Abstract
Several studies have shown that mutations and polymorphisms in clock genes are associated with abnormal circadian parameters in humans and also with more subtle non-pathological phenotypes like chronotypes. However, there have been conflicting results, and none of these studies analyzed the combined effects of more than one clock gene. Up to date, association studies in humans have focused on the analysis of only one clock gene per study. Since these genes encode proteins that physically interact with each other, combinations of polymorphisms in different clock genes could have a synergistic or an inhibitory effect upon circadian phenotypes. In the present study, we analyzed the combined effects of four polymorphisms in four clock genes (Per2, Per3, Clock and Bmal1) in people with extreme diurnal preferences (morning or evening). We found that a specific combination of polymorphisms in these genes is more frequent in people who have a morning preference for activity and there is a different combination in individuals with an evening preference for activity. Taken together, these results show that it is possible to detect clock gene interactions associated with human circadian phenotypes and bring an innovative idea of building a clock gene variation map that may be applied to human circadian biology.
Keywords: clock genes, gene interaction, morningness-eveningness, sleep, circadian rhythm
Introduction
In the last two decades, there has been significant progress in understanding the molecular basis of mammalian circadian rhythmicity (Ko and Takahashi, 2006, Dardente and Cermakian, 2007). Mutations in clock genes are associated with abnormal circadian parameters, including sleep in animals (Ralph and Menaker, 1988; Toh et al., 2001; Iwase et al., 2002; Taheri and Mignot, 2002).
Very similar circadian phenotypes to those observed in animals may be recognized in humans. The first report regarding the influence of a clock gene associated with a circadian rhythm phenotype in humans appeared in 1998. In that study, Katzenberg et al. (1998) reported that a polymorphism in the Clock gene was associated with diurnal preferences. This result was replicated in a Japanese sample (Mishima et al., 2005), but not in a British population sample or in a Brazilian sample (Robiliard et al., 2002; Pedrazzoli et al., 2007).
Polymorphisms in other clock genes were also reported to be associated with circadian phenotypes. Ebisawa et al. (2001) found that polymorphisms in the Per3 gene could be associated with Delayed Sleep Phase Syndrome (DSPS). Archer et al. (2003) reduced the region of influence of the Per3 gene in DSPS to a variable number of tandem repeats (VNTR) (four or five repeats) localized in exon 18, and reported that the shorter allele was associated with DSPS. Pereira et al. (2005) also reported that this VNTR in the Per3 gene was associated with DSPS, but curiously they found that the longer allele was associated with the disease. As the two studies were performed in populations with a Caucasian European genetic background, living at very different latitudes, these conflicting results suggest that latitude can affect the expression of clock genes. Archer et al. (2003) and Pereira et al. (2005) also found that the VNTR polymorphism in the Per3 gene is associated with morning-evening tendencies. Viola et al. (2006) have studied the effects of the Per3 VNTR polymorphism in regulation of diurnal preferences more extensively, under laboratory controlled conditions, and have found that it has predictive value in the response to sleep loss.
Polymorphisms in the Per1 and Per2 genes have also already been associated with morning-evening tendencies in a British population sample (Carpen et al., 2005; Carpen et al., 2006). Additionally, a mutated Per2 allele was shown to be associated with a familial case of Advanced Sleep Phase Syndrome (ASPS) (Toh et al., 2001), but these data were not replicated in a family with ASPS in Japan (Satoh et al., 2003).
As shown above, these previous studies, that have been carried out in several populations around the world, show that polymorphisms in clock genes are independently associated with circadian phenotypes. Nevertheless, they have produced conflicting results, and none of these studies have analyzed the combined effects of two or more genes. The knowledge accumulated up to date shows that the proteins coded by different clock genes interact physically with each other and act as are transcription factors; therefore, it is possible to speculate that combinations of polymorphisms in these genes may influence phenotype, and that a combined analysis of the effects of different clock genes may be more accurate and more informative than single gene analyses.
