Abstract
The karyotype of Amphisbaena ridleyi, an endemic species of the archipelago of Fernando de Noronha, in State of Pernambuco, Brazil, is described after conventional staining, Ag-NOR impregnation and fluorescence in situ hybridization (FISH) with a telomeric probe. The diploid number is 46, with nine pairs of macrochromosomes (three metacentrics, four subtelocentrics and two acrocentrics) and 14 pairs of microchromosomes. The Ag-NOR is located in the telomeric region of the long arm of metacentric chromosome 2 and FISH revealed signals only in the telomeric region of all chromosomes. Further cytogenetic data on other amphisbaenians as well as a robust phylogenetic hypothesis of this clade is needed in order to understand the evolutionary changes on amphisbaenian karyotypes.
Keywords: Amphisbaena ridleyi, karyotype, Fernando de Noronha, Ag-NOR, FISH with telomeric probes
Amphisbaenians, or worm lizards, are a monophyletic group of squamates mostly distributed nowadays in Africa and South America (Gans, 1990, 2005; Kearney, 2003; Kearney and Stuart, 2004; Macey et al., 2004). Due to their fossorial lifestyle and the consequent challenge for collecting them, the group is probably the least-studied group of squamates and many aspects of its biology remain enigmatic.
Although a phylogenetic hypothesis based on morphological and molecular characters for the group was only recently proposed (Kearney, 2003; Kearney and Stuart, 2004; Macey et al., 2004; Vidal et al., 2008), karyological studies on amphisbaenians date back from the 1960's. The karyotypes of 35 out of the 190 recognized amphisbaenian species have been described, mostly including only data on diploid number and chromosomal morphology (Table 1). Amphisbaenian karyotypes present variable diploid number and morphology with distinctive macro and microchromosomes. Diploid numbers range from 2n = 26 in Amphisbaena dubia and Anops kingi to 2n = 50 in Amphisbaena leberi and A. innocens (Huang and Gans, 1971; Beçak et al., 1971a, 1972; Cole and Gans, 1987). This variability is in strong contrast with the conserved karyotype composed by 36 chromosomes (12M + 24m) found in many groups of lizards and considered as the primitive karyotype within Squamata (Olmo, 1986). Except for the study of Hernando (2005) describing the localization of nucleolar organizer regions (NORs) in four South American species, all chromosomal studies in amphisbaenians only presented conventional staining data.
Table 1.
Chromosomal revision of amphisbaenians, with descriptions of diploid number (2n), fundamental number (FN), number and morphology of macrochromosomes, number of microchromosomes, references and occurrence of species.
| Species | 2n | Macro (n. biarmed, n. uniarmed) | micro | FN | Reference1 | Occurrence |
| Amphisbaenidae | ||||||
| Amphisbaena alba | 38 | 22 (14, 8) | 16 | 64 | 4, 5, 6, 7 | South America |
| Amphisbaena angustifrons | 30 | 12 (12, 0) | 18 | 42 | 2 | South America |
| Amphisbaena caeca | 36 | 12 (12, 0) | 24 | 48 | 2 | Central America |
| Amphisbaena camura | 44 | 24 (4, 20) | 20 | 48-502 | 2 | South America |
| Amphisbaena darwini | 30 | 12 (12, 0) | 18 | 46 | 2 | South America |
| Amphisbaena dubia | 25, 26, 27, 28 |
15 (12, 3), 14 (12, 2), 13 (12, 1), 12 (12, 0) |
10, 12, 14, 16 |
- | 3, 6 | South America |
| Amphisbaena fenestrata | 36 | 12 (12, 0) | 24 | 52-562 | 5 | Central America |
| Amphisbaena fuliginosa | 48 | 22 (6, 16) | 26 | 60 | 5 | South America |
