 2009). Splicing enhancers are discrete sequences of 6 or 8 nucleotides that promote both constitutive and regulated splicing, and facilitate exon definition by assisting in the recruitment of splicing factors, especially SR proteins (serine/ arginine-rich proteins) (Chasin, 2007). Ominously, recent studies predict that many human genetic diseases linked to genetic polymorphisms might be caused by the inactivation of splicing enhancers (Wang et al., 2005).
Single nucleotide polymorphisms (SNPs) represent the most frequent type of DNA sequence variations that cause phenotypic variability through multiple mechanisms, this including changes in the encoded protein sequence, the effect on gene regulation, mRNA processing (splicing, mRNA modification and turnover), and translation (Nielsen et al., 2007). The last few years have seen extensive efforts to catalogue human genetic variations and correlate them with phenotypic differences and gene expression. Studies on the impact of genetic variation in mRNA processing have not been sufficiently investigated, especially as it has been documented that a number of neutral genetic variations alter or create essential sequence elements for splicing and mRNA processing (Byrne et al., 2009). These seemingly neutral variations are associated with altered length and/or the steady-state level of cytoplasmic mRNA (Zhang et al., 2007; Hofmann et al., 2008).
We studied the LAMA2 and SGCG genes responsible for the MDC1A and LGMD2C forms of muscular dystrophies, respectively. Limb girdle muscular dystrophies (LGMD(s)) include a heterogeneous group of progressive muscular dystrophy mainly affecting the pelvic and shoulder girdle musculature. 50% of LGMD cases are sarcoglycanopathies related to mutations in SGCA, SGCB, SGCD and SGCG genes, thereby leading to the LGMD2D, LGMD2E, LGMD2F and LGMD2C forms respectively (Guglieri et al., 2005; Daniele et al., 2007). Mutations in LAMA2 gene are known to be involved in the MDC1A form representing that which is most frequent in cases of congenital muscular dystrophy. MDC1A is generally characterized by Total deficiency laminin α2 encoding by the LAMA2 gene, this leading to a severe phenotype marked by neonatal generalized hypotonia and weakness, with no independent ambulation due to severe contracture (Tome et al., 1994; Helbling-Leclerc et al., 1995). Moreover, over the latter years and from recent studies, there is proof of the presence of mutations in the LAMA2 gene in some late-onset LGMD forms, where mutations do not result in the complete absence of laminin α2, but in the production of truncated proteins or in increased proteolytic degradation (Naom et al., 1998).
We report here three novel homozygous sequence variations found in the setting of an extensive sequencing of LAMA2 and SGCG genes in two Tunisian patients with autosomic recessive LGMD. These variations were found in 210 tested controls from five different Mediterranean populations (Tunisian, Moroccan, Algerian, Lebanese and French). Two of these variations modulated cis-acting regulatory elements of LAMA2 and SGCG genes and have potential to modify their RNA secondary structure.
Subjects and Methods
Subjects
New sequence variations were screened in 62 unrelated healthy Tunisian individuals, 50 unrelated controls from Morocco, 45 unrelated healthy controls from Algeria, 35 controls from Lebanon and 20 healthy French individuals. Blood samples were collected after receiving informed consent from all subjects and with the approval of the appropriate Ethic Committees.
DNA extraction
Total genomic DNA was isolated from blood leucocytes samples of the tested individuals, according to the previously described protocol of Kawasaki (1990).
PCR amplification and DNA sequencing of LAMA2 and SGCG genes
PCR amplification of the 65 exons of the LAMA2 gene was performed using appropriately chosen primers, so that at least 30 to 50 bp of flanking intronic sequences became readable. For this, a thermal cycler (GeneAmp PCR system 9700, Applied Biosystem) was used, applying the touchdown method as previously described (Guicheney et al., 1998). The 8 SGCG gene exons were amplified using primers from LMDP (Leiden Muscular Dystrophy Pages), under optimized PCR conditions consisting of 0.1 μg of genomic DNA, 5 μL of 10 X buffer (50 mM Tris-HCl, pH 9.2, 160 mM (NH4)2 SO4, 22.5 mM MgCl2, 2% DMSO, and 1% Tween 20), 10 mM dNTP, 20 pmol of each primer and 2U of Taq DNA polymerase. Direct sequencing of PCR products was performed with the ABI Prism Big Dye terminator cycle sequencing Ready Reaction Kit (ABI Prism/ PE Biosystems), and the products were resolved on ABI Prism 3100-Avant. Blast searches were performed using the NCBI database.
