Abstract
The use of PCR assays with degenerate primers has suggested the existence of numerous as yet uncharacterized bovine papillomaviruses (BPV). Despite the endemic nature of BPV infections, the identification of BPV types in Brazilian cattle is still only sporadic. However, in a recent analysis of a partial segment of the L1 gene, we observed notable diversity among the BPV types detected. The aim of this study was to determine the phylogenetic position of the previously identified wild strain BPV/BR-UEL2 detected in the state of Paraná in Brazil. Since previous analysis of the partial L1 sequence had shown that this strain was most closely related to BPV type 4, genus-specific primers were designed. Phylogenetic analysis using complete L1 ORF sequences revealed that BPV/BR-UEL2 was related to BPV types classified in the genus Xipapillomavirus and shared the highest L1 nucleotide sequence similarity with BPV type 4 (78%). This finding suggests that BPV/BR-UEL2 should be classified as a potential new type of BPV in the genus Xipapillomavirus.
Keywords: BPV, cattle, cutaneous papillomatosis, putative new BPV type, L1 gene
Papillomaviruses (PVs) are a highly diverse group of circular double-stranded DNA viruses that can induce epithelial proliferation in a wide range of vertebrate species. In cattle, the bovine papillomavirus (BPV) has been implicated as the casual agent of cutaneous papillomatosis and cancer of the urinary bladder and upper gastrointestinal tract (Campo, 2002).
Recently, based on a comparison of the entire L1 nucleotide sequence of almost all known PVs, the family Papillomaviridae was found to consist of 18 genera (Alphapapillomavirus to Sigmapapillomavirus), each containing a number of species. The different genera show less than 60% identity in the L1 nucleotide sequence, whereas species within a genus share 60%-70% identity. In addition, traditional types within a species show 71%-89% identity in the same gene (De Villiers et al., 2004).
While more than 100 human papillomavirus (HPV) types have been identified, only six BPV types had been described in cattle before 2007. These BPVs were classified in the genera Deltapapillomavirus (BPV-1 and -2), Xipapillomavirus (BPV-3, -4, and -6) and Epsilonpapillomavirus (BPV-5) (De Villiers et al., 2004). In addition, the most recently characterized BPV types were placed in Epsilonpapillomavirus (BPV-8) and Xipapillomavirus (BPV-9 and -10), with the exception of BPV-7, which belongs to an as yet undesignated PV genus (Ogawa et al., 2007; Tomita et al., 2007; Hatama et al., 2008).
As observed for HPV, PCR assays using degenerate primers that amplify partial fragments of the L1 gene, followed by sequencing, have demonstrated the presence of numerous BPV types in cattle herds from diverse geographical regions. Using the primers FAP59/FAP64 and MY09/MY11, 12 putative new BPV types were detected in teat skin warts and healthy teat skin of cattle from Japan and Sweden (Forslund et al., 1999; Antonsson and Hansson, 2002; Ogawa et al., 2004).
A recent investigation using the strategy above revealed notable diversity among BPV types detected in papillomas of four cattle herds from the state of Paraná in southern Brazil. The study also identified four putative new BPV types designated as BPV/BR-UEL2 to BPV/BR-UEL5 (GenBank accession numbers EU293538 to EU293541, respectively) (Claus et al., 2008). The aim of the current study was to determine the phylogenetic position of BPV/BR-UEL2 in relation to other BPVs.
BPV/BR-UEL2 DNA was isolated from a rice-grain-sized papilloma located in the axillary region of a cow from a dairy cattle herd in Paraná (Claus et al., 2008). The papilloma was homogenized in phosphate-buffered saline (PBS, pH 7.2) and the homogenate (10% w/v) then centrifuged (1500 x g, 15 min, 4 °C). An aliquot (250 μL) of the supernatant was treated with lysis buffer [10 mM Tris, 1 mM EDTA, 0.5% Nonidet P40, 1% SDS and 0.2 mg/mL proteinase K (Invitrogen, Life Technologies, USA)], mixed and incubated at 56 °C for 30 min.
DNA was extracted using a combination of the phenol/chloroform/isoamyl alcohol and silica/guanidine isothiocyanate methods (Alfieri et al., 2006). The DNA was eluted in 50 μL of ultrapure (MilliQ®) sterile water and stored at -20 °C until used. An aliquot of ultrapure sterile water was included as a negative control in the DNA extraction procedure.
