Abstract
The Drosophila-like homolog 1 (DLK1), a transmembrane signal protein similar to other members of the Notch/Delta/Serrate family, regulates the differentiation process in many types of mammalian cells. Callipyge sheep and DLK1 knockout mice are excellent examples of a fundamental role of the gene encoding DLK1 in muscle growth and fat deposition. DLK1 is located within co-regulated imprinted clusters (the DLK1/DIO3 domain), along with other imprinted genes. Some of these, e.g. the RNA coding MEG3 gene, presumedly interfere with DLK1 transcription. The aim of our study was to analyze DLK1 and MEG3 gene expression in porcine tissues (muscle, liver, kidney, heart, brain stem) during postnatal development. The highest expression of both DLK1 and MEG3 variant 1 (MEG3 var.1) was observed in the brain-stem and muscles, whereas that of MEG3 variant 2 (MEG3var.2) was the most abundant in muscles and the heart. During development (between 60 and 210 days of age) expression of analyzed genes was down-regulated in all the tissues. An exception was the brain- stem, where there was no significant change in MEG3 (both variants) mRNA level, and relatively little decline (2-fold) in that of DLK1 transcription. This may indicate a distinct function of the DLK1 gene in the brain-stem, when compared with other tissues.
Keywords: DLK1, imprinting, MEG3, pigs, polar overdominance
The Drosophila-like homolog 1 (DLK1) is a transmembrane signal protein similar to other members of the Notch/Delta/Serrate family (Jensen et al., 1994). This protein controls several cell-differentiation processes throughout embryonic and adult life. A role of DLK1 in adipogenesis has been well documented (Smas and Sul, 1996; Sul , 2009; Nueda et al., 2007). Recently, DLK1 was shown to regulate fate of myogenic cells (Andersen et al., 2009) and human skeletal stem cells (Abdallah et al., 2004). A significant role of DLK1 in maintaining proper organism function was demonstrated by generating DLK1 knockout-mice, which exhibited accelerated obesity, growth disorders and skeletal malformation (Moon et al., 2002). Nevertheless, knowledge about exact function of DLK1 in particular tissues and organs remains fragmentary.
A gene for DLK1 is located in imprinted gene clusters on chromosomes 12, 14 and 7 within the so-called DLK1/DIO3 domain of mice, humans and pigs, respectively. There are few imprinted genes in the neighborhood of DLK1, among which the best studied are paternally expressed PEG11/RTL1, maternally expressed MEG3/GTL2 and MEG8. The full-length PEG11 protein was recently identified in callipyge sheep muscle (Byrne et al., 2010). On the other hand, MEG3 and MEG8 genes produce non-coding RNAs (ncRNA). These play an essential role in growth and differentiation, as the deletion of MEG3/GTL2, together with its differentially methylated region, induces lethal parent-origin-dependent defects in mice (Takahashi et al., 2009). The whole domain has been intensively studied in reference to molecular mechanism of imprinting control, but to date no satisfactory model has been proposed for this cluster.
Among farm animals, the most extensive studies of the DLK1/PEG11 domain have been undertaken in sheep, mainly in an attempt to identify the Single Nucleotide Polymorphism (SNPCLPG) responsible for the so called “callipyge phenotype” in this region. The callipyge phenotype (muscle hypertrophy of the hindquarters) is inherited in a non-Mendelian manner (the so-called polar overdominance), and appears in the offspring only when the mutated allele comes from the father and the wild one from the mother (Cpat/Nmat genotype) (Cockett et al., 1996). Animals with other genotypes (Cpat/Cmat Npat/Nmat, Npat/Cmat) do not exhibit the characteristic phenotype. There is a theory explaining the molecular mechanism underlying the unusual inheritance of callipyge phenotype: CLPG mutation enhances the level of DLK1, MEG3, PEG11 and MEG8 gene expression in cis, probably by modifying the activity of a common regulatory element. The callipyge phenotype results from a DLK1 and/or PEG11 overexpression in skeletal muscle with simultaneous underexpression of MEG3 and MEG8. In Cmat/Cpat individuals, overexpression of MEG8 and MEG3 or other maternally expressed genes interfere in trans with DLK1 and PEG11 and inhibit expression of DLK1 and PEG11 (Charlier et al., 2001; Georges et al., 2003).