Computational analyses
Bioinformatic web-based tools were used for predicting the effect of the new detected variations on enhancer composition and RNA secondary structure of the LAMA2 and SGCG genes. Two distinct softwares for analyzing splicing enhancers in detected polymorphisms were employed. In fact, ESEfinder 3.0 (Cartegni et al., 2003), was specifically designed for identifying exonic splicing enhancers, and we used it for the c.*102A/C situated in the 3'UTR which is an exonic untranslated region. Whereas for the other c.3174+22_23insAT and c.6085 +12delA polymorphisms which are situated in intronic regions (intron 22 and 42 of the LAMA2 gene, respectively); we used the enhancerfinder program (Cartegni et al., 2003). However, the positive and negative results are independent of the software used. Prediction of their effect on the RNA secondary structure was through the MFOLD program (Zuker, 1994).
Results
Novel sequence variations in the LAMA2 and SGCG genes
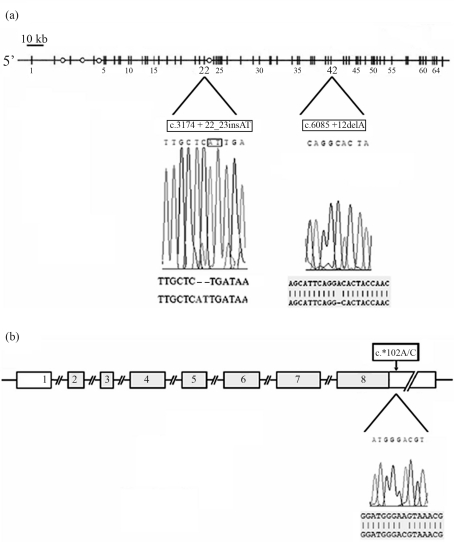
Sequencing of the LAMA2 and SGCG genes was performed with the aim to search for sequence variations in two patients with autosomic recessive LGMD. No mutations were found in either gene in the two patients tested. However, novel sequence variations in the LAMA2 and SGCG genes were detected. We revealed the presence of 2 novel homozygous intronic sequence variations in the LAMA2 gene. The first was an (AT) insertion at position +23 of intron 22 (c.3174+22_23insAT; dbSNP accession number: ss 142460322), whereas the second was a deletion of an adenin at position +12 of intron 42 of the LAMA2 gene (c.6085 +12delA; dbSNP accession number: ss 142460323) (Figure 1a). In the SGCG gene, a new homozygous A to C substitution at position +102 in the 3'UTR region was revealed (c.*102A/C; dbSNP accession number: ss 142460324). These sequences were different from those published in the last release of the NCBI GenBank (Figure 1b). Screening of these variations in the LAMA2 and SGCG genes in 60 unrelated healthy Tunisian controls from different regions revealed their presence in the homozygous state in all the tested subjects (Table 1).
Figure 1.
Novel genetic variations in LAMA2 and SGCG genes. Sequencing of the LAMA2 gene revealed two novel intronic variations: a c.3174+22_23insAT insertion in intron 22 and a c.6085+12delA deletion in intron 42. Sequencing of SGCG gene revealed an A > C substitution at position +102 in the 3'UTR region.
Table 1.
Genotypes of novel SNPs found in the LAMA2 and SGCG genes tested in five ethnic populations.
|
LAMA2 gene (intron 22)
|
LAMA2 gene (intron 42)
|
SGCG gene (3'UTR)
|
||||||
| Original sequence | c.3174+22_23insAT | c.6085 +12A | c.6085 +12delA | c.*102A/C | c.*102A/C | |||
| Tunisian population (n = 60) | 0% | 100% | 0% | 100% | 0% | 100% | ||
| Moroccan population (n = 50) | 0% | 100% | 0% | 100% | 0% | 100% | ||
| Algerian population (n = 45) | 0% | 100% | 0% | 100% | 0% | 100% | ||
| Libanese population (n = 35) | 0% | 100% | 0% | 100% | 0% | 100% | ||
| French population (n = 20) | 0% | 100% | 0% | 100% | 0% | 100% | ||
Note that all the tested individuals are homozygous for the novel sequence variations.
