Abstract
Microsatellite markers, also known as SSRs (Simple Sequence Repeats), have proved to be excellent tools for identifying variety and determining genetic relationships. A set of 127 SSR markers was used to analyze genetic similarity in twenty five Coffea arabica varieties. These were composed of nineteen commercially important Brazilians and six interspecific hybrids of Coffea arabica, Coffea canephora and Coffealiberica. The set used comprised 52 newly developed SSR markers derived from microsatellite enriched libraries, 56 designed on the basis of coffee SSR sequences available from public databases, 6 already published, and 13 universal chloroplast microsatellite markers. Only 22 were polymorphic, these detecting 2-7 alleles per marker, an average of 2.5. Based on the banding patterns generated by polymorphic SSR loci, the set of twenty-five coffee varieties were clustered into two main groups, one composed of only Brazilian varieties, and the other of interspecific hybrids, with a few Brazilians. Color mutants could not be separated. Clustering was in accordance with material genealogy thereby revealing high similarity.
Keywords: SSR, coffee, genetic similarity, molecular marker
Introduction
Coffee is an important crop in several countries. Of all the species, Coffea arabica L. is the most widely grown, due both to the low caffeine content and the smooth final beverage. This species accounts for almost the entire production of Latin American countries (Orozco-Castillo et al., 1994).
Traditionally, morphological and biochemical characteristics have been used to characterize varieties. Although these markers are still important, they are somewhat limited, through the need for physical space for evaluation, the effect of environmental conditions on character expression, and the time required for making a full description, as several characters need to be evaluated during the entire growth period of the plant. For coffee trees, the latter limitation is extremely relevant, through being a perennial crop requiring three-years-growth until full maturity (Mendes and Guimarães, 1998). It takes at least fifteen years to obtain a new variety.
In 2001, C. arabica was included on the Brazilian roll of species from which varieties can be protected, without, however, indicating stable and homogeneous markers required for the effective enforcement of protective measures. In the past, DNA-based markers have been used for studying genetic diversity in many plant species. This type of marker, besides facilitating the analysis of variation present in DNA itself, can also be used for variety identification. In addition, they are environmentally independent, and may be detected in any type of tissue and developmental phase of the plant (Arens et al., 1995; Ferreira and Grattapaglia, 1998). Analysis of C. arabica varieties in Brazil has revealed that the material employed is derived from few ancestral varieties (Typica, Bourbon and Sumatra), which themselves have undergone mutual spontaneous mutations and crossings (Mendes and Guimarães, 1998).
Nuclear DNA variation in coffee has been evaluated by using molecular markers such as RFLP (Lashermes et al., 1999), RAPD (Diniz et al., 2005; Anthony et al., 2002, Silveira et al.., 2003), AFLP (Steiger et al., 2002; Anthony et al., 2002) and SSRs (Combes et al., 2000; Anthony et al., 2002; Moncada and McCouth 2004; Maluf et al., 2005; Poncet et al., 2006; Aggarwal et al., 2007; Silvestrini et al., 2007), whereby it has been shown that genetic variation in the genus Coffea is low, especially among cultivated C. arabica tetraploid varieties. Chloroplast DNA (cpDNA) non-coding regions have been used as a source of molecular markers in studies concerning the relationships within and among species of this genus (Orozco-Castillo et al., 1996; Cros et al., 1998), where only interspecific polymorphism was detected.
Simple sequence repeats (SSR), or microsatellite markers, are very attractive for studies in plant genetics, through their usefulness in evaluating those varieties with a narrow genetic base (Bredemeijer et al., 2002). Furthermore, they can be efficiently analyzed by rapid and simple polymerase chain reactions, besides being co-dominant, highly reproducible and multi-allelic, and capable of being automated (Ferreira and Grattapaglia, 1998).
By using markers developed for C. arabica, Moncada and McCouth (2004) showed the particular value of SSR markers for discriminating closely related commercial varieties of coffee. Maluf et al. (2005) and Silvestrini et al. (2007) confirmed the low genetic diversity in coffee, mainly in tetraploid varieties, although none were related to the varieties under study.
The number of microsatellite markers currently available for coffee remains limited. To date, only 224 genomic SSR markers for species of the Coffea genus have been described (Hendre et al., 2008). Coffea arabica is the most important, and there is an urgent need for additional microsatellite makers for facilitating the identification of closely related varieties. Thus, the aim was to develop and characterize additional microsatellite markers for C. arabica, and evaluate their use in identifying varieties of commercial interest in Brazil.
Material and Methods
Plant material and DNA isolation
A set of 19 Coffea arabica varieties was selected, these representing all the major varieties grown in Brazil (Table 1). The DNA of each genotype was extracted from ground seeds. Six interspecific hybrids of C. arabica, C. liberica and C. canephora from the Centro de Investigação das Ferrugens do Cafeeiro (CIFC) were included in this work. The DNA of this material was extracted from freeze-dried leaves. For the construction of genomic libraries enriched for microsatellites, DNA was extracted from leaves of variety Catuaí Vermelho IAC-44 (C.arabica). In all cases the DNA extraction was carried out using the DNeasy Plant Mini Kit (Qiagen), according to manufacturer's instructions.