| Amphisbaena heterozonota | 30 30 |
12 (12, 0) 12 (12, 0) |
18 18 |
46 60 |
2 12, 13 |
South America |
| Amphisbaena hiata | 30 | 12 (12, 0) | 18 | 60 | 12, 13 | South America |
| Amphisbaena innocens | 50 | 22 (8, 14) | 28 | - | 5 | Central America |
| Amphisbaena leberi | 50 | 22 (8, 14) | 28 | - | 11 | Central America |
| Amphisbaena manni | 36 | 12 (12, 0) | 24 | - | 5 | Central America |
| Amphisbaena mertensi | 40 | 18 (6, 12) | 22 | - | 12 | South America |
| Amphisbaenaridleyi | 48 | 18 (14, 4) | 28 | - | Present work | South America |
| Amphisbaena trachura | 30 | 12 (12, 0) | 18 | 46 | 2 | South America |
| Amphisbaena vermicularis | 44 | 22 (2, 20) | 22 | 46 | 7, 8 | South America |
| Amphisbaena xera | 36 | 12 (12, 0) | 24 | 48 | 2 | Central America |
| Anops kingi | 26 | 12 (12, 0) | 14 | - | 5 | South America |
| Chirindia langi | 30 34 |
12 (12, 0) 12 (12, 0) |
18 22 |
- - |
5 | Africa |
| Chirindia sp | 30 32 |
12 (12, 0) 12 (12, 0) |
18 20 |
46-502 - |
5 | Africa |
| Cynisca leucura | 30 32 |
12 (12, 0) 12 (12, 0) |
18 20 |
5 | Africa | |
| Geocalamus acutus | 38 | 14 (10, 4) | 24 | - | 5 | Africa |
| Leptosternon microcephalum | 34 32 34 |
12 (12, 0) 12 (12, 0) 12 (2,22) |
22 20 22 |
48 44 46 |
4, 6 2 2, 12, 13 |
South America |
| Mesobaena huebneri | 46 | 24 (2, 22) | 22 | - | 10 | South America |
| Monopeltis capensis | 34 | 12 (12, 0) | 22 | 62 | 5 | Africa |
| Zygaspis quadrifrons | 36 36 |
12 (12, 0) 12 (12,0) |
24 24 |
50 72 |
2 5 |
Africa |
| Zygaspis violacea | 36 | 12 (12, 0) | 24 | - | 5 | Africa |
| Bipedidae | ||||||
| Bipes biporus | 40 42 |
20 (20, 0) 20 (20, 0) |
20 22 |
60 66 |
2 5, 9, 11 |
North America |
| Bipes canaliculatus | 46 46 |
22 (16, 6) 22 (20, 2) |
24 24 |
- - |
9 11 |
North America |
| Bipes tridactylus | 46 | 22 (18, 4?) | 24 | - | 11 | North America |
| Blanidae | ||||||
| Blanus cinereus | 32 | 12 (12, 0) | 20 | 44 | 2 | Europe |
| Blanus strauchi | 32 | 12 (12, 0) | 20 | 44 | 2 | Europe |
| Rhineuridae | ||||||
| Rhineura floridana | 46 44 |
20 (2, 18) 24 (16, 8) |
26 20 |
- 54-562 |
1 2 |
North America |
| Trogonophiidae | ||||||
| Diplometopon zarudnyi | 36 | 12 (12, 0) | 24 | 52 | 2, 10 | Africa |
| Trogonophis elegans | 36 | 12 (12, 0) | 24 | 48 | 2 | Africa |
1: 1. Matthey (1933); 2. Huang et al. (1967); 3. Beçak et al. (1971a); 4. Beçak et al. (1971b); 5. Huang and Gans (1971); 6. Beçak et al. (1972); 7. Beçak et al. (1973a); 8. Beçak et al. (1973b); 9. Macgregor and Klosterman (1979); 10. Branch (1980); 11. Cole and Gans (1987); 12. Hernando (2005); 13. Hernando and Alvarez (2005).
2: The variation of FN, according to the authors, is due to the difficulty in determining microchromosome morphology.
Herein we describe the chromosome constitution of Amphisbaena ridleyi, a species endemic to the oceanic archipelago of Fernando de Noronha, Pernambuco, Brazil. Although this species resembles some African members in some external attributes, molecular data indicate that it is closely related to the South American genus Amphisbaena (Gans, 1963; T. Mott, unpublished data). Karyotypic data presented here support the idea that amphisbaenian karyotypes are highly variable and might assemble phylogenetically informative characters. This information allied to phylogenetic hypotheses of amphisbaenian relationships would help to understand the chromosome evolution in this interesting group of fossorial squamates.