This result prompted us to search for these sequence variations on four other ethnic populations. We screened two populations from North Africa, this including 50 healthy Moroccan and 45 Algerian controls, besides individuals from two other Mediterranen populations, namely 35 Libanese and 20 healthy French controls. We found that 100% of the healthy individuals tested were homozygous for the three sequence variations (Table 1).
Effect of sequence variations on cis-acting regulatory elements
The ESEfinder program showed that the c. *102A/C substitution is predicted to create an exon splicing enhancer in the 3'UTR of the SGCG gene. The new exon splicing enhancer is a GGGACGT which is predicted to be recognized by the SF2/ASF SR protein with a score of 3.96, thus significantly higher than the threshold value of 1.956 (Figure 2). c.3174+22_23insAT and c.6085+12delA intronic variations in the LAMA2 gene were predicted not to affect enhancer composition. However, the c.6085+12delA was found to be localised in the donor splicing consensus of intron 42, which mediated a base pairing complementarity with the U1snRNA spliceosomal component (Figure 3b).
Figure 2.
Computational prediction of the effect of c.*102A/C change on enhancer composition. (a) The A at position +102 of the 3'UTR of the SGCG gene does not include an ESE. (b) Note that the A to C substitution creates a new enhancer in the 3'UTR of the SGCG gene recognized by the SF2/ASF SR protein. The arrows indicate the SNP position.
Figure 3.
Proposed secondary-structure models of the new sequence variations in LAMA2 genes. (a) The c.3174+22_23insAT in intron 22 of the LAMA2 gene was predicted not to affect RNA secondary structure. (b) Position of the c.6085delA in LAMA2. The deletion is situated at position +12 in the 5' consensus splice site of intron 42. The c.6085delA is underlined. Position +1 indicates the first nucleotide of the 5' splice site of exon 42. (c) Comparison of RNA secondary structure of [exon 42 + intron 42 + exon 43] between the original sequence and the c.6085+12delA bearing sequence. Several structural changes were predicted. The arrows indicate modifications in folding. (d) Detailed RNA structure analysis of the 5' splicing consensus. The original sequence contained an external closing pair between G12C21 with a hairpin loop between the G13 and C20 positions containing an A at position 6085. The c.6085+12delA deletion shifts the 8 bp external hairpin loop and induced a change in its nucleotide composition. Moreover, orientation of the external loop was modified.
Correlation between cis-acting elements and RNA secondary structure
In order to study whether pre mRNA folding could be influenced by the presence of these sequence variations in the LAMA2 and SGCG genes, we performed bioinformatics analyses using the MFOLD program. The results strongly suggested that the c.3174+22_23insAT in intron 22 of LAMA2 had no marked effect on RNA structure (Figure 3a). However, RNA structure analyses of intron 42 and flanking exons (exons 42 and 43) showed that the c.6085+12delA in the donor splicing consensus of intron 42 of LAMA2 (Figure 3b) was followed by several structural changes (Figure 3c). In fact, detailed RNA structure analyses of the 5' splicing consensus showed that the original sequence contained an external closing pair between G12C21 with a hairpin loop between those G13 and C20 positions containing an A at position 6085. The c.6085+12delA deletion shifts the 8 bp external hairpin loop between C6 and G13 positions, and changes its nucleotide composition, when compared to the original sequence. Moreover, the orientation of the external loop was modified (Figure 3d).
The creation of an exon splicing enhancer following c. *102A/C substitution in 3'UTRs of SGCG is predicted to create many changes in RNA secondary structure, as to the number, position and orientation of the various external loops (Figure 4a, 4b).
Figure 4.