In the present study, we analyzed the possibility of detecting interactions between polymorphisms in different clock genes associated with circadian phenotypes in humans.
Materials and Methods
Subjects
Volunteers
From an initial screening of 1500 volunteers, a total of 98 volunteers of extreme types were selected to participate based on the Horne-Östberg (HO) questionnaire score (1976). A group composed of extreme morning subjects (n = 47) with a mean HO score of 68.1 ± 4.5, 82% Caucasian, 71.3% female, mean age 21.9 ± 2.7 was compared with a group of extreme evening subjects (n = 51), mean HO score of 28.8 ± 5.6, 80% Caucasian, 71.6% female, mean age 22.9 ± 3.3. The study was approved by the Committee on Ethics of the Universidade Federal de São Paulo (# 0790/02, # 0617/02, and # 1453/06). Volunteers were undergraduate students living in their own places, and all were synchronized to have activities during the day and nocturnal rest, attending morning (8:00-12:00) and/or afternoon (14:00-18:00) classes from Monday to Friday. The participants signed an informed consent to participate.
Genotyping clock genes polymorphisms
Blood samples were taken from all participants and DNA was extracted from white blood cells (Miller et al., 1988). Four polymorphisms of four clock genes, Clock, Per3, Per2 and Bmal1, were typed as follows:
For the Clock gene polymorphism T3111C (AF11568), PCR-RFLP was done as described by Pedrazzoli et al. (2007). For the Per3 gene VNTR polymorphism (four or five repeats), PCR was done as described by Ebisawa et al. (2001). For the Per2 C311G polymorphism (rs2304672) an assay-on-demand (assay ID:C2129919_1; Applied Biosystems, Foster City, CA, USA) was used as described by Carpen et al. (2005), and finally we used the DHPLC system using a semi-denaturing temperature of 59.7 °C (Wave system, Transgenomic, Inc. USA) for the Bmal1 promoter region A-1420G polymorphism (rs4757138), which is the only polymorphism in this study not previously reported to be associated with any phenotype. The PCR fragment for DHPLC analysis was obtained using the primers F 5'ggcagagacaaaggagcaat 3' and R 5'cagcgtcttctttttcg 3', designed using the TFSEARCH program ver.1.3 and included the RORα transcription factor binding site.
Statistical Analysis
Initially, all polymorphisms were evaluated with regard to the Hardy-Weinberg equilibrium using ARLEQUIN v. 3.1 software (Excoffier et al., 2005). We designed and implemented an algorithm in PERL script (combination.pl) to generate all possible combinations among the polymorphisms, and also calculated the frequencies of these combinations within the two experimental groups.
In order to test whether differences in combination frequencies between groups were statistically significant, we performed a log ratio test (LRT) using EXACT TEST v. 1.0.0.2 software (EDNData). To avoid bias due to previous gene associations within combinations, we separately performed Fisher's exact test for each gene using the fisher.test function in the R environment software (R Development Core Team 2007). In addition, combinations of two, three and four SNPs were analyzed. Multiple comparisons in all these statistical tests were corrected using a False Discovery Rate (FDR) correction and the p.adjust function in R environment (Storey and Tibshirani, 2003; R Development Core Team 2007). Corrected p-values (pcorr) < 0.05 were considered to be statistically significant (Storey and Tibshirani, 2003).