Three individuals of Amphisbaena ridleyi were collected by two of us (TM, MTR; IBAMA permit number 02010.000240/2007-03), one male (MZUSP 98333) and one female (MZUSP 98335) from the Ilha Rata (3°48'47.6” S, 32°23'21.5” W) and one female (MZUSP 98338) from the Ilha Fernando de Noronha (3°51'21.2” S, 32°26'31.5” W), both in the archipelago of Fernando de Noronha, Pernambuco, Brazil. The animals were brought alive to the Laboratório de Citogenética de Vertebrados, Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, Brazil and after chromosomal preparations were made, the specimens were deposited in the herpetological collection of Museu de Zoologia, Universidade de São Paulo.
The animals were injected with colchicine, according to routine techniques (Kasahara et al., 1987), and chromosomal spreads were obtained from the liver. The diploid number and the localization of Ag-NORs were established after conventional staining and silver staining impregnation (Howell and Black, 1980), respectively. Fluorescence in situ hybridization (FISH) was performed using the Telomere PNA FISH Kit/Cy3 (DAKO, code No. K 5326), according to manufacturer's instructions. FISH signals were visualized using a Zeiss Axiophot microscope equipped with a FITC filter using the softwares Ikaros & Isis v. 5.0 (Zeiss).
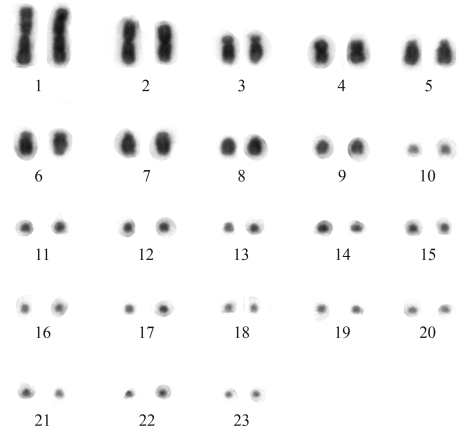
Amphisbaena ridleyi from the Ilhas Rata and Fernando de Noronha had similar karyotype numbers composed by 46 chromosomes, with 9 pairs of macrochromosomes and 14 pairs of microchromosomes (2n = 46, 18M+28m) (Figure 1). The macrochromosomes are three metacentric pairs (1, 2 and 4), two acrocentric pairs (8 and 9) and four subtelocentric pairs (3, 5, 6 and 7), although in some metaphases the short arms of some of these chromosomes was extremely reduced. There was not enough resolution to morphologically identify the 14 pairs of microchromosomes. No secondary constrictions or heteromorphic sex chromosomes were observed.
Figure 1.
Conventionally stained karyotype of Amphisbaena ridleyi, female, 2n = 46 (18M + 28m), from Fernando de Noronha, Pernambuco, Brazil.
The karyotype described for A. ridleyi (2n = 46, 18M + 28m) is unique among its congeners (Huang et al., 1967; Huang and Gans, 1971; Beçak et al., 1972, 1973a; Cole and Gans, 1987; Hernando, 2005). Furthermore, the comparison of the karyotype of A. ridleyi with those of other amphisbaenian genera with the same diploid number, such as Bipes canaliculatus, Bipes tridactylus (22M + 24m), Mesobaena huebneri (24M + 22m) and Rhineura floridana (26M +20m), revealed that the number of macro and microchromosomes and the number of biarmed chromosomes were very distinct among different genera, (Table 1) (Matthey, 1933; Huang et al., 1967; Macgregor and Klosterman, 1979; Cole and Gans, 1987).