Effect of the c.*102A/C in the 3'UTR region of the SGCG gene on RNA secondary structure. (a) Analyses of the RNA folding of 500 bp of intron 8 with the whole 3'UTR region of SGCG. We marked structural modifications between the c.*102A and the c.*102C alleles. The arrows indicate modifications in RNA folding. (b) Detailed RNA analysis showed that c.*102A/C is predicted to create many changes regarding the number, position and orientation of the different external loops.
Discussion
The regulatory mechanisms involved in gene expression became an important aspect of genomics revealed by the completed genome sequencing of many organisms.
Most common SNPs have now been assessed in genome-wide studies for statistical associations with many complex traits, these including many important and common diseases (Roeder and Luca, 2009). Although these studies have provided new biological insights, only a limited knowledge of SNPs within regulatory gene elements affecting mRNA post-transcriptional processing has been acquired.
In this study, we discovered three novel sequence variations in the LAMA2 and SGCG genes present in 100% of the 155 healthy tested individuals from North Africa, including Tunisian, Algerian and Moroccan populations and in the 35 and 20 controls from the Lebanon and France, respectively. These results indicate that the c.3174+22_23insAT insertion and c.6085+12delA deletion in LAMA2, as well as the c.*102A/C substitution in SGCG were not only specific to North African, but also apply to other Mediterranean populations. We thus emphasize that these novel sequence variants represent novel polymorphisms in the GenBank sequences of the LAMA2 and SGCG genes.
Intron removal during pre-mRNA splicing in higher eukaryotes requires the accurate identification of the two splice sites at the ends of the exons or in the intron: the exon/intron definition (House and Lynch, 2008). This important task is executed in the nucleus by the spliceosome which is assembled at the correct donor and acceptor splice sites. Moreover, additional information is provided by cis-acting regulatory sequences that serve to enhance or repress splicing, the splicing enhancers and the splicing silencers, respectively (Cartegni et al., 2003).
In our study, the ESEfinder program predicted that c.*102A/C created a new enhancer in the 3'UTR of SGCG. In the LAMA2 gene, the c.3174+22_23insAT did not affect the regulatory elements of the gene itself. On the other hand, c.6085+12delA may disrupt base pairing between the donor splicing consensus of intron 42 of LAMA2 pre mRNA and the 5' end of U1snRNA, this recognition being crucial for U6snRNP recruitment (Lund and Kjems, 2002). The importance of this region was also confirmed in our previous studies which demonstrated that a 7 bp deletion from position +5 to +11 in the donor splicing consensus of intron 17 of the LAMA2 gene triggered total skipping of exon 17 (Siala et al., 2008). Moreover, the position +12 in the 5' splicing consensus has already been demonstrated to be determinant for the correct mRNA processing of the ATM gene through its interaction with the U1snRNA (Lewandowska et al., 2005).
The 3' UTR contains regulatory elements that are essential for appropriate expression of many genes. These regulatory elements are involved in the control of nuclear transport (Ding and Lipshitz, 1993), the polyadenylation status and sub-cellular targeting (St Johnston, 1995), as well as in the rates of translation and degradation of mRNA by the NMD (nonsense mediated mRNA decay) system (Sachs, 1993). These processes are mediated by the interaction of specific sequences in 3' UTRs with specific RNA binding proteins. The creation of an enhancer in the 3'UTR also emphasizes alterations in the splicing pattern and mRNA stability of the SGCG transcript. In fact, it could also be responsible of the synthesis of an alternative isoform of SGCG mRNA. This idea was already reported in HLA-G transcript (Rousseau et al., 2003) and in the mRNA encoding the β-catalytic subunit of the mitochondrial H+-ATP synthase, where translation enhancing activity generates a new isoform, depending upon the cell type analysed (Di Liegro et al., 2000).
These results suggest a possible functional role for both c.6085+12delA and c.*102A/C in pre mRNA stability and splicing processing of the LAMA2 and SGCG genes, respectively. The localisation of human coding and non-coding variations in splicing consensus or enhancers, as well as their effect on mRNA splicing has already been reported in several studies (Fairbrother et al., 2004).