Results
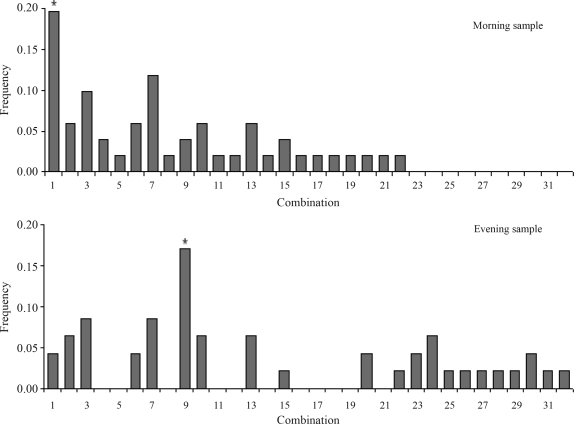
No associations were observed when each polymorphism was considered individually (Clock T311C pcorr = 0.7839; Per3 pcorr = 0.4113; Per2 pcorr = 1.0000; Bmal1 pcorr = 0.4113), or when combinations of two or three polymorphisms were tested. Nevertheless, when analyzing the combination of four polymorphisms it became evident that there were statistically significant differences in the frequency distribution (pcorr = 0.0330) between combination #1 (morning sample) and combination #9 (evening sample) (Figure1). We found 31 different four polymorphism combinations in our sample out of the 81 that are mathematically possible, obviously the combinations involving low frequency alleles are much less likely to be found (Table 1).
Figure 1.
Frequency distribution of the polymorphism combinations in the morning and evening samples, when considering combinations of four polymorphisms. Significant difference between groups, *p < 0.05.
Table 1.
Genotype combinations of polymorphisms in the Per3, Per2, Clock and Bmal1 genes.
| GC # | Per3 | Bmal1 | Per2 | Clock | N | % |
| 1 | 4/5 | G/g | C/C | T/T | 12 | 12.24 |
| 2 | 4/4 | G/g | C/C | C/T | 6 | 6.12 |
| 3 | 4/4 | A/g | C/C | C/T | 10 | 9.8 |
| 4 | 4/4 | G/g | C/C | C/C | 2 | 2.04 |
| 5 | 4/4 | A/A | C/C | T/T | 1 | 1.02 |
| 6 | 4/4 | G/g | C/C | T/T | 5 | 5.10 |
| 7 | 4/4 | A/g | C/C | T/T | 10 | 10.20 |
| 8 | 4/5 | A/A | G/C | C/T | 1 | 1.02 |
| 9 | 4/5 | A/g | C/C | T/T | 10 | 10.20 |
| 10 | 4/5 | A/g | C/C | C/T | 6 | 6.12 |
| 11 | 4/4 | A/g | C/C | C/C | 1 | 1.02 |
| 12 | 4/4 | A/g | G/C | T/T | 1 | 1.02 |
| 13 | 4/5 | G/g | C/C | C/T | 6 | 6.12 |
| 14 | 4/4 | A/A | C/C | C/C | 1 | 1.02 |
| 15 | 4/5 | A/A | C/C | C/T | 3 | 3.06 |
| 16 | 4/5 | A/A | C/C | T/T | 1 | 1.02 |
| 17 | 4/5 | A/g | G/C | C/T | 1 | 1.02 |
| 18 | 5/5 | A/g | G/C | T/T | 1 | 1.02 |
| 19 | 4/4 | G/g | G/g | T/T | 1 | 1.02 |
| 20 | 4/4 | A/g | G/C | C/T | 3 | 3.06 |
| 21 | 4/4 | G/g | G/C | T/T | 1 | 1.02 |
| 22 | 4/4 | A/A | C/C | C/T | 2 | 2.04 |
| 23 | 4/4 | G/g | G/C | C/T | 2 | 2.04 |
| 24 | 5/5 | A/g | C/C | T/T | 3 | 3.06 |
| 25 | 5/5 | A/g | C/C | C/C | 1 | 1.02 |
| 26 | 4/4 | A/A | G/C | T/T | 1 | 1.02 |
| 27 | 4/5 | G/g | G/C | C/T | 1 | 1.02 |
| 28 | 5/5 | G/g | C/C | T/T | 1 | 1.02 |
| 29 | 4/5 | G/g | C/C | C/C | 1 | 1.02 |
| 30 | 5/5 | G/g | C/C | C/T | 2 | 2.04 |
| 31 | 4/5 | G/g | G/C | T/T | 1 | 1.02 |
| Total | 98 | 100.00 |
GC#, Genotype combination number.
Interestingly, combinations #1 #9 only differed in their Bmal1 genotypes, which are homozygous G and heterozygous A/G, respectively. They shared the same genotypes for the other clock genes analyzed, i.e., they were heterozygous for the Per3 VNTR, homozygous C for the Per2 SNP and homozygous T for the Clock SNP.
Discussion
This study was designed to evaluate the possibility of observing the combined effects of polymorphisms in four different clock genes, two in the positive arm (Clock and Bmal1) and the other two in the negative arm (Per3 and Per2) of the main molecular loop of circadian rhythms.
Statistical analysis demonstrated that there could be an interaction among the gene polymorphisms, which would give a tendency to determined chronotypes. This hypothesis is supported by the fact that the only difference between combinations #1 and #9 is the Bmal1 polymorphism, but this polymorphism alone did not provide any evidence of association. In fact, it seems that specific combinations of polymorphisms among different clock genes are stronger markers to chronotypes than single polymorphisms.
The cellular clock function hypothesis proposes that the molecular mechanism responsible for clock function operates as a negative feedback loop in which the clock encoded proteins physically interact with each other, whereby Clock and Bmal1 also act as transcription factors within and outside of the loop (Lowrey et al., 2000). It is therefore reasonable to hypothesize that subtle changes in protein structure, due to non-synonymous polymorphisms, could alter the dimerization rates of proteins, thus causing subtle alterations in the circadian regulation and leading to slightly different phenotypes, as seen with the different chronotypes. In addition, subtle changes in the promoter or regulatory regions could affect rates of gene expression, thus changing the phase relationship between clock components and leading to subtle differential regulation of circadian rhythms.
At this stage of the research it is still difficult to propose a mechanism by which these specific combinations of polymorphisms affect circadian phenotypes. The only genotype that differed between the two relevant combinations was the Bmal1 genotype. This polymorphism is localized in the promoter region that includes a RORα transcription factor binding site, where the REV-ERB protein binds (Preitner et al., 2002) to repress Bmal1 expression. Therefore one can hypothesize that mRNA availability or the transcription rate of the Bmal1 gene may be affected by the polymorphism. This differential expression would affect the amount of BMAL1 protein, consequently changing its rate of dimerization with the CLOCK protein, thus leading to different phases and/or amplitudes of expression to the other clock genes that could affect the regulatory loop timing. In studies on humans, differences in Bmal1 phase of expression have been observed in blood cells (Teboul et al., 2005), and animal studies have shown that Bmal1 gene expression regulates other clock gene expression profiles, such as Per1 and 2 (Bunger et al., 2002).
Viola et al. (2006) demonstrated strong effects of the Per3 VNTR polymorphism in the sleeping and waking EEG structure and in the response to sleep loss. However, they mainly studied homozygotes and did not report upon the heterozygous phenotype for this polymorphism. In the context of the present results, it will be informative to test Per3 VNTR heterozygotes under the same laboratory conditions but in combination with the Bmal1 polymorphisms that we have described here.
The hypothesis of differential interactions between clock genes alleles associated with morningness-eveningness tendencies can be reasonably tested in our experimental set, which consisted of young adults selected for extreme diurnal preferences who were tested for the presence of specific combinations of clock gene polymorphisms. In this simple experimental setting it was possible to detect combined, but not individual polymorphism effects. It may be argued that our sample is small, which would decrease the power of the analyses, but we have chosen young subjects who are extreme morning or evening types, and selecting those individuals with extreme phenotypes for a quantitative trait, such as HO score, from a larger population, increases the power per individual genotyped (Ronin et al.,1998).
In spite of the logical rationale that the products of the clock genes interact to form the molecular clock machinery and that interactions of different alleles could therefore result in altered working of the machinery, very few studies have reported approaches to deal with this problem. In general, in animal studies, circadian behavior and circadian expression patterns of clock genes have been investigated in knockout mice with the absence of one or two components (double mutants) of the clock machinery. Data from these studies have shown that there is no circadian rhythmicity in the absence of more than one component (van der Horst et al., 1999; Vitaterna et al., 1999; Zheng et al., 2001; Oster et al., 2003). These data showed that the interaction between the clock components is necessary for molecular clock function, but failed to show how minor modifications, which still retain the function of the gene, may modify the regulation of circadian rhythms and the entrainment to environmental time cues. Thus, animal model studies (which are still lacking at the moment) that deal with naturally occurring polymorphisms would have a potential to clarify this question of clock gene interactions in regulating circadian behavior.
To our knowledge, this type of interaction has not previously been reported in humans, although several articles have reported associations between clock gene polymorphisms associated with circadian phenotypes related to the sleep/wake cycle (Katzenberg et al., 1988; Ebisawa et al., 2001; Archer et al., 2003, Carpen et al., 2005; Pereira et al., 2005; Pedrazzoli et al., 2007).
The present results demonstrate that interactions are accessible to study and should be expanded to search for haplotype interactions between clock genes, which should be more informative than single SNPs per gene. Taken together, the results show that it is possible to detect interactions between clock genes associated with human circadian phenotypes, thus pointing a way towards the construction of a clock gene polymorphism map applicable to human circadian biology.
Acknowledgments
We thank FAPESP (grant # 98/14303-3), CNPq (grant # 49028/2004-6), CAPES (PAEP grant # 0674/07-9) and the AFIP for financial support.
Footnotes
Associate Editor: Peter Pearson
References
- Albrecht U., Zheng B., Larkin D., Sun Z.S., Lee C.C. mPer1 and mPer2 are essential for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- Archer S.N., Robilliard D.L., Skene D.J., Smits M., Williams A., Arendt J., von Schantz M. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–415. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bunger M.K., Wilsbacher L.D., Moran S.M., Clendenin C., Radcliffe L.A., Hogenesch J.B., Simon M.C., Takahashi J.S., Bradfield C.A. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpen J.D., Archer S.N., Skene D.J., Smits M., von Schantz M. A single-nucleotide polymorphism in the 5'-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:237–293. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- Carpen J.D., von Schantz M., Smits M., Skene D.J., Archer S.N. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet. 2006;51:1122–1125. doi: 10.1007/s10038-006-0060-y. [DOI] [PubMed] [Google Scholar]
- Dardente H., Cermakian N. Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol Int. 2007;24:195–213. doi: 10.1080/07420520701283693. [DOI] [PubMed] [Google Scholar]
- Ebisawa T., Uchiyama M., Kajimura N., Mishima K., Kamei Y., Katoh M., Watanabe T., Sekimoto M., Shibui K., Kim K., et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L.G.L., Schneider S. Arlequin v. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Horne J.A., Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- Iwase T., Kajimura N., Uchiyama M., Ebisawa T., Yoshimura K., Kamei Y., Shibui K., Kim K., Kudo Y., Katoh M., et al. Mutation screening of the human Clock gene in circadian rhythm sleep disorders. Psychiatry Res. 2002;109:121–128. doi: 10.1016/s0165-1781(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Katzenberg D., Young T., Finn L., Lin L., King D.P., Takahashi J.S., Mignot E.A. CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- Ko C.H., Takahashi J.S. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Lowrey P.L., Shimomura K., Antoch M.P., Yamazaki S., Zemenides P.D., Ralph M.R., Menaker M., Takahashi J.S. Positional synthetic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–491. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.S., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima K., Tozawa T., Satoh K., Saitoh H., Mishima Y. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med Genet B Neuropsychiatr Genet. 2005;133:101–104. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- Oster H., van der Horst G.T., Albrecht U. Daily variation of clock output gene activation in behaviorally arrhythmic mPer/mCry triple mutant mice. Chronobiol Int. 2003;20:683–695. doi: 10.1081/cbi-120022408. [DOI] [PubMed] [Google Scholar]
- Pedrazzoli M., Louzada F.M., Pereira D.S., Benedito-Silva A.A., Lopez A.R., Martynhak B.J., Korczak A.L., Koike B.V., Barbosa A.A., et al. Clock polymorphisms and circadian rhythms phenotypes in a sample of the Brazilian population. Chronobiol Int. 2007;24:1–8. doi: 10.1080/07420520601139789. [DOI] [PubMed] [Google Scholar]
- Pereira D.S., Tufik S., Louzada F.M., Benedito-Silva A.A., Lopez A.R., Lemos N.A., Korczak A.L., D'Almeida V., Pedrazzoli M. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: Does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERB alpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Ralph M.R., Menaker M. A mutation of the ircadian system in golden hamsters. Science. 1988;241:1225–1227. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- Robilliard D.L., Archer S.N., Arendt J., Lockley S.W., Hack L.M., English J., Leger D., Smits M.G., Williams A., Skene D.J., et al. The 3111 Clock gene polymorphism is not associated with sleep and circadian rhythmicity in phenotypically characterized human subjects. J Sleep Res. 2002;11:305–312. doi: 10.1046/j.1365-2869.2002.00320.x. [DOI] [PubMed] [Google Scholar]
- Ronin Y.I., Korol A.B., Weller J.I. Selective genotyping to detect quantitative trait loci affecting multiple traits: Interval mapping analysis. Theor Appl Genet. 1998;97:1169–1178. [Google Scholar]
- Satoh K., Mishima K., Inoue Y., Ebisawa T., Shimizu T. Two pedigrees of familial advanced sleep phase syndrome in Japan. Sleep. 2003;26:416–417. doi: 10.1093/sleep/26.4.416. [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Jin X., Lee C., Reppert S.M., Weaver D.R. Targeted disruption of the mPer3 gene: Subtle effects on circadian clock function. Mol Cell Biol. 2000;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significancee for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri S., Mignot E. The genetics of sleep disorders. Lancet Neurol. 2002;1:242–250. doi: 10.1016/s1474-4422(02)00103-5. [DOI] [PubMed] [Google Scholar]
- Teboul M., Barrat-Petit M.A., Li X.M., Claustrat B., Formento J.L., Delaunay F., Lévi F., Milano G. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J Mol Med. 2005;83:693–699. doi: 10.1007/s00109-005-0697-6. [DOI] [PubMed] [Google Scholar]
- Toh K.L., Jones C.R., He Y., Eide E.J., Hinz W.A., Virshup D.M., Ptácek LJ nd Fu Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- van der Horst G.T.J., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., Wit J., Verkerk A., Eker A.P.M., Leenen D., et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Viola A.U., Archer S.N., James L.M., Groeger J.A., Lo J.C., Skene D.J., von Schantz M., Dijk D.J. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2006;17:613–618. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- Vitaterna M.H., King D.P., Chang A.M., Kornhauser J.M., Lowrey P.L., McDonald J.D., Dove W.F., Pinto L.H., Turek F.W., Takahashi J.S. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna M.H., Selby C.P., Todo T., Niwa H., Thompson C., Fruechte E.M., Hitomi K., Thresher R.J., Ishikawa T., Miyazaki J., et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Larkin D.W., Albrecht U., Sun Z.S., Sage M., Eichele G., Lee C.C., Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zheng B., Albrecht U., Kaasik K., Sage M., Lu W., Vaishnav S., Li Q., Sun Z.S., Eichele G., Bradley A., et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Internet Resources
- R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2007. [November 2008 ]. R: A language and environment for statistical computing. Available from: http://www.r-project.org. [Google Scholar]
- TFSEARCH program v. 1.3. Available from: http://mbs.cbrc.jp/research/db/TFSEARCH.html.