Despite the fact that only 20% of amphisbaenian species have had their karyotypes studied, a great variability of diploid numbers has been observed. There are 77 described species of Amphisbaena (Gans, 2005; Mott et al. 2008, 2009) from which 18 had their karyotypes described, including A. ridleyi from the present study. This is the genus that exhibits the higher variability in chromosome number and morphology, including all the range of variation found in amphisbaenians, such as 2n = 26 (14M + 12m) in males and 2n = 25, 26, 27 and 28 in females of A. dubia; 2n = 30 (12M + 18m) in A. angustifrons, A. darwini, A. heterozonata, A. hiata, A. trachura; 2n = 36 (12M + 24m) in A. caeca, A. fenestrata, A. manni, A. xera; 2n = 38 (22M + 16m) in A. alba; 2n = 40 (18M + 22m) in A. mertensi; 2n = 44 in A. camura (24M + 20m) and A. vermicularis (22M + 22m); 2n = 48 (22M + 26m) in A. fuliginosa and 2n = 50 (22M + 28m) in A. leberi and A. innocens (Table 1). Probably fusion/fission rearrangements occurred in the karyotypic diversification of amphisbaenians (Cole and Gans, 1987; Hernando, 2005), but the number of taxa studied and the absence of differential staining do not allow more detailed hypotheses on the karyotypic evolution of this group.
Some karyotypes reported in the literature do not allow to determine the fundamental number due to the difficulty of identifying the morphology of the microchromosomes. The species A. dubia showed an intraindividual variation of the diploid number, involving macro-and microchromosomes, and the authors suggested that this would be due to fusions/fissions of microchromosomes (Beçak et al. 1971a, 1972). However, the polymorphism detected in A. dubia should be viewed with reservations due to the low quality of the chromosome preparations.
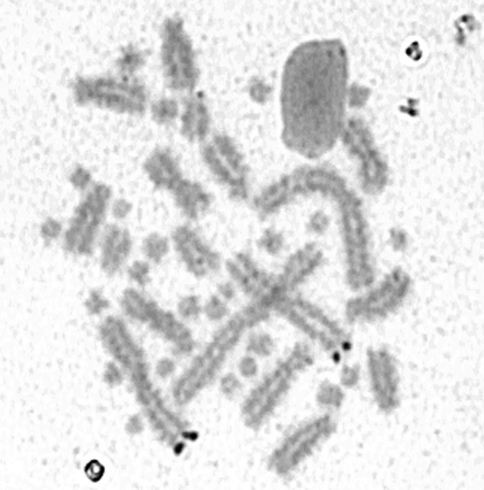
The Ag-NORs of all specimens of A. ridleyi were located in the telomeric region of the long arm of the metacentric pair 2 (Figure 2) in 17 metaphases on specimens from Ilha Rata and in 13 metaphases on the specimen from Fernando de Noronha, differing from all four South American amphisbaenian species previously studied (Hernando, 2005). In Leposternum microcephalum (2n = 34, 12M + 22m), Ag-NORs were detected in the telomeric region of the long arm of pair 3; in A. hiata (2n = 30, 12M +18m) it was located in the subterminal portion of the short arm of pair 4; in A. mertensi (2n = 40, 18M + 22m) a medium acrocentric macrochromosome was the Ag-NOR-bearing pair, and in A. heterozonata (2n = 30, 12M + 18m), Ag-NORs were found either in pair 2 or in pairs 1, 3 and 4 (Hernando, 2005).
Figure 2.
Incomplete metaphase after silver staining showing the Ag-NORs on the telomeric region of chromosome 2 of Amphisbaena ridleyi from Fernando de Noronha (Pernambuco, Brazil).
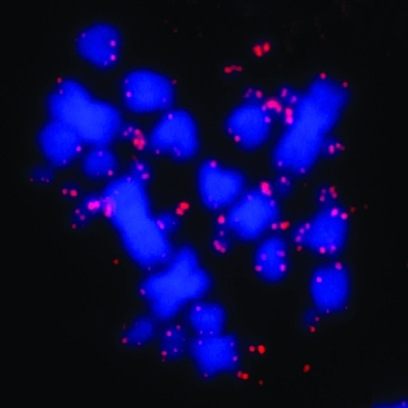
Fluorescence in situ hybridization using the (TTAGGG)n sequence detected signals on the telomeric regions of all chromosomes of A. ridley (Figure 3). Some of the signals were tiny and sometimes it was difficult to visualize them in the photographs. Despite the small number of studies using fluorescence in situ hybridization in Squamata, different patterns of distribution of telomeric sequences were observed. In Leposoma scincoides (Gymnophthalmidae), Polychrus marmoratus (Polychrotidade) and Phrynosoma cornutum (Phrynosomatidae) only telomeric signals were detected, while the chromosomes of Cnemidophorus sexlineatus, C. guturalis (Teiidae), Scelophorus olivaceus, Cophosaurus texanus (Phrynosomatidae), Gonatodes taniae (Gekkonidae), Leposoma guianense, Leposoma osvaldoi (Gymnophthalmidae) and Polychrus acutirostris (Polychrotidade) presented additional interstitial telomeric sites (Meyne et al., 1989, 1990; Schmid et al., 1994; Pellegrino et al., 1999; Bertolotto et al., 2001). The exclusive telomeric pattern observed in A. ridleyi is the first report of FISH for amphisbaenids.
Figure 3.
Distribution of the (TTAGGG)n sequence in chromosomes ofAmphisbaena ridleyi, from Fernando de Noronha (Pernambuco, Brazil).
The New World amphisbaenids form a monophyletic group within the paraphyletic radiation of African amphisbaenids (Kearney and Stuart, 2004; Vidal et al., 2008). African members of Amphisbaenidae show a lower range of variation in diploid number, like Cynisca leucura (2n = 30) and Geocalamus acutus (2n = 38) (Huang and Gans, 1971), when compared to South American congeners. Nevertheless, a more complete taxonomic sampling, including cytogenetic data with differential staining analyses, is needed in order to obtain a better picture of karyotype evolution in amphisbaenids. Despite the scarce information about Ag-NORs location on amphisbaenian karyotypes, the preliminary data available suggest that this marker is phylogenetically informative. We strongly recommend that further studies on amphisbaenian karyotypes include this information.
Acknowledgments
We thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), CNPq, CAPES for the financial support and IBAMA for collection permits.
Footnotes
Associate Editor: Fausto Foresti
References
- Beçak M.L., Beçak W., Denaro L. Polimorfismo cromossômico intraindividual em Amphisbaena dubia (Sauria) Cienc Cult. 1971a;23:123. [Google Scholar]
- Beçak M.L., Beçak W., Denaro L. Cariologia comparada em oito espécies de lacertílios. Cienc Cult. 1971b;23:124. [Google Scholar]
- Beçak M.L., Beçak W., Denaro L. Chromosome polymorphism, geographical variation and karyotypes in Sauria. Caryologia. 1972;25:313–326. [Google Scholar]
- Beçak M.L., Beçak W., Napoleone L.F. Amphisbaena vermicularis. In: Benirschke K., Hsu T.C., editors. Chromosome Atlas: Fish, Amphibians, Reptiles and Birds. v. 2. Berlin and Heidelberg, New York: Springer-Verlag; 1973a. p. R-25. [Google Scholar]
- Beçak M.L., Beçak W., Napoleone L.F., Reis L. Contribuição ao estudo cariotípico e variação de DNA em lacertílios. Cienc Cult. 1973b;25:219. [Google Scholar]
- Bertolotto C.E.V., Rodrigues M.T., Yonenaga-Yassuda Y. Banding patterns, multiple sex chromosome system and localization of telomeric (TTAGGG)n sequences by FISH on two species of Polychrus (Squamata, Polychrotidae) Caryologia. 2001;54:217–226. [Google Scholar]
- Branch W.R. Chromosome morphology of some reptiles from Oman and adjacent territories. J Oman Stud Spec Rep. 1980;2:333–345. [Google Scholar]
- Cole C.J., Gans C. Chromosomes of Bipes, Mesobaena, and other amphisbaenians (Reptilia), with comments on their evolution. Am Mus Nov. 1987;2869:1–9. [Google Scholar]
- Gans C. Redescription of Amphisbaena ridleyi Boulenger. Copeia. 1963;1963:102–107. [Google Scholar]
- Gans C. Patterns in amphisbaenian biogeography. A preliminary analysis. In: Peters G., Hutterer R., editors. Vertebrates in the Tropics. Bonn: Alexander Koenig Zoological Research Institute and Zoological Museum; 1990. pp. 133–143. [Google Scholar]
- Gans C. Checklist and bibliography of the amphisbaenia of the world. Bull Am Mus Nat Hist. 2005;289:130. [Google Scholar]
- Hernando A. Cytogenetic study of Leposternon and Amphisbaena (Amphisbaenia, Squamata) Caryologia. 2005;58:178–182. [Google Scholar]
- Hernando A., Alvarez B. Estudios cromosómicos en saurios y anfisbénidos del litoral fluvial argentino y área de influencia. Estado del conocimiento. INSUGEO Miscelánea. 2005;14:427–440. [Google Scholar]