Since FAP sequence analysis of the wild type BPV/BR-UEL2 (GenBank accession number EU293538) has shown that this isolate is most closely (77%) related to BPV type 4, sequences from the L2, L1 and LCR regions of Xipapillomavirus representatives (BPV-3, -4 and -6) were aligned and used to design degenerate primers to obtain the entire L1 nucleotide sequence.
Combinations of the two primer sets (L2Bf/L1Br and L1Bf/LCRBr) and the previously described FAP primer pair were tested (see Table 1 for primer features) (Forslund et al., 1999). In addition, the same primer sets were also tested on DNA samples known to harbor BPV type 6, a Xipapillomavirus representative. Sequence alignment and primer design were done using the CLUSTAL W Multiple Alignment program and Gene Runner v 3.05 (Hastings Software Inc., Hastings, NY), respectively (Thompson et al., 1994).
Table 1.
Sequences and features of primers used in the polymerase chain reactions.
| Primer | Genomic region targeted | Polarity | Sequence1 | Nucleotide positions2 | Degree of degeneracy |
| L2Bf | L2 | + | 5'GTIAARYTITTYATHAAYGAYGC3' | 5385-5407 | 96 |
| FAP593 | L1 | + | 5'TAACWGTIGGICAYCCWTATT3' | 5729-5749 | 8 |
| L1Br | L1 | - | 5'AASACTCTGAATTGACTGCC3' | 5794-5813 | 2 |
| L1Bf | L1 | + | 5'GRGAGCAYTGGGAYAAAG3' | 6089-6106 | 8 |
| FAP643 | L1 | - | 5'CCWATATCWVHCATITCICCATC3' | 6175-6197 | 36 |
| LCRBr | LCR | - | 5'CWRCATTTTATTKSSAASATTC3' | 7181-7202 | 64 |
1Degenerate nucleotides: I = inosine; R = G, A; Y = T, C; H = A, T, C; W = A, T; S = G, C; V = G, A, C; K = G, T.
2Relative position in the BPV-4 genome.
The PCR reactions contained 2.5 μL of extracted DNA, 20 pmol of each primer, 200 μM of each dNTP, 2.5 U of Platinum Taq DNA polymerase (Invitrogen), 1 x PCR buffer (20 mM Tris-HCl, pH 8.4 and 50 mM KCl), 1.5 mM MgCl2 and ultrapure sterile water in a final volume of 25 μL. The reactions were run in a PTC-200 thermocycler (MJ Research Co., USA) using the following times and temperatures: 10 min at 94 °C followed by 40 cycles of 1 min at 94 °C, 1 min at an optimum temperature for primer annealing, 1 min at 72 °C, and a final extension of 10 min at 72 °C. The annealing temperatures for primer pairs L2Bf/FAP64, L2Bf/L1Br and L1Bf/LCRBr were 50 °C, 54 °C and 57 °C, respectively. The amplified products were analyzed by electrophoresis in 1.5% agarose ethidium bromide stained gels in TBE buffer, pH 8.4 and examined under UV light.
Initially, all PCR products were purified using a PureLink quick gel extraction kit (Invitrogen) and then cloned using TOPO TA cloning kit for sequencing (Invitrogen), according to the manufacturer's instructions. The inserts from two clones selected for each PCR amplicon were then sequenced using DYEnamic ET dye terminator cycle sequencing kit (GE Healthcare, Little Chalfont, UK) with M13 forward and reverse primers in a MegaBACE 1000/Automated 96 Capillary DNA sequencer (GE Healthcare), according to the manufacturer's instructions. The sequences obtained were examined with the PHRED application for quality analysis of chromatogram readings. The sequences were accepted if base quality was ≥ 20. The consensus sequence was determined using CAP3 software and the sequence identity was verified against all sequences deposited in GenBank using the BLAST application. The L1 ORF of Brazilian wild type BPV was predicted based on analysis with the ORF Finder tool. The alignment and degree of similarity among sequences at the nucleotide and amino acid levels were determined using BIOEDIT v 5.0.9 software (Hall, 1999). The phylogenetic tree was constructed using MEGA v. 3.1 software (Kumar et al., 2004) and the neighbor-joining method with the Kimura two-parameter distance estimate (Kimura, 1980). Bootstrap support values were determined for 1000 replications.