Table 1.
Genealogy of the studied coffee varieties.
| Number | Origin | Name | Background |
| 1 | Brazil | Acaiá Cerrado MG1474 | Selection from Mundo Novo |
| 2 | Brazil | Mundo Novo IAC 376-4 | Sumatra X Bourbon Vermelho |
| 3 | Brazil | Obatã IAC1669-20 | Selection from Sarchimor1 |
| 4 | Brazil | Oeiras MG 6851 | Selection from Caturra Vermelho X Hibrido do Timor |
| 5 | Brazil | Ouro Verde IAC H5010-5 | Selection from Catuaí Amarelo and Mundo Novo |
| 6 | Brazil | Rubi MG1192 | BC Catuaí Vermelho X Mundo Novo |
| 7 | Brazil | Topázio MG1190 | BC Catuaí Amarelo X Mundo Novo |
| 8 | Brazil | Bourbon Amarelo IAC J22 | Típica |
| 9 | Brazil | Bourbon Vermelho IAC 662 | Típica |
| 10 | Brazil | Catuaí Amarelo IAC 62 | Selection Mundo Novo X Caturra Amarelo |
| 11 | Brazil | Catuaí Vermelho IAC 99 | Mundo Novo X Caturra Amarelo |
| 12 | Brazil | Catucaí Amarelo 2015/ cova 479 | Icatu Amarelo X Catuaí Vermelho |
| 13 | Brazil | Catucaí Vermelho 2015/cova 476 | Icatu Vermelho X Catuaí Amarelo |
| 14 | Brazil | Caturra Amarelo IAC 476 | Mutant of Bourbon Vermelho |
| 15 | Brazil | Caturra Vermelho IAC 477 | Mutant of Bourbon Vermelho |
| 16 | Brazil | IAPAR 59 | Selection from Sarchimora |
| 17 | Brazil | Tupi IAC 1669-33 | Selection from Sarchimora |
| 18 | Brazil | Icatu Amarelo IAC 2944 | Bourbon Amarelo X Icatu Vermelho |
| 19 | Brazil | Icatu Vermelho IAC 2945 | Bourbon Vermelho X C. canephora |
| 20 | Portugal | CIFC H147/1 | C. arabica X C. liberica |
| 21 | Portugal | CIFC 34/13 (S353-4/5) | C. arabica X C. liberica |
| 22 | Portugal | CIFC 832/1 | Híbrido do Timor (C. arabica X C. canephora) |
| 23 | Portugal | CIFC 1343/269 | Híbrido do Timor clone (C. arabica X C. canephora) |
| 24 | Portugal | CIFC 110/5 | C. arabica X C. Arabica |
| 25 | Portugal | CIFC H539/8 | C. arabica X C. Canephora |
1Sarchimor = Villa Sarchi X Híbrido do Timor (= C. arabica X C. canephora).
SSR sequences from public databases and primer design
Coffee microsatellite sequences were extracted from the NCBI database. Those containing di-nucleotide (n > 10) or tri-nucleotide (n > 6) repeats were selected for primer design. PCR primers flanking the repeat sequence were designed using the primer select module of the DNAstar Lasergene package. Six SSRs for C. arabica described by Combes et al. (2000) and 13 cpDNA SSRs (Taberlet et al., 1991; Orozco-Castillo et al., 1996) were also tested.
Microsatellite isolation
Additional microsatellites were isolated from enriched small-insert genomic libraries constructed according to Van de Wiel et al. (1999), with a minor modification. DNA of the Catuaí Vermelho IAC-44 variety was digested with AluI, RsaI, MboI or TaqI enzymes instead of being sonicated. After digestion, the DNA fragments were hybridized to filters containing the following synthetic oligonucleotides: (TCT)10, (TGT)9, (GAG)8, (GTG)8, (TGA)9, (AGT)10, (CGT)8, (GCT) 8, (CT)12 and (GT)12. Filters, on being washed with 0.5xSSC 1% SDS (low stringency wash) and 0.2xSSC 1% SDS (high stringency wash), gave rise to two genomic libraries.
Nomenclature
The newly developed markers were named according to the nomenclature proposed by Hendre et al. (2008) for C.canephora SSR markers. Each marker was identified by the suffix CarM, indicating C. arabica microsatellite marker, followed by a number.
Microsatellite analysis
Microsatellites were amplified by PCR in a 20 μL reaction volume, containing 10 mM of Tris-HCl pH 9.0, 20 mM of (NH4)2SO4, 0.01% Tween 20, 1.5 mM MgCl2, 0.1 mM of each dNTP, 4 pmol of each primer, 0.2 units of Goldstar Taq DNA polymerase (Eurogentec, Maastricht, The Netherlands), and 16 ng of genomic DNA. The amplifications were performed in a PTC-200 MJ Research Thermal Cycler, programmed for one step at 94 °C for 3 min, followed by 30 cycles (30 s at 94 °C, 30 s at the annealing temperature determined for each primer pair, and 45 s at 72 °C), and a final extension at 72 °C for 3 min. All primers were synthesized by Eurogentec (Maastricht, The Netherlands) (Table S1).
The PCR products were separated on 6% polyacrylamide gels, by using a Sequi-Gen Sequencing Cell (Bio-Rad) apparatus at 110W for 1-3 h in 1x TBE buffer. After electrophoresis, the products were visualized through silver staining as described by Van de Wiel et al. (1999), and the patterns analyzed for the presence of polymorphism and the quality of the banding pattern, according to Arens et al. (1995).
Data analysis
For polymorphic microsatellite loci, the number of alleles per locus and allelic phenotypes were counted. Considering that C. arabica is a tetraploid species, assessing the actual genotype itself based on band intensity is unreliable. Therefore, banding patterns were observed for each polymorphic locus and recorded as allelic phenotypes (Becher et al., 2000). In order to quantify the discrimination power of the microsatellite markers, the number of effective alleles (ne) for each marker was calculated according to the formula (Hartl and Clark, 1997): ne = 1/Σ (E/F)2, where E is the total number of genotypes with each allele of locus i, and F is the total number of alleles of the locus i in all genotypes.
A presence/absence (1/0) allele matrix was built, and Jaccard similarity was calculated by using the NTSYS (version 2.1) computer program. UPGMA dendrogram was calculated using the SHAN algorithm of the NTSYS package. Bootstrapping was applied to evaluate the degree of association between the genetic similarity matrix and dendrogram, using the BOOD software version 3.0 (Coelho, 2001). Pearson correlationship was calculated using the GENES software Windows version (Cruz, 2001) to indicate the extent to which the clustering of genotypes demonstrated in the dendogram accurately represents the estimates of genetic similarity.
Results
Microsatellite enrichment from C. arabica
An overview of the results obtained with microsatellite enrichment procedures is given in Table 2. An arbitrary number of positive clones were sequenced. Of the 135 recombinant clones obtained from the first enrichment (low stringency library), 110 were sequenced, with 41% (45) containing a microsatellite sequence, 2 of which redundant. Twenty microsatellite sequences had perfect repeats, 22 had imperfect repeats and three were compound repeats. Flanking regions in eighteen inserts were large enough for primer design. From the 397 recombinant clones characterized in the second enrichment (high stringency library), 192 were sequenced, with 46% (89) containing microsatellite sequences, 14 of which redundant. For this set, 51 microsatellite sequences were perfect repeats and 29 imperfect and nine were compound repeats. Flanking regions in 35 inserts were suitable for primer design.
Table 2.
Results from microsatellite cloning and sequencing of two enriched libraries (EL) of C. arabica. Two elution conditions were used: 1) low-stringency (0.5xSSC) and 2) high-stringency (0.2xSSC). Positive clone indicates the number of clones hybridizing to a labeled oligo probe mixture. SSR indicates the number of clones containing a microsatellite. Designed primers indicate the number of clones on the basis of which primers could be designed for amplification of the microsatellite.
| EL | Screened clones | Positive clones | Sequenced clones | SSR | Primers designed | Polymorphic markers |
| 1 | 3572 | 135 | 110 | 45 | 18 | 2 |
| 2 | 3840 | 397 | 192 | 89 | 35 | 5 |
In total, 53 primer pairs could be developed, 23 for di-, 24 for tri-, 3 for tetra-, 2 for penta- and 1 for compound-nucleotide repeats. The latter two were found only in the first enriched library. The sequences of all markers obtained are shown in Table S1. Compound microsatellite sequences consisting of CCA/TCA and TGA/GAA repeats were found in both libraries, whereas clones containing microsatellite sequences which were not specifically searched for, as GGA, were found in the first. GT and TGA were the most common di- and tri-nucleotide motifs encountered (results not shown). All 53 primer pairs produced a clear PCR fragment.
Markers from public database sequences and literature
The screening of a public (NCBI) database for microsatellite sequences resulted in 56 accessions which met the set criteria (more than 6 repeat units per tri-nucleotide repeat and 10 repeat units per di-nucleotide). Some of these sequences had already been used for marker development. Nevertheless, as primer design was undertaken independently, there are differences in the primers used to amplify SSR markers in our study and theirs (Coulibaly et al., 2003; Poncet et al., 2006). An additional 6 primer pairs were available from literature (Combes et al., 2000). These sequences are also incorporated in Table S1. Beside the nuclear DNA markers, 13 cpDNA primers were tested (Taberlet et al., 1991; Orozco-Castillo et al., 1996).
Marker characterization and allelic variation
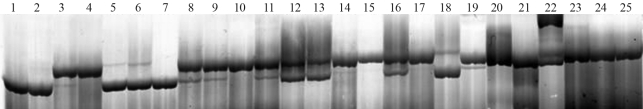
A total of 127 primer pairs were tested for pattern quality and degree of polymorphism using a set of 19 coffee-varieties and 6 inter-specific hybrids. 125 primers amplified the expected DNA fragments, although only 22 were polymorphic. Most markers contained a GT repeat. All polymorphic markers gave a pattern quality of 1 or 2 (Arens et al., 1995) and could be scored unambiguously. An example of the molecular pattern obtained with the CarM092 marker is shown in Figure 1. A total of 55 alleles were detected using the 22 polymorphic SSR loci, the number of alleles per locus ranging from 1 to 7, an average of 2.5 alleles per locus (Table 3).
Figure 1.
Molecular pattern obtained with the marker CarM092 (1: Acaiá Cerrado MG1474; 2:Mundo Novo IAC 376-4; 3:Obatã IAC 1669-20; 4: Oeiras MG6851; 5: Ouro Verde IAC H5010-5; 6: Rubi MG1192; 7: Topázio MG1190; 8: Bourbon Amarelo IAC J22; 9: Bourbon Vermelho IAC 662; 10: Catuaí Amarelo IAC 62; 11: Catuaí Vermelho IAC 99; 12:Catucaí Amarelo 2015/cova479; 13: Catucaí Vermelho 2015/cova476; 14: Caturra Amarelo IAC 476; 15: Caturra Vermelho IAC 477; 16: IAPAR 59; 17: Tupi IAC 1669-33; 18: Icatu Amarelo IAC2944; 19: Icatu Vermelho IAC 2945; 20: CIFC H147/1; 21: CIFC 34/13(S353-4/5); 22: CIFC 832/1; 23: CIFC1343/269; 24: CIFC 110/5; 25: CIFC H539/8.
Table 3.
Number of alleles per locus, number of effective alleles (ne) and number of allelic phenotypes from the 22 polymorphic microsatellite markers.
| SSR marker | N. of alleles per locus | N. of effective alleles (ne) | N. of allelic phenotype |
| M201 | 3 | 1.3 | 4 |
| M24 | 4 | 1.9 | 5 |
| CarM0652 | 1 | 1 | 1 |
| CarM070 | 1 | 1 | 1 |
| CarM069 | 2 | 1.1 | 2 |
| CarM068 | 1 | 1 | 1 |
| CarM0863 | 1 | 1 | 1 |
| CarM092 | 3 | 1.9 | 4 |
| CarM096 | 3 | 1.8 | 2 |
| CarM101 | 7 | 3.9 | 4 |
| CarM105 | 2 | 1.4 | 3 |
| CarM0014 | 1 | 1 | 1 |
| CarM002 | 1 | 1 | 1 |
| CarM0485 | 5 | 1.8 | 6 |
| CarM049 | 2 | 1.2 | 3 |
| CarM050 | 2 | 1.5 | 2 |
| CarM051 | 5 | 3.4 | 6 |
| CarM052 | 7 | 1.8 | 7 |
| Ccmp36 | 1 | 1 | 1 |
| Ccmp6 | 2 | 1.2 | 2 |
| Ccmp10 | 1 | 1 | 1 |
| NTCP8 | 1 | 1 | 1 |
| Total number of alleles | 55 | ||
| Average alleles/locus | 2.5 | ||
1Primer sequences published by Combes et al. (2000), 2Primers developed from clone sequences published by Rovelli et al. (2000) in the NCBI database; 3Primers developed from clone sequences published in the NCBI database; 4Primers obtained in the first genomic library; 5Primers obtained in the second genomic library; 6Chloroplast markers.
Variety identification
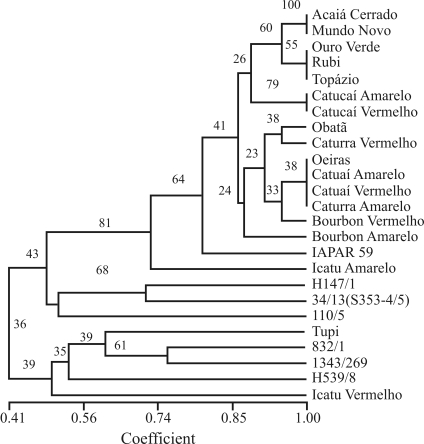
The 22 polymorphic SSR markers were used to group the set of varieties and inter-specific hybrids. The UPGMA dendrogram revealed that most of the Brazilian varieties were placed in a group with high bootstrap value (81.7%), thereby indicating reliable clustering (Figure 2). The interspecific hybrids and two Brazilian varieties (Tupi and Icatu Vermelho) were placed in groups with bootstrap values below 50%. Pearson correlation was 0.958, thus indicating that the observed clustering of varieties in the dendrogram accurately represented the estimates of genetic similarity.
Figure 2.
UPGMA dendrogram obtained using the Jaccard similarity of 19 coffee varieties and 6 interspecific hybrids, with data from 22 polymorphic microsatellite loci (Bootstrap values are in percentages).
The allelic profiles of all the varieties used in this study can be seen in Table S2. As regards Brazilian material, variety-specific alleles were detected with the markers CarM101, CarM051 and CarM052 for the varieties Bourbon Vermelho, Icatu Amarelo and Vermelho.
The loci CarM068, CarM086, CarM002, ccmp3, ccmp10 and NTCP8 amplified only in interspecific hybrids.
The CarM051 locus was the most discriminating, with six allelic phenotypes and 3.4 effective alleles (Table 3). Although for CarM092 the effective alleles count was low, with only four allelic phenotypes, it was, together with CarM101, CarM051 and CarM052, one of the more discriminating markers for Brazilian varieties.
Discussion
Enriched libraries
Enrichment based on the hybridization of genomic DNA fragments to filters containing synthetic oligonucleotide repeats, has been shown to be an efficient way of microsatellite retrieval in several species like tomato, lettuce, roses and C. canephora (Vosman and Arens, 1997; Van de Wiel et al., 1999; Esselink et al., 2003; Hendre et al., 2008). Results presented in this paper indicated efficacy also for C. arabica.
In terms of efficiency, the second library (high stringency filter washing) generated a higher frequency of clones containing microsatellites, which, besides being longer, consisted of a higher percentage of perfect repeats. Most of the di-nucleotide microsatellite repeats were of the GT motif, which is in agreement with previous microsatellite retrieval efforts in coffee (Vascotto et al., 1999), and with the C. arabica microsatellite sequences present in the NCBI database. In contrast, Ruas et al. (2003) showed that the GA-nucleotide motif, combined with other di, tri and tetra-nucleotide motifs, produced a high number of DNA fragments, thereby inferring a high frequency of poly GA microsatellite motifs in the coffee genome. It is well known that di-nucleotide repeats are very common in plants (Morgante and Olivieri, 1993). In C. canephora, the most common was di-nucleotide repeats (AT and AG) (Hendre et al., 2008), which is in agreement with our results. The TGA motif was the most common among the tri-nucleotides, whereas in C. canephora, this was AGC (Hendre et al. 2008). Clones containing the TGA motif were also found by Vascotto et al. (1999) in coffee and other species, such as tomatoes and roses (Esselink et al., 2003; He et al., 2003). In Arabidopsis thaliana (Depeiges et al., 1995), sugarcane (Cordeiro et al., 2000) and black poplar (Van der Schoot et al., 2000), the frequency of this motif was lower. In general SSR markers developed for Coffea sp. are mainly comprised of di and tri-nucleotide repeats (Poncet et al., 2006; Aggarwal et al., 2007).
Allele variation
In the set of coffee varieties and interspecific hybrids, only 22 (17%) out of the 127 markers tested were polymorphic, thereby clearly revealing the narrow genetic base of coffee. There was little diversity among the material tested, especially among the Brazilian varieties. The number of alleles per locus ranged from 1 to 7, which is in agreement with previous studies (Vascotto et al., 1999; Anthony et al., 2002; Moncada and McCouth 2004; Aggarwal et al., 2007; Hendre et al., 2008).
Moncada and McCouch (2004) used a set of 34 SSR markers to distinguish closely related commercial varieties of C. arabica, thereby confirming the need for working with SSR marker sets, in the case of crops with a narrow genetic base.
On considering the low level of polymorphism detected with isolated microsatellites, an attractive strategy could be to first try selecting SSRs with a high chance of being polymorphic. Recently, a software tool for identifying such SSRs in EST sequences was developed (Tang et al., 2008). With more than 55,000 ESTs in the database (NCBI, December 2008), it was possible to identify several promising SSRs based on coding regions (Poncet et al., 2006; Aggarwal et al., 2007).
Variety identification
As already mentioned, C. arabica varieties are highly similar to each other. This high genetic similarity is possibly a consequence of the self-pollinating nature of C. arabica, as well as the breeding strategies used for coffee (Lashermes et al., 1999; Combes et al., 2000; Anthony et al., 2002; Steiger et al., 2002; Ruas et al., 2003; Moncada and McConch 2004; Maluf et al., 2005).
The interspecific hybrids clustered far from most of the Brazilian varieties probably because of the presence of C. canephora and C. liberica in the genealogy of these genotypes. This could be confirmed by using chloroplast markers, which detect only inter-specific variation (Table S2). The same was shown by Orozco-Castillo et al. (1996) in a study of taxonomic relationships within the genus Coffea, when using chloroplast DNA markers. Taberlet et al. (1991) also demonstrated that the sequence of chloroplast DNA intergenic spacers can be used for phylogenetic studies of closely related species. The removal of chloroplast data from the analysis did not alter the dendrogram obtained in the present study, thereby showing that the clustering obtained was really based on the presence of other Coffea species. This is also the case for Brazilian coffee variety clustering with interspecific genotypes. The Tupi variety is a hybrid between C. arabica and Híbrido do Timor. Icatu Vermelho comes from a cross between C. canephora and Bourbon Vermelho. Even though C. canephora is present in the genealogies of Oeiras, Catucaí Amarelo, Catucaí Vermelho and Obatã, these genotypes were grouped separate from interspecific hybrids. This might be due to differences in the background of the material used, or to the size of C. canephora introgressions in the varieties. The clustering of Brazilian varieties is in accordance with genealogical data. The varieties Acaiá Cerrado and Mundo Novo showed 100% similarity, which can be explained by Acaiá Cerrado being a selection inside Mundo Novo. In spite of the high genetic similarity among varieties, they are phenotypically different. In Acaiá Cerrado, tree-tops are cylindrical and diameter reduced when compared to Mundo Novo. Maluf et al. (2005) also found these two varieties to be identical. Genetic similarity among Ouro Verde, Rubi and Topázio is most probably due to the latter two having Catuaí as a parent, whereas Ouro Verde is a selection from Catuaí Amarelo. In Ouro Verde and Rubi, fruits are red and in Topázio yellow, whereas the young leaves of Ouro Verde are green and those of Rubi tanned.
The impossibility of separating color mutants, such as Catucaí Amarelo and Vermelho and Catuaí Amarelo and Vermelho, is to be expected, as mutants are usually the result of very few mutations that are difficult to spot with molecular markers (Weising et al., 1995; Vosman and Arens, 1997). No polymorphism caused by mutation was observed with microsatellites in peaches (Testolin et al., 2000), Pelagonium (Becher et al., 2000) and roses (Esselink et al., 2003; Vosman et al., 2004). When a mutation occurs in any of the genes involved in the synthesis of color components, a color-mutant might be generated. In coffee, one gene involved in fruit-color formation is known, and two alleles (Xc and xc) have been identified (Mendes and Guimarães, 1998). Among the commercial arabica varieties, there are other morphological differences, such as plant height, leaf shape and size, leaf-color, branch-angle and stature. However, the differences among varieties at the DNA level are limited, probably due to several commercial arabica varieties originating either from single mutations or few ancestors.
According to the pairwise similarity matrix, genetic similarity was at least 0.860 in Obatã, Caturra Amarelo and Vermelho, Oeiras, Catuaí Amarelo and Vermelho and Bourbon Vermelho. Many of these varieties are known mutants or were obtained from selections or crosses between these varieties, as is shown in Table 1. Similar results have been recorded by many authors. Steiger et al. (2002) and Maluf et al. (2005) also observed high genetic similarity between Caturra and Catuaí. The Obatã variety is a selection from Sarchimor, itself originating from crossing Villa Sarchi with Híbrido do Timor. High genetic similarity was also observed between the varieties Villa Sarchi and Caturra (Anthony et al., 2002). Thus Obatã was clustered with the above mentioned varieties.
The distance between Bourbon Vermelho and Amarelo is probably due to the latter being a natural cross between Bourbon Vermelho and the variety Amarelo de Botucatu (Mendes and Guimarães, 1998; Maluf et al., 2005). Anthony et al. (2002) noted that the M-24 primer was useful for discriminating Bourbon from other varieties. In the present work the same primer amplified an allele that also facilitated the separation of Bourbon Amarelo from all other Brazilian varieties.
Probably through being derived from Sarchimor with C.canephora ancestry, IAPAR 59 remained clustered close to interspecific hybrids. Likewise, the varieties Icatu Amarelo, Icatu Vermelho and Tupi, also with C.canephora as a common ancestor, were clustered among interspecific material.
Irrespective of the high genetic similarity, a certain level of polymorphism is still to be found among C. arabica varieties, whereby hybrids with better performances have been obtained in Brazilian breeding programs. Heterosis reached 25% in hybrids between the varieties IAPAR 59 and Mundo Novo (Diniz et al., 2005). This could be the result of the complementary action of simply a few genes.
The present fingerprint data generated for Brazilian varieties could be used to construct a DNA reference database for the molecular identification of varieties, as previously suggested (Bredemeijer et al., 2002; Aggarwal et al., 2004; Hendre et al., 2008).
Supplementary Material
The following online material is available for this article
Sequences of the developed primers.
Allelic profiles of the 19 coffee varieties and 6 interspecific hybrids obtained with polymorphic markers.
This material is made available as part of the online article from http://www.scielo.br.gmb.
Acknowledgments
The authors wish to thank Martijn van Kaauwen, Yolanda Noordijk and Wendy Van't Westende for technical support, Paul Arens for helpful discussions, the Instituto Agronômico do Paraná (IAPAR), the Universidade Federal de Lavras (UFLA), Epamig and Sjaak van Heusden for providing material, and Antônio Nazareno Guimarães Mendes for providing information on varieties. This research received financial support from CAPES (Coordenação de Aperfeiçoamento de Nível Superior, Brazil).
Footnotes
Associate Editor: Everaldo Gonçalves de Barros
References
- Aggarwal R.K., Rajkumar R., Rajendrakumar P., Hendre P.S., Baruah A., Phanindranath R., Annapurna V., Prakash N.S., Santaram A., Sreenivarsan C.S., et al. Fingerprint of Indian coffee selections and development of reference DNA polymorphism panels for creating molecular IDs for variety identification. Proceedings of 20th International Conference on Coffee Science; India: Bangalore; 2004. pp. 751–755. [Google Scholar]
- Aggarwal R.K., Hendre P.S., Varshney R.K., Bhat P.R., Krishnakumar V., Singh L. Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet. 2007;114:359–372. doi: 10.1007/s00122-006-0440-x. [DOI] [PubMed] [Google Scholar]
- Anthony F., Quiros O., Ropart P., Bertrand B., Lashermes P. Detection by simple sequence repeat markers of introgression from Coffea canephora in Coffea arabica varieties. Plant Breed. 2002;121:542–544. [Google Scholar]
- Arens P., Bredemeijer G., Smulders M., Vosman B. Identification of tomato varieties using microsatellites. Acta Hort. 1995;412:49–57. [Google Scholar]
- Becher S.A., Steinmetz K., Weising K., Boury S., Peltier D., Renou J.P., Kahl G., Wolff K. Microsatellites for variety identification in Pelargonium. Theor Appl Genet. 2000;101:643–651. [Google Scholar]
- Bredemeijer G.M.M., Cooke R.J., Ganal M.W., Peeters R., Isaac P., Noordijk Y., Rendell S., Jackson J., Roder M.S., Wendehake K., et al. Construction and testing of microsatellite database containing more than 500 tomato varieties. Theor Appl Genet. 2002;105:1019–1026. doi: 10.1007/s00122-002-1038-6. [DOI] [PubMed] [Google Scholar]
- Coelho A.S.G. BOOD - Avaliação de dendrogramas baseada em estimativas de distancias/similaridades genéticas através do procedimento de bootstrap, v. 3.0. Goiânia: Universidade Federal de Goiás; 2001. [Google Scholar]
- Combes M., Andrzejewski S., Anthony F., Bertrand B., Rovelli P., Graziosi G., Lashermes P. Characterization of microsatellite loci in Coffea arabica and related coffee species. Mol Ecol. 2000;9:1171–1193. doi: 10.1046/j.1365-294x.2000.00954-5.x. [DOI] [PubMed] [Google Scholar]
- Cordeiro G.M., Taylor G.O., Henry R.J. Characterization of microsatellite markers from sugarcane (Saccharum sp). A highly polyploid species. Plant Sci. 2000;155:161–168. doi: 10.1016/s0168-9452(00)00208-9. [DOI] [PubMed] [Google Scholar]
- Coulibaly I., Revol B., Noirot M., Poncet V., Lorieux M., Carasco-Lacombe C., Minier J., Dufour M., Hamon P. AFLP and SSR polymorphism in a Coffea interspecific backcross progeny [(C heterocalyx x C. canephora) x C. canephora] Theor Appl Genet. 2003;107:1148–1155. doi: 10.1007/s00122-003-1355-4. [DOI] [PubMed] [Google Scholar]
- Cros J., Combes M.C., Trouslot P. Phylogenetic analysis of chloroplast DNA variation in Coffea L. Mol Phylogenet Evol. 1998;9:109–117. doi: 10.1006/mpev.1997.0453. [DOI] [PubMed] [Google Scholar]
- Cruz C.D. Programa GENES. Viçosa: Universidade Federal de Viçosa; 2001. [Google Scholar]
- Depeiges A., Golbely C., Lenoir A., Cocherel S., Picard G., Raynal M., Grellet F., Delseny M. Identification of the most represented repeated motif in Arabidopsis thaliana microsatellite loci. Theor Appl Genet. 1995;91:160–168. doi: 10.1007/BF00220873. [DOI] [PubMed] [Google Scholar]
- Diniz L.E.C., Ruas C.F., Carvalho V.P., Torres F.M., Ruas E.A., Santos M.O., Sera T. Genetic diversity among 40 coffee varieties accessed by RAPD markers associated with restriction digestion. Arq Biol Tecnol. 2005;4:511–521. [Google Scholar]
- Esselink G.D., Smulders M.J.M., Vosman B. Identification of cut rose (Rosa hybrida) and rootstock varieties using robust sequence tagged microsatellite site markers. Theor Appl Genet. 2003;106:277–286. doi: 10.1007/s00122-002-1122-y. [DOI] [PubMed] [Google Scholar]
- Ferreira M.E., Grattapaglia D. Introdução ao Uso de Marcadores Moleculares em Análises Genéticas. 2nd edition. Brasília: Embrapa-Cenargen; 1998. [Google Scholar]
- Hartl L., Clark A.G. Principles of Population Genetics. 3rd edition. Sunderland: Sinauer Associates Inc; 1997. [Google Scholar]
- He C., Poysa V., Yu K. Development and characterization of simple sequence repeat markers and their use in determining relationships among Lycopersicon esculentum varieties. Theor Appl Genet. 2003;106:363–373. doi: 10.1007/s00122-002-1076-0. [DOI] [PubMed] [Google Scholar]
- Hendre P.S., Phanindranath R., Annapurna V., Lalremruata A., Aggarwal K. Development of new genomic microsatellite markers from robusta coffee (Coffea canephora Pierre ex A. Froehner) showing broad cross-species transferability and utility in genetic studies. BMC Plant Biol. 2008;8:e51. doi: 10.1186/1471-2229-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashermes P., Combes M.C., Robert J., Trouslot P., D'hont A., Anthony F., Charrier A. Molecular characterization and origin of the Coffea arabica L. genome. Mol Gen Genet. 1999;261:259–266. doi: 10.1007/s004380050965. [DOI] [PubMed] [Google Scholar]
- Maluf M.P., Silvestrini M., Ruggiero L.M.C., Guerreiro-Filho O., Colombo C.A. Genetic diversity of cultivated Coffea arabica inbred lines assessed by RAPD, AFLP and SSR marker systems. Sci Agric. 2005;62:366–373. [Google Scholar]
- Mendes A.N.G., Guimarães R.J. Genética e Melhoramento do Cafeeiro. Lavras: UFLA/FAEPE; 1998. [Google Scholar]
- Moncada P., McCouch S. Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome. 2004;47:501–509. doi: 10.1139/g03-129. [DOI] [PubMed] [Google Scholar]
- Morgante M., Olivieri A.M. PCR-amplified microsatellite markers in plant genetics. Plant J. 1993;3:175–182. [PubMed] [Google Scholar]
- Orozco-Castillo C., Chalmers K.J., Waugh R., Powell W. Detection of genetic diversity and selective gene introgression in coffee using RAPD markers. Theor Appl Genet. 1994;87:934–940. doi: 10.1007/BF00225787. [DOI] [PubMed] [Google Scholar]
- Orozco-Castillo C., Chalmers K.J., Powell W., Waugh R. RAPD and organelle specific PCR re-affirms taxonomic relationships within the genus Coffea. Plant Cell Rep. 1996;15:337–341. doi: 10.1007/BF00232367. [DOI] [PubMed] [Google Scholar]
- Poncet V., Rondeau M., Tranchant C., Cayrel A., Hamon S., Kochko A., Hamon P. SSR mining in coffee tree EST databases: Potential use of EST-SSRs as markers for the Coffea genus. Mol Genet Genomics. 2006;276:436–449. doi: 10.1007/s00438-006-0153-5. [DOI] [PubMed] [Google Scholar]
- Rovelli P., Mettulio R., Anthony F., Anzuetto F., Lashermes P., Graziosi G. Microsatellites in Coffea arabica L. In: Coffee Biotechnology and Quality. Dordrecht: Kluwer Academic Publishers; 2000. pp. 123–133. [Google Scholar]
- Ruas P.M., Ruas C.F., Pampim L., Carvalho V.P., Ruas E.A., Sera T. Genetic relationship in Coffea species and parentage determination of interspecific hybrids using ISSR markers. Genet Mol Biol. 2003;26:319–327. [Google Scholar]
- Silveira S.R., Ruas P.M., Ruas C.F., Sera T., Carvalho V.P., Coelho A.S.G. Assessment of genetic variability within and among coffee progenies and varieties using RAPD markers. Genet Mol Biol. 2003;26:329–336. [Google Scholar]
- Silvestrini M., Junqueira M.G., Favarin A.C., Guerreiro-Filho O., Maluf M.P., Silvarolla M.B., Colombo C.A. Genetic diversity and structure of Ethiopian, Yemen and Brazilian Coffea arabica L. accessions using microsatellite markers. Genet Resour Crop Evol. 2007;54:1367–1379. [Google Scholar]
- Steiger D.L., Nagai C., Moore P.H., Morden C.W., Osggod F.V., Ming R. AFLP analysis of genetic diversity within and among Coffea arabica varieties. Theor Appl Genet. 2002;105:209–215. doi: 10.1007/s00122-002-0939-8. [DOI] [PubMed] [Google Scholar]
- Taberlet P., Ludovic G., Pantou G., Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tang J., Baldwin S., Jacobs J., Van der Linden G.C., Voorrips R.E., Leunissen J.A.M., Van Eck H.J., Vosman B. Large-scale identification of polymorphic microsatellites using an in silico approach. BMC Bioinformatics. 2008;9:374. doi: 10.1186/1471-2105-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testolin R., Marrazzo T., Cipriani G., Quarta R., Verde I., Dettori M.T., Pancaldi M., Sansavini S. Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of varieties. Genome. 2000;43:512–520. [PubMed] [Google Scholar]
- Van de Wiel C., Arens P., Vosman B. Microsatellite retrieval in lettuce (Lactuca sativa L. ) Genome. 1999;42:139–149. doi: 10.1139/g98-119. [DOI] [PubMed] [Google Scholar]
- Van der Schoot J., Pospíkova M., Vosman B., Smulders M.J.M. Development and characterization of microsatellite markers in black popular (Populus nigra L. ) Theor Appl Genet. 2000;101:317–322. [Google Scholar]
- Vascotto F., Degli Ivanissevich S., Rovelli P., Anthony R., Anzueto F., Lashermes P., Graziosi G. Microsatellite in Coffea arabica: Construction and selection of two genomic libraries. Proceedings of the International Seminar on Biotechnology in the Coffee Agroindustry; Londrina: IAPAR/IRD; 1999. pp. 125–128. [Google Scholar]
- Vosman B., Arens P. Molecular characterization of GATA/GACA microsatellite repeats in tomato. Genome. 1997;40:25–33. doi: 10.1139/g97-004. [DOI] [PubMed] [Google Scholar]
- Vosman B., Visser D., Voort J.R., Smulders M.J.M., Eeuwijk F. Detection of mutants and the establishment of essential derivation among rose varieties using AFLP. Theor Appl Genet. 2004;109:1718–1725. doi: 10.1007/s00122-004-1809-3. [DOI] [PubMed] [Google Scholar]
- Weising K., Nybom H., Wolff K., Meyer W. DNA Fingerprint in Plants and Fungi. Boca Raton: CRC Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of the developed primers.
Allelic profiles of the 19 coffee varieties and 6 interspecific hybrids obtained with polymorphic markers.