The bioinformatics analyses performed to evaluate the effect of these sequence variations on RNA secondary structure showed that the c.3174+22_23insAT is predicted not to affect the RNA structure, whereas the c.6085+12delA insertion in intron 42 of the LAMA2 gene, as well as the c.*102A/C in the 3'UTR of the SGCG gene is predicted to generate several alterations in RNA secondary structure compared to the original sequences. This correlation between RNA structure and cis-acting regulatory elements can be explained by pre-mRNA folding within these regions being crucial for their binding to SR proteins (serine/arginine rich proteins), and may be important for the recruitment of spliceosomal components mediating splicing. Indeed, a growing body of evidence has shown that mRNA folding influenced a wide range of transcription events, such as mRNA splicing (Sheng and Tinoco, 1995), processing (Allain et al., 1996), translational control (Pelletier and Sonenberg, 1987) and regulation (Addess et al., 1997).
In conclusion, in this study we reported 3 novel sequence variations in the LAMA2 and SGCG gene sequences, two of which predicted to modulate cis-acting regulatory elements and RNA secondary structure. Sequence variations were retrieved in all the healthy controls tested, thereby indicating their not being involved in disease susceptibility or phenotypic variability. Indeed, the contribution of common sequence variation to mRNA structural and functional diversity could provide an insight into the fundamental mechanisms of gene expression.
Acknowledgments
We thank Mr. Jamil JAOUA, Head of the English Unit at the Sfax Faculty of Science, for proofreading this paper. This work was supported by the Ministry of Higher Education, Scientific Research and Technology of Tunisia.
Footnotes
Associate Editor: Sandro José de Souza
References
- Addess K.J., Basilion J.P., Klausner R.D., Rouault T.A., Pardi A. Structure and dynamics of the iron responsive element RNA: Implications for binding of the RNA by iron regulatory binding proteins. J Mol Biol. 1997;274:72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- Allain F.H., Gubser C.C., Howe P.W., Nagai K., Neuhaus D., Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- Byrne J.A., Strautnieks S.S., Ihrke G., Pagani F., Knisely A.S., Linton K.J., Mieli-Vergani G., Thompson R.J. Missense mutations and single nucleotide polymorphisms in ABCB11 impair bile salt export pump processing and function or disrupt pre-messenger RNA splicing. Hepatology. 2009;49:553–567. doi: 10.1002/hep.22683. [DOI] [PubMed] [Google Scholar]
- Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L.A. Searching for splicing motifs. Adv Exp Med Biol. 2007;623:85–106. doi: 10.1007/978-0-387-77374-2_6. [DOI] [PubMed] [Google Scholar]
- Danièle N., Richard I., Bartoli M. Ins and outs of therapy in limb girdle muscular dystrophies. Int J Biochem Cell Biol. 2007;39:1608–1624. doi: 10.1016/j.biocel.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Di Liegro C.M., Bellafiore M., Izquierdo J.M., Rantanen A., Cuezva J.M. 3'-untranslated regions of oxidative phosphorylation mRNAs function in vivo as enhancers of translation. Biochem J. 2000;352:109–115. [PMC free article] [PubMed] [Google Scholar]
- Ding D., Lipshitz H.D. Localized RNAs and their functions. Bioessays. 1993;15:651–658. doi: 10.1002/bies.950151004. [DOI] [PubMed] [Google Scholar]
- Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother W.G., Holste D., Burge C.B., Sharp P.A. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:1388–1395. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglieri M., Magri F., Comi G.P. Molecular etiopathogenesis of limb girdle muscular and congenital muscular dystrophies: Boundaries and contiguities. Clin Chim Acta. 2005;361:54–79. doi: 10.1016/j.cccn.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Guicheney P., Vignier N., Zhang X., He Y., Cruaud C., Frey V., Helbling-Leclerc A., Richard P., Estournet B., Merlini L., et al. PCR based mutation screening of the laminin alpha2 chain gene (LAMA2): Application to prenatal diagnosis and search for founder effects in congenital muscular dystrophy. J Med Genet. 1998;35:211–217. doi: 10.1136/jmg.35.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbling-Leclerc A., Zhang X., Topaloglu H., Cruaud C., Tesson F., Weissenbach J., Tomé F.M., Schwartz K., Fardeau M., Tryggvason K., et al. Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin- deficient congenital muscular dystrophy. Nat Genet. 1995;11:216–218. doi: 10.1038/ng1095-216. [DOI] [PubMed] [Google Scholar]