- Howell W.M., Black D.A. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia. 1980;36:1014–1015. doi: 10.1007/BF01953855. [DOI] [PubMed] [Google Scholar]
- Huang C.C., Gans C. The chromosomes of 14 species of amphisbaenians (Amphisbaenia, Reptilia) Cytogenetics. 1971;10:10–22. doi: 10.1159/000130122. [DOI] [PubMed] [Google Scholar]
- Huang C.C., Clark H.F., Gans C. Karyological studies on fifteen forms of amphisbaenians (Amphisbaenia, Reptilia) Chromosoma. 1967;22:1–15. [Google Scholar]
- Kasahara S., Yonenaga-Yassuda Y., Rodrigues M.T. Geographical karyotypic variations and chromosome banding patterns in Tropidurus hispidus (Sauria, Squamata) from Brazil. Caryologia. 1987;40:3–57. [Google Scholar]
- Kearney M. Systematics of the Amphisbaenia (Lepidosauria, Squamata) based on morphological evidence from recent and fossil forms. Herp Monogr. 2003;17:1–74. [Google Scholar]
- Kearney M., Stuart L. Repeated evolution of limblessness and digging heads in worm lizards revealed by DNA from old bones. Proc R Soc B. 2004;271:1677–1683. doi: 10.1098/rspb.2004.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey J.R., Papenfuss T.J., Kuel J.V., Fourcade H.M., Boore J.L. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genome sequences. Mol Phylogenet Evol. 2004;33:2–31. doi: 10.1016/j.ympev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Macgregor H., Klosterman L. Observations on the cytology of Bipes (Amphisbaenia) with special reference to its lampbrush chromosomes. Chromosoma. 1979;72:67–87. [Google Scholar]
- Matthey R. : Nouvelle contribution a l'etude des chromosomes chez les Sauriens. Rev Suisse Zool. 1933;40:281–316. [Google Scholar]
- Meyne J., Ratliff R.L., Moyzis R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyne J., Baker R.J., Hobart H.H., Hsu T.C., Ryder O.A., Ward O.A., Wiley J.E., Wurster-Hill D.H., Yates T.L., Moyzis R.K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;99:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- Mott T., Rodrigues M.T., Freitas M.A., Silva T.F.S. New species of Amphisbaena with a nonautotomic and dorsally tuberculate blunt tail from state of Bahia, Brazil (Squamata, Amphisbaenidae) J Herpetol. 2008;42:172–175. [Google Scholar]
- Mott T., Rodrigues M.T., Santos E.M. A new Amphisbaena with chevron-shaped anterior body annuli from state of Pernambuco: Brazil (Squamata, Amphisbaenidae) Zootaxa. 2009;2165:52–58. [Google Scholar]
- Olmo E. Animal Cytogenetics. Chordata 3. Reptilia. Berlin: Gebrüder Borntraeger; 1986. [Google Scholar]
- Pellegrino K.C.M., Rodrigues M.T., Yonenaga-Yassuda Y. Chromosomal evolution in Brazilian lizards of genus Leposoma (Squamata, Gymnophthalmidae) from Amazon and Atlantic rain forest: Banding patterns and FISH of telomeric sequences. Hereditas. 1999;131:15–21. doi: 10.1111/j.1601-5223.1999.00015.x. [DOI] [PubMed] [Google Scholar]
- Schmid M., Feichtinger W., Nanda I., Schakowski R., Garcia R.V., Puppo J.M., Badillo A.F. An extraordinary low diploid chromosome number in the reptile Gonatodes taniae (Squamata, Gekkonidae) J Hered. 1994;85:255–260. doi: 10.1093/oxfordjournals.jhered.a111452. [DOI] [PubMed] [Google Scholar]
- Vidal N., Azvolinsky A., Cruaud C., Hedges S.B. Origin of tropical American burrowing reptiles by transatlantic rafting. Biol Lett. 2008;4:115–118. doi: 10.1098/rsbl.2007.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]