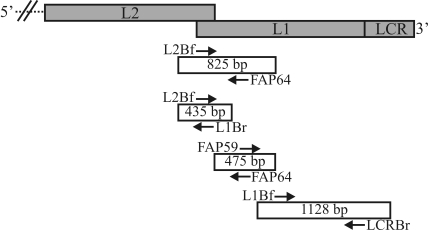
The first L1 segment of the Brazilian wild strain BPV was obtained using a semi-nested PCR assay (SN-PCR) with the primer pair L2Bf/FAP64 in the first round and the primer pair L2Bf/L1Br in the second round, which yielded an amplicon of 435 bp. Use of the FAP59/FAP64 (475 bp) and L1Bf/LCRBr (1128 bp) primer sets allowed amplification of the remaining portions of the same gene (Figure 1).
Figure 1.
Schematic diagram showing the relative positions of overlapping PCR fragments in the L2, L1, and LCR regions of the BPV/BR-UEL2 isolate. The length of each amplicon is indicated in the white box while the primer sets are shown as arrows.
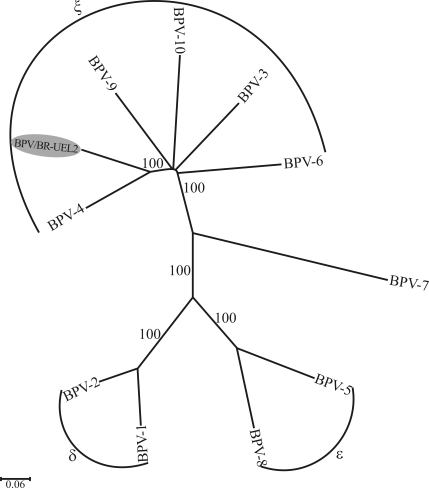
A consensus sequence of 1804 nt (GenBank accession number GQ471901) spanning nt 5385 to nt 7184 of BPV-4 was obtained with the L2Bf/L1Br, FAP59/FAP64 and L1Bf/LCRBr overlapping amplicons of the BPV/BR-UEL2 isolate. Phylogenetic analysis using complete L1 ORF sequences revealed that the BPV/BR-UEL2 isolate was related to BPV types in the genus Xipapillomavirus (Figure 2). In addition, pairwise DNA sequence alignments showed that this isolate shared the highest L1 nucleotide sequence similarity with BPV type 4 (78%), which again suggested its classification in the genus Xipapillomavirus; at the amino acid level, the identity with BPV-4 was 83.6%. Finally, DNA samples known to contain BPV-6 were amplified and the three overlapping amplicons (L2Bf/FAP64, FAP59/FAP64 and L1Bf/LCRBr) were obtained in all of the samples tested.
Figure 2.
Neighbor-joining phylogenetic tree of L1 ORFs (nt) of BPVs, including that of the BPV/BR-UEL2 isolate (indicated by shading). The tree is divided into the previously determined genera Deltapapillomavirus (BPV-1 and -2), Epsilonpapillomavirus (BPV-5 and -8), Xipapillomavirus (BPV-3, -4, -6, -9 and -10) and an undesignated PV genus (BPV-7). The numbers at the internal nodes represent the bootstrap support values determined for 1000 replications.
The L1 nucleotide sequence of our isolate showed 71%-78% identity with Xipapillomavirus representatives whereas with representatives of other genera the similarity was 55.6-55.9%. A phylogenetic analysis based on amino acid sequence alignments yielded a tree with a topology very similar to that based on nucleic acid sequence alignments (data not shown).
Compared with other BPV genera, Xipapillomavirus contains a great diversity of BPV types, as shown by studies in which several putative new BPV types have been identified, e.g., BAA-1, BAPV-3, -8, -9 and -10 and BPV/BR-UEL3 (Antonsson and Hansson, 2002; Ogawa et al., 2004; Claus et al., 2008).
BPV type 4 has traditionally been associated with cattle alimentary tract tumors (Campo et al., 1994; Borzacchiello et al., 2003). However, our Brazilian BPV isolate was detected in a skin wart located in the axillary region of a dairy cow and was thus associated with cutaneous papillomatosis.
Despite the similarity with BPV-4, the L1 ORF-encoded protein of this Brazilian isolate consisted of 532 amino acids, similar to the L1 protein of BPV-9, a newly described Xipapillomavirus type, which contains 531 amino acids. In contrast, the BPV-4 L1 ORF codes for a 506 amino acid protein. ORF analysis of the consensus sequence revealed an overlap between L2 and L1, also seen in BPV-9 and -10, which differed from that observed in BPV-4.
In contrast to our putative novel BPV type, the upstream fragment of the L1 gene of BPV-6 could be obtained by PCR using the L2Bf/FAP64 primer set whereas the subsequent L1 segments were obtained using the same primer pairs as those used for the BPV/BR-UEL2 isolate (data not shown). The specificity of the amplified products was confirmed for one of these samples by analyzing the resulting consensus sequence.
The strategy developed in this study involved the use of three pairs of degenerate primers, two of them specifically designed to amplify the L1 gene of certain Xipapillomavirus representatives. This approach was very useful since it allowed us to easily and unambiguously obtain the entire L1 gene sequence and determine the correct phylogenetic position of an uncharacterized BPV type.
Despite the relatively common occurrence of BPV infections in Brazil, the identification of BPV types in cattle herds is still sporadic. In addition, the few studies done have usually involved the use of type-specific primers and have determined the presence of BPV types 1 and 2 (Dos Santos et al., 1998; De Freitas et al., 2003; Wosiacki et al., 2005, 2006). Recently, use of the FAP59/FAP64 primer pair allowed the identification of previously described BPV types (BPV-1, -2, -6 and -8) and four putative new BPV types in skin warts of cattle from Paraná (Claus et al., 2007, 2008, 2009a,b,c).
In Brazil, cattle diseases caused by BPV, such as chronic enzootic hematuria, cancer of the upper gastrointestinal tract and teat papillomatosis, have prevented the economic exploitation of livestock in specific geographic areas. However, few studies have sought to characterize these pathogens and those aiming at developing means of controlling and treating these diseases have been neglected in Brazil. The importance of the molecular characterization of wild type BPVs isolated from Brazilian cattle rests with the fact that, as in humans, the identification of viral types associated with particular clinical manifestations and the definition of the most prevalent types could spur the development of prophylactic and therapeutic vaccines as means of controlling diseases in the field.
Acknowledgments
This work was supported by the Brazilian funding agencies CAPES, CNPq, FINEP and Fundação Araucária (FAP/PR). A.F.A., A.A.A. and M.H.P.F. are supported by research fellowships from CNPq.
Footnotes
Associate Editor: Sérgio Olavo Pinto da Costa
References
- Alfieri A.A., Parazzi M.E., Takiuchi E., Médici K.C., Alfieri A.F. Frequency of group A rotavirus in diarrhoeic calves in Brazilian cattle herds, 1998-2002. Trop Anim Health Prod. 2006;38:521–526. doi: 10.1007/s11250-006-4349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A., Hansson B.G. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J Virol. 2002;76:12537–12542. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borzacchiello G., Ambrosio V., Roperto S., Poggiali F., Tsirimonakis E., Venuti A., Campo M.S., Roperto F. Bovine papillomavirus type 4 in oesophageal papillomas of cattle from the south of Italy. J Comp Pathol. 2003;128:203–206. doi: 10.1053/jcpa.2002.0626. [DOI] [PubMed] [Google Scholar]
- Campo M.S. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- Campo M.S., O'Neil B.W., Barron R.J., Jarrett W.F. Experimental reproduction of the papilloma-carcinoma complex of the alimentary canal in cattle. Carcinogenesis. 1994;15:1597–1601. doi: 10.1093/carcin/15.8.1597. [DOI] [PubMed] [Google Scholar]