- Hofmann M.H., Blievernicht J.K., Klein K., Saussele T., Schaeffeler E., Schwab M., Zanger U.M. Aberrant splicing caused by single nucleotide polymorphism c. 516G > T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325:284–292. doi: 10.1124/jpet.107.133306. [DOI] [PubMed] [Google Scholar]
- House A.E., Lynch K.W. Regulation of alternative splicing: More than just the ABCs. J Biol Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- Kawasaki E. Sample preparation from blood, cells and other fluids. In: M Innis, D Gelfand, J Snisky., editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. pp. 146–152. [Google Scholar]
- Lewandowska M.A., Stuani C., Parvizpur A., Baralle F.E., Pagani F. Functional studies on the ATM intronic splicing processing element. Nucleic Acids Res. 2005;33:4007–4015. doi: 10.1093/nar/gki710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M., Kjems J. Defining a 50 splice site by functional selection in the presence and absence of U1 snRNA 50 end. RNA. 2002;8:166–179. doi: 10.1017/s1355838202010786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naom I., D'Alessandro M., Sewry C.A., Jardine P., Philpot J., Manzur A.Y., Dubowitz V., Muntoni F. Laminin alpha 2-chain gene mutations in two siblings presenting with limb-girdle muscular dystrophy. Neuromusc Disord. 1998;8:495–501. doi: 10.1016/s0960-8966(98)00065-0. [DOI] [PubMed] [Google Scholar]
- Nielsen K.B., Sørensen S., Cartegni L., Corydon T.J., Doktor T.K., Schroeder L.D., Reinert L.S., Elpeleg O., Krainer A.R., Gregersen N., et al. Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: A synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer. Am J Hum Genet. 2007;80:416–432. doi: 10.1086/511992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. The involvement of mRNA secondary structure in protein synthesis. Biochem Cell Biol. 1987;65:576–581. doi: 10.1139/o87-074. [DOI] [PubMed] [Google Scholar]
- Roeder K., Luca D. Searching for disease susceptibility variants in structured populations. Genomics. 2009;93:1–4. doi: 10.1016/j.ygeno.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth F.P., Hughes J.D., Estep P.W., Church G.M. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Rousseau P., Le Discorde M., Mouillot G., Marcou C., Carosella E.D., Moreau P. The 14 bp deletion-insertion polymorphism in the 3_ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol. 2003;64:1005–1010. doi: 10.1016/j.humimm.2003.08.347. [DOI] [PubMed] [Google Scholar]
- Sachs A.B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Sheng L.X., Tinoco J.R. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Siala O., Louhichi N., Triki C.H., Morinière M., Fakhfakh F., Baklouti F. LAMA2 mRNA processing alterations generate a complete deficiency of laminin alpha2 protein and a severe congenital muscular dystrophy. Neuromusc Disord. 2008;18:137–145. doi: 10.1016/j.nmd.2007.09.003. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- Tome F.M., Evangelista T., Leclerc A., Sunada Y., Manole E., Estournet B., Barois A., Campbell K.P., Fardeau M. Congenital muscular dystrophy with merosin deficiency. C R Acad Sci III. 1994;317:351–357. [PubMed] [Google Scholar]
- Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Wang J., Smith P.J., Krainer A.R., Zhang M.Q. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Arias M.A., Ke S., Chasin L.A. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009;15:367–376. doi: 10.1261/rna.1498509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Bertolino A., Fazio L., Blasi G., Rampino A., Romano R., Lee M.L., Xiao T., Papp A., Wang D., et al. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci USA. 2007;104:20552–20557. doi: 10.1073/pnas.0707106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Prediction of RNA secondary structure by energy minimization. Meth Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
Internet Resources
- ESEfinder 3.0. [July 28, 2009]. Available from: http://rulai.cshl.edu/tools/ESE/
- Enhancerfinder program. [August 3, 2009]. Available from: http://violin.genet.sickkids.on.ca/~ali/enhancerfinder.html.
- MFOLD program. [August 3, 2009]. Available from: http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi.