Abstract
Rhagomys rufescens (Rodentia: Sigmodontinae) is an endemic species of the Atlantic forest from Southern and Southeastern Brazil. Some authors consider Rhagomys as part of the tribe Thomasomyini; but its phylogenetic relationships remain unclear. Chromosomal studies on eight specimens of Rhagomys rufescens revealed a diploid number of 2n = 36 and a number of autosome arms FN = 50. GTG, CBG and Ag-NOR banding and CMA3 /DAPI staining were performed on metaphase chromosomes. Eight biarmed and nine acrocentric pairs were found in the karyotype of this species. The X and Y chromosomes were both acrocentric. Most of the autosomes and the sex chromosomes showed positive C-bands in the pericentromeric region. The X chromosome showed an additional heterochromatic block in the proximal region of the long arm. Nucleolus organizer regions (NORs) were located in the pericentromeric region of three biarmed autosomes (pairs 4, 6 and 8) and in the telomeric region of the short arm of three acrocentrics (pairs 10, 12 and 17). CMA 3 /DAPI staining produced fluorescent signals in many autosomes, especially in pairs 4, 6, and 8. This study presents cytogenetic data of Rhagomys rufescens for the first time.
Keywords: Rhagomys rufescens, Thomasomyini, Rodentia, Atlantic forest, karyotype
The subfamily Sigmodontinae (Wagner 1843) comprises 74 genera and 377 species (Musser and Carleton, 2005) and includes predominantly South American Cricetidae rodents, such as Rhagomys rufescens. This species is endemic to the Atlantic forest from Southern and Southeastern Brazil, and has already been reported in Rio de Janeiro, Minas Gerais, São Paulo, Espírito Santo and Santa Catarina (Moojen, 1952; Emmons and Feer, 1997; Eisenberg and Redford, 1999; Nowak, 1999; Percequillo et al., 2004; Pinheiro et al., 2004; Metzger et al., 2006; Pardini and Umetzu, 2006; Steiner-Souza et al., 2008). The first record of R. rufescens in Southern Brazil was recently obtained at the Parque Natural Municipal Nascentes do Garcia (PNMNG), in the state of Santa Catarina (Steiner-Souza et al., 2008).
Rhagomys rufescens was originally described as Hesperomys rufescens, based on a female collected in Rio de Janeiro, southeastern Brazil. In the beginning of the 20th century, Thomas collected another specimen from an unknown locality, which was used for the description of the genus Rhagomys (Thomas 1917) (Percequillo et al., 2004). A second species, Rhagomys longilingua, was recently described based on a male collected in the Montana forests in southern Peru (Luna and Patterson, 2003), that was later found to reach as far as Bolivia (Villalpando et al., 2006).
Rhagomys is considered incertae sedis (Reig, 1980, 1984; McKenna and Bell, 1997; Smith and Patton, 1999; Musser and Carleton, 2005) or a “plesiomorphic Neotropical muroid”, according to Voss (1993) and Steppan (1995), and there is no consensus regarding its tribal position (Percequillo et al., 2004). Nevertheless, some authors included Rhagomys in the tribe Thomasomyini based on morphological characters (Pacheco, 2003) or on nuclear IRBP gene sequences (D'Elia et al., 2006, 2007). Cytogenetic studies on species of Thomasomyini have shown significant variation both in diploid number (2n = 20-82) and in the number of autosome arms (FN = 34-114) (Table 1). The species in Table 1 are grouped in “Andean” Thomasomyini, which includes genera with a predominantly Andean distribution (sensu Pacheco, 2003), and “Endemic Atlantic” Thomasomyini, an informal group named by Oliveira and Bonvicino (2002).
Table 1.
Diploid numbers (2n) and number of autosome arms (FN) of Thomasomyini species.
| Species | 2n | FN | Authors |
| “Andean” Thomasomyini species | |||
| Aepeomys fuscatus | 54 | 62 | Gardner and Patton (1976) |
| Aepeomyslugens | 28 | 48 | Aguilera et al. (2000) |
| Aepeomyslugens | 44 | 46 | Gómez-Laverde et al. (1997) |
| Aepeomys sp. | 44 | 46 | Aguilera et al. (1994) |
| Aepeomysreigi | 44 | 46 | Ochoa et al. (2001) |
| Rhipidomys cearanus | 44 | - | Zanchin et al. (1992a) |
| Rhipidomys latimanus | 44 | 48 | Gardner and Patton (1976) |
| Rhipidomys leucodactylus | 44 | 48 | Zanchin et al. (1992a) |
| Rhipidomys leucodactylus | 44 | 48 | Andrades-Miranda et al. (2002) |
| Rhipidomys leucodactylus cytotype 1 | 44 | 52 | Andrades-Miranda et al. (2002) |
| Rhipidomys mastacalis | 44 | 74 | Zanchin et al. (1992a) |
| Rhipidomys mastacalis cytotype 1 | 44 | 80 | Andrades-Miranda et al. (2002) |
| Rhipidomys mastacalis cytotype 2 | 44 | 76 | Andrades-Miranda et al. (2002) |
| Ripidomys cf. mastacalis | 44 | 52 | Silva and Yonenaga-Yassuda (1999) |
| Rhipidomys nitela | 48 | 68 | Andrades-Miranda et al. (2002) |
| Rhipidomys sclateri | 44 | 48 | Aguilera et al. (1994) |
| Rhipidomys sp. | 44 | 48 | Svartman and Almeida (1993) |
| Rhipidomys sp. | 44 | 49 | Svartman and Almeida (1993) |
| Rhipidomys sp. | 44 | 50 | Zanchin et al. (1992a) |
| Rhipidomys sp. A | 44 | 61 | Silva and Yonenaga-Yassuda (1999) |
| Rhipidomys sp. B | 50 | 71,72 | Silva and Yonenaga-Yassuda (1999) |
| Thomasomys andersoni | 44 | 42 | Salazar-Bravo and Yates (2007) |
| Thomasomys aureus | 44 | 42 | Gardner and Patton (1976) |
| Thomasomys kalinowskii | 44 | 44 | Gardner and Patton (1976) |
| Thomasomys laniger | 42 40 |
40 40 |
Aguilera et al. (2000) Gómez-Laverde et al. (1997) |
| Thomasomys monochromos | 42 | 42 | Gardner and Patton (1976) |
| Thomasomys niveipes | 24 | 42 | Gomez-Laverde et al. (1997) |
| Thomasomys notatus | 44 | 44 | Gardner and Patton (1976) |
| Thomasomys sp. | 44 | 42 | Gardner and Patton (1976) |
| Thomasomys taczanowskii | 44 | 44 | Gardner and Patton (1976) |
| Thomasomys vestitus | 44 | 42 | Aguilera et al. (2000) |
| “Endemic Atlantic” Thomasomyini species | |||
| Delomys collinus | 82 | 86 | Bonvicino and Geise (1995) |
| Delomys dorsalis | 82 | 80 | Zanchin et al. (1992b) |
| Delomys sublineatus | 72 | 90 | Zanchin et al. (1992b) |
| Phaenomys ferrugineus | 78 | 114 | Bonvicino et al. (2001) |
| Juliomys ossitenuis | 20 | 36 | Costa et al. (2007) |
| Juliomys pictipes | 36 | 34 | Bonvicino and Otazu (1999) |
| Juliomys rimofrons | 20 | 34 | Oliveira and Bonvicino (2002) |
| Juliomys sp. | 32 | 48 | Paresque et al. (2009) |
| “Other” Thomasomyini species | |||
| Abrawayaomys ruschii | 58 | - | Pereira et al. (2008) |
| Andinomy edax | 56 | 56 | Spotorno et al. (2001) |
| Irenomys tarsalis | 64 | 98 | Ojeda et al. (2004) |
| Rhagomys rufescens | 36 | 50 | Present report |
| Wiedomys cerradensis | 60 | 88 | Gonçalves et al. (2005) |
| Wiedomys pyrrhorhinos | 62 | 86 | Maia and Langguth (1981) |
The objective of this study was to describe the karyotype of Rhagomys rufescens from southern Brazil after conventional and CMA3/DAPI staining, and GTG, CBG and Ag-NOR banding. The chromosomal data presented in this work can provide additional information for studies on both taxonomic and phylogenetic relationships.
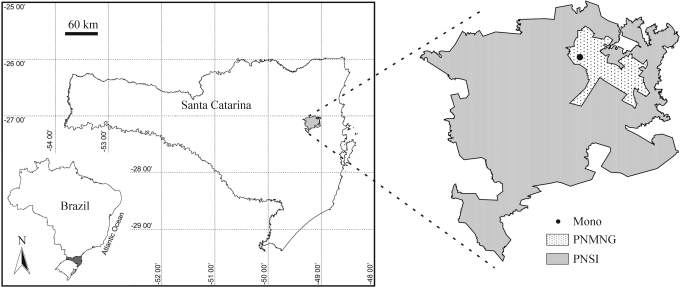
Eight specimens (five males and three females) were analyzed. They were captured at PNMNG, at “Mono” locality (27°02'59” S, 49°08'57” W), in Indaial city, in the state of Santa Catarina, southern Brazil. This park is now part of Parque Nacional da Serra do Itajaí (PNSI) (Figure 1). The animals were caught in Sherman traps placed at 3 m from the ground, according to Kierulff et al. (1991), with adaptations.
Figure 1.
“Mono” locality, data collection site of specimens at Parque Natural Municipal Nascentes do Garcia (PNMNG), part of Parque Nacional Serra do Itajaí (PNSI), state of Santa Catarina, Southern Brazil.
Chromosomes were obtained directly from bone marrow according to the method of Ford and Hamerton (1956), modified by Sbalqueiro and Nascimento (1996). Conventional Giemsa staining (5%) was used to determine diploid number (2n), chromosomal morphology and the number of autosomal arms (FN). At least 20 metaphase plates per individual were examined. GTG, CBG and Ag-NOR banding were performed according to Seabright (1971), Sumner (1972), and Howell and Black (1980), respectively. Chromomycin A3 (CMA3), and 4,6-diamidino-2-phenylindole (DAPI) were used according to Schweizer (1976). Chromosomes were classified as metacentric (M), submetacentric (SM), and acrocentric (A).
Skins and skulls of specimens were deposited at the Coleção Zoológica da Fundação Universidade Regional de Blumenau (CZFURB), in Blumenau, State of Santa Catarina, Brazil.
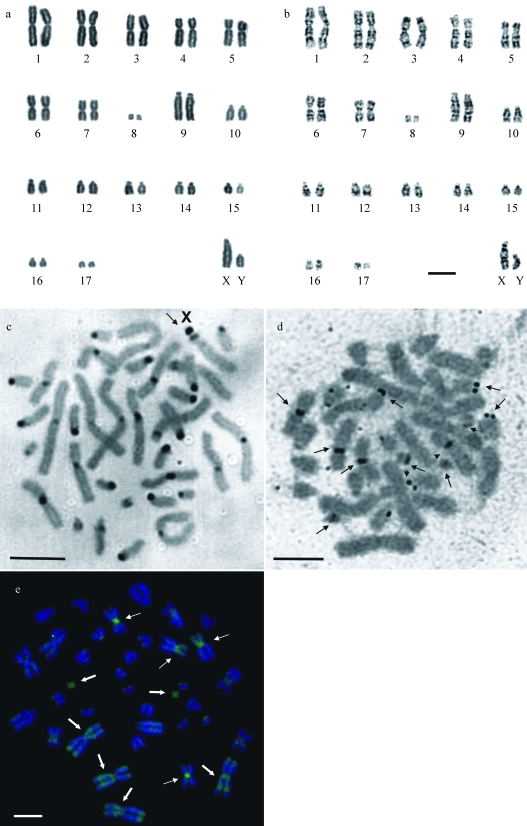
Analyses after conventional staining showed 2n = 36 and FN = 50 in all specimens (Figure 2a), with five metacentric pairs (1, 3, 6 and 8), three submetacentric (pairs 2, 4 and 5) and nine acrocentric pairs (pairs 9 to 17), decreasing gradually in size. The X chromosomes were acrocentric, indistinguishable from pair 9, whereas the Y chromosome was also acrocentric and similar in size to pair 10. All chromosome pairs, including the sex chromosomes, could be identified after G-banding. The X chromosome showed two positive bands in the medium portion of the long arm and the Y chromosome had one proximal band in the long arm (Figure 2b).
Figure 2.
Karyotype of Rhagomys rufescens (male) after (a) conventional staining and (b) G-banding. Bar = 2,5 μm. (c) Metaphase of Rhagomys rufescens after C-banding. The arrow points to the X chromosome. (d) Metaphase after Ag-NOR staining. The arrows point to the nucleolus organizing regions. (e) Metaphase after CMA3/DAPI staining. The arrows point to the chromosomes with intense fluorescent CMA3 signals in the “p” and “q” arms (thick arrow) and pericentromeric region (thin arrow).
C-banding revealed pericentromeric constitutive heterochromatic blocks in most autosomes and also in the sex chromosomes. An additional interstitial C-band was present in the proximal region of the long arm of the X chromosome (Figure 2c).
NORs were detected in the pericentromeric region of pairs 4, 6 and 8, and in the telomeric region of the short arm of acrocentric pairs 10, 12 and 17 (Figure 2d). Two to twelve NORs were observed, with a mean of 7.33 ± 3.19 per cell (N = 39).
The double staining with the GC- and AT-specific fluorochromes, CMA3 and DAPI, respectively, showed intense fluorescent CMA3 signals in the pericentromeric region of pairs 4 and 6, and throughout the length of pair 8. Less intense signals were observed in other pairs (Figure 2e).
Rhagomys is a polytypic genus composed by R. longilingua and R. rufescens. After comparative morphological analyses, Pacheco (2003) considered it monophyletic, although the two forms show a discontinuous distribution: R. longilingua can be found in Peru and Bolivia, whereas R. rufescens occurs in southern and southeastern Brazil. The phylogenetic relationships of this genus with other Sigmodontinae are controversial and uncertain and it has been previously included in different tribes of this subfamily.
Pacheco (2003) compared morphological characters of R. rufescens to those of various species of Sigmodontinae. This author suggested a phylogenetic relationship with the tribe Thomasomyini: Abrawayaomys, Aepeomys, Chilomys, Delomys, Juliomys, Phaenomys, Rhipidomys, Thomasomys (including Erioryzomys and Inomys), Wiedomys, and Wilfredomys. R. rufescens appeared as a sister group of Abrawayaomys ruschii or within the “Andean” Thomasomyine group (Thomasomys, Aepeomys, Chilomys and Rhipidomys).
After analyses of the nuclear IRBP (Interphotoreceptor Retinoid Binding Protein) gene sequences, D'Elia et al. (2006) suggested grouping Rhagomys longilingua with the Thomasomyini species as a sister-group of Thomasomys and as part of a larger clade that also includes Aepeomys and Rhipidomys.
On the other hand, Percequillo et al. (2004), based on mitochondrial cytochrome B sequences, concluded that the position of R. rufescens within Sigmodontinae was uncertain and that Rhagomys was either closely associated to Andinomys, followed by a Thomasomys-Rhipidomys group, or closer to Juliomys, followed by Andinomys.
Therefore, all these studies suggested a phylogenetic relationship of Rhagomys with Thomasomyini species. So far, cytogenetic data have shown a significant variation in both diploid number (2n = 20-82) and FN (34-114) (Zanchin et al., 1992b Bonvicino and Geise, 1995; Bonvicino and Otazu, 1999; Oliveira and Bonvicino, 2002; Costa et al., 2007). Nevertheless, most species presented 2n = 44 and a predominance of acrocentric chromosomes, which were the cases of the species of Rhipidomys and Thomasomys (Table 1), possibly a sister group of Rhagomys (Pacheco, 2003; Percequillo et al., 2004, D'Elia et al., 2006). These results differ from our chromosome data of Rhagomys rufescens (2n = 36 and FN = 50), which had eight biarmed and nine acrocentric autosomal pairs.
The cytogenetic data of Rhagomys rufescens (2n = 36 and FN = 50) described herein are the first for this genus. Pair 9 and the X chromosome were undistinguishable after conventional staining because of their similar sizes and morphologies. However, GTG and CBG banding patterns showed significant differences allowing their individual identification. The two interstitial G-bands in the long arm of the X chromosome, characteristic of mammalian X chromosomes (Pathack and Stock, 1974), could be observed. Furthermore, an additional block of interstitial constitutive heterochromatin was present in the proximal region of the long arm of X chromosome, whereas pair 9 only presented a pericentromeric heterochromatic block. The Y chromosome, which is almost completely heterochromatic in many species of South-American rodents (Sbalqueiro et al. 1991; Andrades-Miranda et al., 2001), only presented a positive C-band in the pericentromeric region in Rhagomys rufescens.
After double fluorochrome staining, CMA3-positive and DAPI-negative signals were present in sites coincident with all AgNORs. The correlation of NORs with GC-rich sites is relatively common among vertebrates (Schmid, 1982; Amemiya and Gold, 1986; Artoni et al., 1999, among others), although the reverse correlation is not always valid. Additional GC-rich sites were also observed, mainly in the first three chromosome pairs. These sites were euchromatic domains adjacent to G-bands, known to correspond to GC-rich isochores (R-bands), especially close to the telomeric region (Bernardi, 1993; Holmquist and Ashley, 2006). However, several authors suggested the use of the silver staining technique in conjunction with FISH (rDNA probes) to confirm the number and location of NORs (Santos et al., 2001; Fagundes et al., 2003; Leite-Silva et al., 2003).
The comparison of the chromosome data presented herein to those of the other Thomasomyini species mentioned above does not allow to determine the taxonomic relationship of Rhagomys rufescens. The scarcity of cytogenetic data of a larger number of species and the lack of techniques that could show more details about chromosome structure makes further taxonomic analysis a difficult task. It is evident that several chromosome rearrangements have contributed to the karyotypic variability observed in Thomasomyini. Complementary data obtained from differential staining associated with FISH techniques, such as ZOO-FISH (Hass et al., 2008), is necessary for clarifying the mechanisms of karyotypic evolution in this group, and hence contribute to determine the taxonomic position of this genus. The data reported herein are important as a first characterization of the chromosome complement of R. rufescens because it allows the identification of some primary features of its karyotype.
Acknowledgments
The authors would like to thank Dr. Iris Hass (UFPR), Dr. Edivaldo Herculano Correa de Oliveira (UFPA), Dr. Rafael Noleto (UFPR), and Dr. Geraldo Moretto (FURB) for their critical reading of the manuscript; Rafael Pasold and Esmeralda Dávida Ferreira da Silva for their help with the fieldwork; Guilherme Pereira Rabelo for technical assistance; Instituto Parque das Nascentes (IPAN) and IBAMA (Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis) for logistic support with data collection at the Parque Natural Municipal Nascentes do Garcia (PNMNG).
Footnotes
Associate Editor: Yatiyo Yonenaga-Yassuda
References
- Aguilera M., Pérez-Zapata M., Martino A., Barros M.A., Patton J. Karyosystematics of Aepeomys and Rhipidomys (Rodentia, Cricetidae) Acta Cient Venez. 1994;48:247–248. [Google Scholar]
- Aguilera M., Pérez-Zapata A., Achoa J., Soriano P. Karyology of Aepeomys and Thomasomys (Rodentia, Muridae) from the Venezuelan Andes. J Mammal. 2000;8:52–58. [Google Scholar]
- Amemiya C.T., Gold J.R. Chromomycin A3 stains nucleolus organizer regions of fish chromosomes. Copeia. 1986;1986:226–231. [Google Scholar]
- Andrades-Miranda J., Oliveira L.F.B., Lima-Rosa C.A.V., Nunes A.P., Zanchin N.I.T., Mattevi M.S. Chromosomes studies in seven species of the genus Oligoryzomys (Rodentia, Sigmodontinae) from Brazil. J Mammal. 2001;82:1080–1091. [Google Scholar]
- Andrades-Miranda J., Oliveira L.F.B., Lima-Rosa C.A.V., Sana D.A., Nunes A.P., Mattevi M.S. Genetic studies in representatives of genus Rhipidomys (Rodentia, Sigmodontinae) from Brazil. Acta Theriol. 2002;47:125–135. [Google Scholar]
- Artoni R.F., Molina W.F., Bertollo L.A.C., Galetti P.M., Jr Heterochromatin analysis in the fish species Liposarcus anisitsi and Leporinus elongatus (Characiformes) Genet Mol Biol. 1999;22:1–6. [Google Scholar]
- Bernardi G. The vertebrate genome: Isochores and evolution. Mol Biol Evol. 1993;10:186–204. doi: 10.1093/oxfordjournals.molbev.a039994. [DOI] [PubMed] [Google Scholar]
- Bonvicino C.R., Geise L. Taxonomic status of Delomys dorsalis collinus Thomas, 1917 (Rodentia, Cricetidae) and description of a new karyotype. Mamm Biol. 1995;60:124–127. [Google Scholar]
- Bonvicino C.R., Otazu I. The Wilfredomys pictipes (Rodentia, Sigmodontinae) karyotype with comments on the karyosystematics of Brazilian Thomasomyini. Acta Theriol. 1999;44:329–332. [Google Scholar]
- Bonvicino C.R., Oliveira J.A., D'Andrea O.S., Carvalho R.W. The endemic Atlantic Forest rodent Phaenomys ferrugineus (Thomas, 1894): New data on its morphology and karyology. Bol Mus Nac. 2001;467:1–12. [Google Scholar]
- Costa L.P., Pavan S.E., Leite Y.L.R., Fagundes V. A new species of Juliomys (Mammalia, Rodentia, Cricetidae) from the Atlantic forest of southeastern Brazil. Zootaxa. 2007;1463:21–37. [Google Scholar]
- D'Elia G., Luna L., Gonzalez E.M., Patterson B.D. On the Sigmodontinae radiation (Rodentia, Cricetidae): An appraisal of the phylogenetic position of Rhagomys. Mol Phylogenet Evol. 2006;38:558–564. doi: 10.1016/j.ympev.2005.08.011. [DOI] [PubMed] [Google Scholar]
- D'Elia G., Pardiñas U.F.J., Teta P., Patton J.L. Defination and diagnosis of new tribe os Sigmodontinae rodents (Cricetidae, Sigmodontinae), and a revised classification of the subfamily. Gayana. 2007;71:187–194. [Google Scholar]
- Eisenberg J.F., Redford K.H. Mammals of the Neotropics: The Central Neotropics. v. 3. Chicago: University of Chicago Press; 1999. [Google Scholar]
- Emmons L.H., Feer F. Neotropical Rainforest Mammals: A Field Guide. 2nd edition. Chicago: University of Chicago Press; 1997. [Google Scholar]
- Fagundes V., Christoff A.U., Amaro-Ghilard R.C., Scheibler D.R., Yonenaga-Yassuda Y. Multiple interstitial ribosomal sites in the Brazilian squirrel Sciurus aestuans ingrami (Rodentia, Sciuridae) with 2n = 40: An overview of Sciurus cytogenetics. Genet Mol Biol. 2003;26:253–257. [Google Scholar]
- Ford C.E., Hamerton J.L. A colchicine hypotonic citrate squash sequence for mammalian chromosome. Stain Technol. 1956;31:247–251. doi: 10.3109/10520295609113814. [DOI] [PubMed] [Google Scholar]
- Gardner A.L., Patton J.L. Karyotypic variation in oryzomyine rodents with comments on chromosomal evolution in the Neotropical cricetine complex. Occas Pap Mus Zool. 1976;49:1–48. [Google Scholar]
- Gomez-Laverde M., Montenegro-Diaz O., Lopez-Arevalo H., Cadena A., Bueno M.L. Karyology, morphology, and ecology of Thomasomys laniger and T. niveipes in Colombia. J Mammal. 1997;78:1282–1289. [Google Scholar]
- Gonçalves P.R., Almeida F.C., Bonvicino C.R. A new species of Wiedomys (Rodentia, Sigmodontinae) from Brazilian Cerrado. Mamm Biol. 2005;70:46–60. [Google Scholar]
- Hass I., Sbalqueiro I.J., Müller S. Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetidae) from Southern Brazil established by ZOO-FISH using Mus musculus painting probes. Chromosome Res. 2008;16:75–88. doi: 10.1007/s10577-007-1211-5. [DOI] [PubMed] [Google Scholar]
- Holmquist G.P., Ashley T. Chromosome organization and chromatin modification: Influence on genome function and evolution. Cytogenet Genome Res. 2006;114:96–125. doi: 10.1159/000093326. [DOI] [PubMed] [Google Scholar]
- Howell W.M., Black D.A. Controlled silver staining of nucleolus organizer region with protective colloidal developer: A 1-step method. Experientia. 1980;36:1014–1015. doi: 10.1007/BF01953855. [DOI] [PubMed] [Google Scholar]
- Kierulff M.C., Stalling J.R., Sabato E.L. A method to capture the bamboo rat (Kannabateomys amblyonyx) in bamboo forests. Mammalia. 1991;55:633–635. [Google Scholar]
- Leite-Silva C., Santos N., Fagundes V., Yonenaga-Yassuda Y., de Souza M.J. Karyotypic characterization of the bat species Molossus ater, M. molossus and Molossops planirostris (Chiroptera, Molossidae) using FISH and banding techniques. Hereditas. 2003;138:94–100. doi: 10.1034/j.1601-5223.2003.01693.x. [DOI] [PubMed] [Google Scholar]
- Luna L., Patterson B. A remarkable new mouse (Muridae, Sigmodontinae) from southeastern Peru: With comments on the affinities of Rhagomys rufescens (Thomas, 1886) Fieldiana Zool. 2003;101:1–24. [Google Scholar]
- Maia V., Langguth A. New karyotypes of Brazilian Akodont rodents with notes on taxonomy. Mamm Biol. 1981;46:241–249. [Google Scholar]
- McKenna M.C., Bell S.K. Classification of Mammals Above the Species Level. New York: Columbia University Press; 1997. [Google Scholar]
- Moojen J. Os Roedores do Brasil. Rio de Janeiro: Instituto Nacional do Livro e Biblioteca Científica Brasileira; 1952. [Google Scholar]
- Musser G.G., Carleton M.D. Superfamily Muroidea. In: Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd edition. Baltimore: The Johns Hopkins University Press; 2005. pp. 894–1531. [Google Scholar]
- Nowak R.M. Walker's Mammals of the World. 6th edition. v. 2. Baltimore: Johns Hopkins University Press; 1999. [Google Scholar]
- Ochoa G.L., Aguilera M., Pacheco V., Soriano O.J. A new species of Aepeomys Thomas, 1898 (Rodentia, Muridae) from the Andes of Venezuela. Mamm Biol. 2001;66:228–237. [Google Scholar]
- Ojeda A.A., Ríos C.A., Gallardo M.H. Chromosomal characterization of Irenomys tarsalis (Rodentia, Cricetidae, Sigmodontinae) Mastozool Neotrop. 2004;11:95–98. [Google Scholar]
- Oliveira J.A., Bonvicino C.R. A new species of sigmodontine rodent from the Atlantic forest of eastern Brazil. Acta Theriol. 2002;47:307–322. [Google Scholar]
- Paresque R., Christoff A.U., Fagundes V. Karyology of the Atlantic forest rodent Juliomys: A new karyotype from southern Brazil. Genet Mol Biol. 2009;32:301–305. doi: 10.1590/S1415-47572009005000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathack S., Stock A.D. The X chromosome of mammals: Karyological homology as revealed by banding techniques. Genetics. 1974;78:703–714. doi: 10.1093/genetics/78.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percequillo A.R., Gonçalves P.R., Oliveira J.A. The rediscovery of Rhagomys rufescens (Thomas, 1886), with a morphological rediscription and comments on its systematic relationships based on morphological and molecular characters. Mamm Biol. 2004;69:238–257. [Google Scholar]
- Pereira L.G., Geise L., Cunha A.A., Cerqueira R. Abrawayaomys ruschii Cunha & Cruz, 1979 (Rodentia, Cricetidae) no Estado do Rio de Janeiro, Brasil. Pap Avulsos Zool. 2008;48:33–40. (Abstract in English) [Google Scholar]
- Pinheiro P.S., Hartmann P.A., Geise L. New records of Rhagomys rufescens (Thomas 1886) (Rodentia, Muridae, Sigmodontinae) in Atlantic forest of southeastern Brazil. Zootaxa. 2004;431:1–11. [Google Scholar]
- Reig O.A. A new fossil genus of South American cricetid rodents allied to Wiedomys, with an assessment of the Sigmodontinae. J Zool. 1980;192:257–281. [Google Scholar]
- Reig O.A. Distribuição geográfica e história evolutiva dos roedores muroideos Sulamericanos (Cricetidae, Sigmodontinae) Braz J Genet. 1984;8:333–365. [Google Scholar]
- Salazar-Bravo J., Yates T.L. A new species of Thomasomys (Cricetidae, Sigmodontinae) from central Bolivia. In: The Quintessential Naturalist: Honoring the Life and Legacy of Oliver P. Pearson. Berkeley: University of California Publications in Zoology; 2007. pp. 747–774. [Google Scholar]
- Santos N., Fagundes V., Yonenaga-Yassuda Y., Souza M.J. Comparative karyology of Brazilian vampire bats Desmodus rotundus and Diphylla ecaudata (Phyllostomidae, Chiroptera): Banding patterns, base-specific fluorochromes and FISH of ribosomal genes. Hereditas. 2001;134:189–194. doi: 10.1111/j.1601-5223.2001.00189.x. [DOI] [PubMed] [Google Scholar]
- Sbalqueiro I.J., Mattevi M.S., Oliveira L.F.B., Solano M.J.V. B chromosome system in populations of Oryzomys flavescens (Rodentia, Cricetidae) from southern Brazil. Acta Theriol. 1991;36:193–199. [Google Scholar]
- Sbalqueiro I.J., Nascimento A.P. Occurrence of Akodon cursor (Rodentia, Cricetidae) whith 14, 15 and 16 chromosome cytotypes in the same geographic area in southern Brazil. Braz J Genet. 1996;19:565–569. [Google Scholar]
- Schmid M. Chromosome banding in Amphibia. VII. Analysis of the structure and variability of NORs in Anura. Chromosoma. 1982;87:327–444. [Google Scholar]
- Schweizer D. Reverse fluorescent chromosome banding with chromomycin an DAPI. Chromosoma. 1976;58:307–324. doi: 10.1007/BF00292840. [DOI] [PubMed] [Google Scholar]
- Seabright M. A rapid technique for human chromosomes. Lancet. 1971;2:971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Silva M.J.J., Yonenaga-Yassuda Y. Autossomal and sex chromossomal polymorphisms with multiple rearrangements and a new karyotype in the genus Rhipidomys (Sigmodontinae, Rodentia) Hereditas. 1999;131:211–220. doi: 10.1111/j.1601-5223.1999.00211.x. [DOI] [PubMed] [Google Scholar]
- Smith M.F., Patton J.L. Phylogenetic relationships and the radiation of sigmodontine rodents in South America: Evidence from cytochrome b. J Mammal. 1999;6:89–128. [Google Scholar]
- Spotorno A.E., Walker L.I., Flores S.V., Yevenes M., Marín J.C., Zuleta C. Evolución de los filotinos (Rodentia, Muridae) en los Andes del Sur. Rev Chil Hist Nat. 2001;74:151–166. [Google Scholar]
- Steiner-Souza F., Cordeiro-Estrela P., Percequillo A.R., Testoni A.F., Althoff S.L. New records of Rhagomys rufescens (Rodentia, Sigmodontinae) in the Atlantic forest of Brazil. Zootaxa. 2008;1824:28–34. [Google Scholar]
- Steppan S.J. Revision of the tribe Phyllotini (Rodentia, Sigmodontinae), with a phylogenetic hypothesis for the Sigmodontinae. Fieldiana Zool. 1995;80:1–112. [Google Scholar]
- Sumner A.T. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Svartman M., Almeida E.J.C. Pericentric inversion and X chromosome polymorphism in Rhipidomys sp. (Cricetidae, Rodentia) from Brazil. Caryologia. 1993;46:219–225. [Google Scholar]
- Thomas O. On small mammals from the Delta of the Parana. Ann Mag Nat Hist. 1917;8:96–97. [Google Scholar]
- Villalpando G., Vargas J., Salazar-Bravo J. First record of Rhagomys (Mammalia, Sigmodontinae) in Bolivia. Mastozool Neotrop. 2006;13:43–149. [Google Scholar]
- Voss R.S. A revision of the Brazilian muroid rodent genus Delomys with remarks on “Thomasomyine” characters. Am Mus Novit. 1993;3073:1–44. [Google Scholar]
- Zanchin N.I.T., Langguth A., Mattevi M.S. Karyotypes of Brazilian species of Rhipidomys (Rodentia, Cricetidae) J Mammal. 1992a;73:120–122. [Google Scholar]
- Zanchin N.I.T., Sbalqueiro I.J., Langguth A., Bossle R.C., Castro E.C., Oliveira L.F.B., Mattevi M.S. Karyotype and species diversity of genus Delomys (Rodentia, Cricetidae) in Brazil. Acta Theriol. 1992b;37:163–169. [Google Scholar]
Internet Resources
- Metzger J.P., Alves L.F., Pardini R., Dixo M., Nogueira A.A., Negrao M.F.F., Montesen A.C., Catharino E.L.M. Caracteristicas ecologicas e implicacoes para a conservacao da Reserva Florestal do Morro Grande. [February 24, 2010];Biota Neotrop. 2006 6 Available from: http://www.biotaneotropica.org.br/v6n2/pt/abstract?article+bn01006022006. [Google Scholar]
- Pacheco V.R. Phylogenetic analyses of the Thomasomyini (Muroidea, Sigmodontinae) based on morphological data [PhD Thesis] New York.: The City University of New York; 2003. [February 24, 2010]. at Available from: https://sites.google.com/site/departamentodemastozoologia/publicaciones. [Google Scholar]
- Pardini R., Umetsu F. Pequenos mamiferos nao-voadores da Reserva Florestal do Morro Grande - Distribuicao das especies e da diversidade em uma area de Mata Atlantica. [February 24, 2010];Biota Neotrop. 2006 6 Available from: http://www.biotaneotropica.org.br/v6n2/pt/abstract?article+bn01006022006. [Google Scholar]