- Claus M.P., Vivian D., Lunardi M., Alfieri A.F., Alfieri A.A. Phylogenetic analysis of bovine papillomavirus associated with skin warts in cattle herds from the state of Paraná. Pesq Vet Bras. 2007;27:314–318. [Google Scholar]
- Claus M.P., Lunardi M., Alfieri A.F., Ferracin L.M., Fungaro M.H.P., Alfieri A.A. Identification of unreported putative new bovine papillomavirus types in Brazilian cattle herds. Vet Microbiol. 2008;132:396–401. doi: 10.1016/j.vetmic.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Claus M.P., Lunardi M., Alfieri A.F., Sartori D., Fungaro M.H.P., Alfieri A.A. Identification of the recently described new type of bovine papillomavirus (BPV-8) in a Brazilian beef cattle herd. Pesq Vet Bras. 2009a;29:25–28. [Google Scholar]
- Claus M.P., Lunardi M., Alfieri A.A., Otonel R.A.A., Ferracin L.M., Fungaro M.H.P., Alfieri A.F. A bovine teat papilloma specimen harboring Deltapapillomavirus (BPV-1) and Xipapillomavirus (BPV-6) representatives. Braz Arch Biol Technol. 2009b;52:87–91. [Google Scholar]
- Claus M.P., Lunardi M., Alfieri A.A., Otonel R.A.A., Sartori D., Fungaro M.H.P., Alfieri A.F. Multiple bovine papillomavirus infections associated with cutaneous papillomatosis in Brazilian cattle herds. Braz Arch Biol Technol. 2009c;52:93–98. [Google Scholar]
- De Freitas A.C., De Carvalho C., Brunner O., Birgel-Junior E.H., Dellalibera A.M.M.P., Benesi F.J., Gregory L., Beçak W., Dos Santos R.C.S. Viral DNA sequences in peripheral blood and vertical transmission of the virus: A discussion about BPV-1. Braz J Microbiol. 2003;34:76–78. [Google Scholar]
- De Villiers E.M., Fauquet C., Broker T.R., Bernard H.U., Zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dos Santos R.C.S., Lindsey C.J., Ferraz O.P., Pinto J.R., Mirandola R.S., Benesi F.J., Birgel E.H., Pereira C.A., Beçak W. Bovine papillomavirus transmission and chromosomal aberrations: An experimental model. J Gen Virol. 1998;79:2127–2135. doi: 10.1099/0022-1317-79-9-2127. [DOI] [PubMed] [Google Scholar]
- Forslund O., Antonsson A., Nordin P., Stenquist B., Hansson B.G. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80:2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hatama S., Nobumoto K., Kanno T. Genomic and phylogenetic analysis of two novel bovine papillomaviruses, BPV-9 and BPV-10. J Gen Virol. 2008;89:158–163. doi: 10.1099/vir.0.83334-0. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Tomita Y., Okada M., Shinozaki K., Kubonoya H., Kaiho I., Shirasawa H. Broad-spectrum detection of papillomaviruses in bovine teat papillomas and healthy teat skin. J Gen Virol. 2004;85:2191–2197. doi: 10.1099/vir.0.80086-0. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Tomita Y., Okada M., Shirasawa H. Complete genome and phylogenetic position of bovine papillomavirus type 7. J Gen Virol. 2007;88:1934–1938. doi: 10.1099/vir.0.82794-0. [DOI] [PubMed] [Google Scholar]
- Thompson J., Higgins D., Gibson T. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y., Literak I., Ogawa T., Jin Z., Shirasawa H. Complete genomes and phylogenetic positions of bovine papillomavirus type 8 and a variant type from a European bison. Virus Genes. 2007;35:243–249. doi: 10.1007/s11262-006-0055-y. [DOI] [PubMed] [Google Scholar]
- Wosiacki S.R., Barreiro M.A.B., Alfieri A.F., Alfieri A.A. Semi-nested PCR for detection and typing of bovine papillomavirus type 2 in urinary bladder and whole blood from cattle with enzootic haematuria. J Virol Meth. 2005;126:215–219. doi: 10.1016/j.jviromet.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Wosiacki S.R., Claus M.P., Alfieri A.F., Alfieri A.A. Bovine papillomavirus type 2 detection in the urinary bladder of cattle with chronic enzootic haematuria. Mem Inst Oswaldo Cruz. 2006;101:635–638. doi: 10.1590/s0074-02762006000600009. [DOI] [PubMed] [Google Scholar]
Internet Resources
- PHRED. [November 5, 2009]. Available from: http://bioinformatica.ucb.br/electro.html.
- Basic Local Alignment Search Tool (BLAST) [November 5, 2009]. Available from: http://www.ncbi.nlm.nih.gov/BLAST.
- ORF Finder Tool. [November 5, 2009]. Available from: http://www.ncbi.nlm.nih.gov/gorf/gorf.html.