The considerable effect of SNPCLPG on sheep musculature has made DLK1 an interesting candidate gene for marker-assisted selection in other farm animals. Unfortunately, SNPCLPG appeared to be a private allele, encountered exclusively in callipyge flocks (Smit et al., 2003). Nevertheless, Kim et al. (2004), on identifying a DLK1 polymorphism (silent SNP mutation) associated with growth, fatness and body composition in pigs, proved its polar-overdominant inheritance. These results have been confirmed through QTL (Quantitative Trait Locus) analysis, although the molecular mechanism involved has, as yet, not been stated (Li et al., 2008). Recently, gDLK1 has been proposed as a novel selection-marker for high muscle growth in chickens, since observed DLK1 mRNA expression was greater in the muscles of broilers than in layers (Shin et al., 2009).
In pigs, four different DLK1 and two MEG3 transcripts have been identified, so far (Deiuliis et al., 2006; Li et al., 2008; Samulin et al., 2009). In the pigs, the short form (DLK1 C2) is the most abundant transcript in all analyzed tissues (Deiuliis et al., 2006, Samulin et al., 2009). MEG3 expression has recently been observed in the liver, heart, spleen, fat, kidney and skeletal muscle of two-month old pigs. Nevertheless, relative abundance of different MEG3 variants has not been studied so far (Jiang and Yang, 2009).
Since DLK1 appears to be an interesting candidate gene for marker-assisted selection, and as little is known about postnatal gene expression within the DLK1/DIO3 domain in pigs, we decided to analyze relative mRNA abundance of DLK1 and MEG3 genes in various porcine tissues (muscle, liver, brain, kidney, heart), as well as changes in the expression of these genes during postnatal development.
Animals for the study were kept in the Pilot Plant of the National Research Institute of Animal Production in Pawlowice under identical housing and feeding conditions. They were divided into 6 age-groups, according to the day of slaughter (60-, 90-, 120-, 150-, 180- and 210-days-old pigs). In the first part of the experiment, two batches each of the breeds Large White and Duroc, with 4 to 6 pigs of both in each age group, were analyzed separately. As no significant differences between breeds were found, we decided to create only age groups consisting of 8 to12 animals (4 to 6 Large White and 4-6 Duroc). Animals were related – all pigs within the breed had the same father, and their mothers were sisters. Fragments of muscle (longissimus dorsi) and the liver were collected from animals of all the age groups, whereas fragments of the brain-stem, heart and kidney were collected only from the 60 and 210 days groups. Tissues were collected immediately after slaughter, and kept in liquid nitrogen during transportation. All animals were stress-resistant (RYR1 C/C).
Total RNA was extracted using TRI-Reagent (Sigma) and a Silent Crusher S homogenizer (Heidolph), according to the method described by Chomczyski (1993). RNA from the brain-stem was isolated by using the SV Total RNA System (Promega). The amount of extracted RNA was estimated by BioPhotometer (Eppendorf), and its quality evaluated by gel electrophoresis.
The RNA (1 μg) was reverse transcribed into cDNA at 37 °C by using a High Capacity cDNA Reverse Transcription Kit with random primers (Applied Biosystems), according to the manufacturer's protocol.
Primers and probes for all the genes were designed with Primer Express software (Applied Biosystems). MEG3 primers were designed to distinguish alternative isoforms, whereas DLK1 primers covered exon-exon junction and amplified all transcripts. The sequences of probes and primers were as follows: DLK1 for-5'AGGACGGCT GGGATGGA, rev-5'CGAGGTTCGCGCAGGTT, probe-5'TCTGTGACCTAGACATC MEG3 var. 1 for-5'GG AAGGGACCTCAGACATGTG, rev-5'CCTAGCCTGC CGATTTCAGA, probe-5'CTCCTGCACCCTCC, MEG3 var. 2 for- 5'TGAGGCTGGAGGAGCGTTAG, rev-5'CCAACTTGGACCCCTTCTCTT, probe-5'ATCTTGT CGCATTGCT.
Relative quantification of expression was performed on a 7500 Real-Time PCR System using labeled TaqMan® Tamra and MGB probes and TaqMan® Universal PCR Master Mix with UNG AmpErase (Applied Biosystems). Reactions, in a total volume of 25 μL were done in triplicate and according to the TaqMan® Universal PCR Master Mix protocol. GAPDH was used as endogenous control to compare expression changes of DLK1 and MEG3 during development in muscle and liver. RPL27 was used for between-tissue comparison, due to variations in GAPDH expression among the analyzed tissues. The results were analyzed using Sequence Detection System software v. 2.0 (Applied Biosystems). Statistical analysis was performed using One Way Anova, Tukey test (SAS Institute).
In our investigation we compared relative expression of DLK1 and MEG3 genes in the brain stem, muscle, heart, kidney and liver of pigs. For the muscle (longissimus dorsi) and liver, six developmental stages (60, 90, 120, 150, 180 and 210 days) were taken into consideration, whereas for the brain-stem, heart and kidney only two were (60 and 210 days).
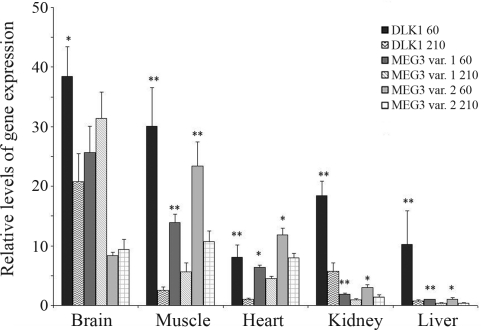
The lowest expression of DLK1 and MEG3 var. 1 was noted in the liver, and the highest in the brain-stem (DLK1 – 3.8-fold higher than in liver, MEG3 var.1 - 26-fold higher). An exception was the MEG3 var.2 transcript, which was the most abundant in the muscle (23-fold higher than in liver). In the muscle, abundance of other transcripts was also relatively high (DLK1 - 3-fold higher than in liver, MEG3 var. 1 – 14-fold higher). The level of expression was intermediate in the heart and kidneys, when compared with other tissues, although MEG3 transcripts were more abundant in the heart than in the kidney, whereas in DLK1 transcripts this was to the contrary (Figure 1).
Figure 1.
DLK1, MEG3 var.1 and MEG3 var. 2 gene expession in various porcine tissues at 60 and 210 days after birth. Values are presented as mean ± SEM. Asterisks indicate significant differences between 60 and 210 days for each gene. *p < 0.05, **p < 0.01.
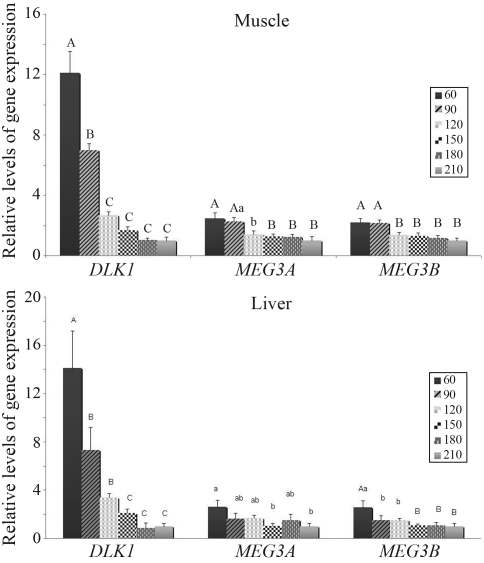
We also analyzed alteration in the expression of these genes during postnatal development. In the brain stem, kidney and heart DLK1 and MEG3 mRNA abundance was compared between 60 and 210 days of age, while in the muscle and liver additional developmental periods were analyzed (90, 120, 150 and 180 days of age). The expression level of all transcripts decreased with age in all the tissues investigated (Figures 1 and 2), with the exception of MEG3 (both variants) in the brain, where there was no significant change. DLK1 gene expression declined much more intensively (2-fold in the brain between 60 and 210 days, 8-fold in the heart, 14-fold in the liver and 12-fold in the muscle) than MEG3 (both transcripts, 1.4-, 2.6-, 2,2-fold higher in the respective tissues). In the brain-stem, expression of the analyzed genes seemed to be more stable during aging, when compared to the other analyzed tissues (Figure 1). In the muscle, where more developmental stages were analyzed, no fluctuation during the 90-180-days interval was observed. DLK1 expression decreased gradually, whereas that of MEG3 remained similar at 60 and 90 days to afterwards enter in sharp decline (Figure 2a). The pattern of DLK1 expression in the liver was similar to that observed in the muscle, whereas that of MEG3 was more pronounced only at 60 days (Figure 2b).
Figure 2.
DLK1, MEG3 var.1 and MEG3 var. 2 gene expression in the longissimus dorsi muscle (a) and liver (b) of pigs at 60, 90, 120, 150, 180 and 210 days after birth. Values are presented as mean ± SEM. Different letters indicate significant differences. Capital letters – p < 0.01, small letters – p < 0.05.
Previous studies have shown that in humans, DLK1 gene expression is high during prenatal development, whereas after birth this is restricted to hormone-secreting cells and monoaminergic neurons in the central nervous system (Abdallah et al., 2007). In contrast, postnatal DLK1 expression in pigs was observed in the kidneys, heart, spleen, fat and muscles (Deiuliis et al., 2006), as confirmed herein. Moreover, high mRNA abundance of DLK1 and MEG3 in the brain-stem was detected, with the MEG3 var.1 being the predominant variant of the latter. To date, DLK1 expression has not been studied in the brain-stem of farm animals, although in adult mice, high DLK1 and MEG3 expression has been observed in certain regions of the brain, e.g. hypothalamus, medulla and cerebral cortex (www.brain-map.org) (Labialle et al., 2008). A semi-quantitative analysis of tissue-specific expression of porcine MEG3 gene has been performed previously by RT-PCR (Jiang and Yang, 2009).The authors noted the highest expression of this gene in the brain and lungs, followed by the tongue, spleen, stomach, fat, kidneys, liver, skeletal muscles, heart and small intestine. Contrarily, our results suggest a much higher expression of MEG3 (both variants) in skeletal muscles than in the liver or kidneys. However, the results are difficult to compare due to the multiple isoforms of the MEG3 gene.
Declining expression of eleven imprinted genes (including DLK1 and MEG3) during postnatal development in multiple tissues (heart, lung and kidney) of mice at 1, 4 and 8 weeks of age has been previously reported (Lui et al., 2008). The authors inferred that the down-regulation of imprinted genes contributes to a deceleration in organ growth. Recently DLK1 has been shown to regulate growth hormone (GH) expression (Ansell et al., 2007). This, as well as our results, strongly supports the hypothesis for the kidney, liver and heart. In the present study, the decrease in DLK1 gene expression was also very pronounced in muscles. This may reflect a deceleration in muscle-mass growth. In contrast, the decline of DLK1 transcript abundance in the brain was relatively low (2-fold), with no significant change in MEG3 transcripts, thereby implying the maintenance of high expression levels even at 210 days. This may indicate a distinct function of the DLK1 gene in the brain-stem.
Although the biological consequences remain unknown, it is evident that expression of the DLK1/DIO3 domain in pigs, as in mice, is regulated in a tissue-specific manner. A full understanding of the complex mechanism involved in gene expression control in the DLK1/DIO3 domain, requires an analysis of the imprinting status of genes in various tissues during development. DLK1 is generally considered as a paternally expressed gene and MEG3 as a maternally expressed one. An increasing number of experiments, however, imply that the imprinting status of many genes, besides changing during development, differs among tissues (Khatib, 2007). Further studies of expression and the methylation status of this important domain in a wider range of tissues and developmental stages are planned in the future.
Acknowledgments
This work was supported by the National Research Institute for Animal Production. Statutory activity No. 1130.1.
Footnotes
Associate Editor: Ricardo Guelerman P. Ramos
References
- Abdallah B.M., Ding M., Jensen C.H., Ditzel N., Flyvbjerg A., Jensen T., Dagnaes-Hansen F., Gasser J.A., Kassem M. Dlk1/FA1 is a novel endocrine regulator of bone and fat mass and its serum level is modulated by growth hormone. Endocrinology. 2007;148:3111–3121. doi: 10.1210/en.2007-0171. [DOI] [PubMed] [Google Scholar]
- Abdallah B.M., Jensen C.H., Gutierrez G., Leslie R.G., Jensen T.G., Kassem M. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res. 2004;19:841–852. doi: 10.1359/JBMR.040118. [DOI] [PubMed] [Google Scholar]
- Andersen D.C., Petersson S.J., Jørgensen L.H., Bollen P., Jensen P.B., Teisner B.D., Schroeder H., Jensen C.H. Characterization of DLK1+ cells emerging during skeletal muscle remodeling in response to myositis, myopathies, and acute injury. Stem Cells. 2009;27:898–908. doi: 10.1634/stemcells.2008-0826. [DOI] [PubMed] [Google Scholar]
- Ansell P.J., Zhou Y., Schjeide B.M., Kerner A., Zhao J., Zhang X., Klibanski A. Regulation of growth hormone expression by Delta-like protein 1 (Dlk1) Mol Cell Endocr. 2007;271:55–63. doi: 10.1016/j.mce.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K., Colgrave M.L., Vuocolo T., Pearson R., Bidwell C.A., Cockett N.E., Lynn D.J., Fleming-Waddell J.N., Tellam R.L. The imprinted retrotransposon-like gene PEG11 (RTL1) is expressed as a full-length protein in skeletal muscle from Callipyge sheep. PLoS One. 2010;5:e8638. doi: 10.1371/journal.pone.0008638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C., Segers K., Karim L., Shay T., Gyapay G., Cockett N., Georges M. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet. 2001;27:367–369. doi: 10.1038/86856. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–537. [PubMed] [Google Scholar]
- Cockett N.E., Jackson S.P., Shay T.L., Farnir F., Berghmans S., Snowder G.D., Nielsen D.M., Georges M. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- Deiuliis J.A., Li B., Lyvers-Peffer P.A., Moeller S.J., Lee K. Alternative splicing of delta-like 1 homolog (DLK1) in the pig and human. Comp Biochem Physiol B. 2006;145:50–59. doi: 10.1016/j.cbpb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Georges M., Charlier C., Cockett N. The callipyge locus: Evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 2003;19:248–252. doi: 10.1016/S0168-9525(03)00082-9. [DOI] [PubMed] [Google Scholar]
- Jensen C.H., Krogh T.N., Højrup P., Clausen P.P., Skjødt K., Larsson L.I., Enghild J.J., Teisner B. Protein structure of fetal antigen 1 (FA1). A novel circulating human epidermal-growth-factor-like protein expressed in neuroendocrine tumors and its relation to the gene products of dlk and pG2. Eur J Biochem. 1994;225:83–92. doi: 10.1111/j.1432-1033.1994.00083.x. [DOI] [PubMed] [Google Scholar]
- Jiang C., Yang Z. Characterization, imprinting status and tissue distribution of porcine GTL2 gene. Agric Sci China. 2009;8:216–222. [Google Scholar]
- Khatib H. Is it genomic imprinting or preferential expression? Bioessays. 2007;29:1022–1028. doi: 10.1002/bies.20637. [DOI] [PubMed] [Google Scholar]
- Kim K.S., Kim J.J., Dekkers J.C., Rothschild M.F. Polar overdominant inheritance of a DLK1 polymorphism is associated with growth and fatness in pigs. Mamm Genome. 2004;15:552–559. doi: 10.1007/s00335-004-2341-0. [DOI] [PubMed] [Google Scholar]
- Labialle S., Yang L., Ruan X., Villemain A., Schmidt J.V., Hernandez A., Wiltshire T., Cermakian N., Naumova A.K. Coordinated diurnal regulation of genes from the Dlk1-Dio3 imprinted domain: Implications for regulation of clusters of non-paralogous genes. Hum Mol Genet. 2008;17:15–26. doi: 10.1093/hmg/ddm281. [DOI] [PubMed] [Google Scholar]
- Li X.P., Do K.T., Kim J.J., Huang J., Zhao S.H., Lee Y., Rothschild M.F., Lee C.K., Kim K.S. Molecular characteristics of the porcine DLK1 and MEG3 genes. Anim Genet. 2008;39:189–192. doi: 10.1111/j.1365-2052.2007.01693.x. [DOI] [PubMed] [Google Scholar]
- Lui J.C., Finkielstain G.P., Barnes K.M., Baron J. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol – Regul Integr Comp Physiol. 2008;295:R189–R196. doi: 10.1152/ajpregu.00182.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon Y.S., Smas C.M., Lee K., Villena J.A., Kim K.H., Yun E.J., Sul H.S. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nueda M.L., Baladrón V., Sánchez-Solana B., Ballesteros M.A., Laborda J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J Mol Biol. 2007;367:1281–1293. doi: 10.1016/j.jmb.2006.10.043. [DOI] [PubMed] [Google Scholar]
- Samulin J., Berg P.R., Sundvold H., Grindflek E., Lien S. Expression of DLK1 splice variants during porcine adipocyte development in vitro and in vivo. Anim Genet. 2009;40:239–241. doi: 10.1111/j.1365-2052.2008.01812.x. [DOI] [PubMed] [Google Scholar]
- Shin J., Velleman S.G., Latshaw J.D., Wick M.P., Suh Y., Lee K. The ontogeny of delta-like protein 1 messenger ribonucleic acid expression during muscle development and regeneration: Comparison of broiler and Leghorn chickens. Poultry Sci. 2009;88:1427–1437. doi: 10.3382/ps.2008-00529. [DOI] [PubMed] [Google Scholar]
- Smas C.M., Sul H.S. Characterization of Pref-1 and its inhibitory role in adipocyte differentiation. Int J Obesity Rel Metabol Disorders. 1996;20(Suppl 3):65–72. [PubMed] [Google Scholar]
- Smit M., Segers K., Carrascosa L.G., Shay T., Baraldi F., Gyapay G., Snowder G., Georges M., Cockett N., Charlier C. Mosaicism of solid gold supports the causality of a noncoding A-to-G transition in the determinism of the callipyge phenotype. Genetics. 2003;163:453–456. doi: 10.1093/genetics/163.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul H.S. Minireview: Pref-1: Role in adipogenesis and mesenchymal cell fate. Mol Endocrinol. 2009;11:1717–1725. doi: 10.1210/me.2009-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Okamoto A., Kobayashi R., Shirai M., Obata Y., Ogawa H., Sotomaru Y. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]