SUMMARY
Gram-negative pathogens commonly exhibit adhesive pili on their surfaces that mediate specific attachment to the host. A major class of pili is assembled via the chaperone/usher pathway. Here, the structural basis for pilus fiber assembly and secretion performed by the outer membrane assembly platform— the usher—is revealed by the crystal structure of the translocation domain of the P pilus usher PapC and single particle cryo-electron microscopy imaging of the FimD usher bound to a translocating type 1 pilus assembly intermediate. These structures provide molecular snapshots of a twinned-pore translocation machinery in action. Unexpectedly, only one pore is used for secretion, while both usher protomers are used for chaperone-subunit complex recruitment. The translocating pore itself comprises 24 β strands and is occluded by a folded plug domain, likely gated by a conformationally constrained β-hairpin. These structures capture the secretion of a virulence factor across the outer membrane of Gram-negative bacteria.
INTRODUCTION
The chaperone/usher (CU) pathway is responsible for the assembly of a major class of adhesive fibers on the outer membrane (OM) of a diverse group of Gram-negative bacteria, including important human and animal pathogens (Sauer et al., 2004). These filamentous extracellular organelles, called pili or fimbriae, form a class of virulence factors responsible for specific host recognition and attachment, invasion, and biofilm formation. CU pilus biogenesis encompasses the ordered noncovalent polymerization of periplasmic chaperone-bound pilus subunits at an OM assembly platform termed the usher (Figure 1) (Sauer et al., 2004). The usher catalyzes polymerization and facilitates the translocation of folded subunits across the bacterial OM in the absence of accessory proteins and in a process independent of cellular ATP or the proton motive force (Jacob-Dubuisson et al., 1994).
Figure 1. Pilus Assembly by the Chaperone/Usher Pathway.
(A) Multisubunit structure of E. coli P pili (left) and type 1 pili (right). Subunits are represented as oval shapes colored by subunit type. The subunit’s N-terminal extensions are represented as short rectangular protrusions.
(B) Schematic diagram of the CU assembly pathway. The outer membrane, extracellular, and periplasmic spaces are labeled OM, E, and P, respectively. The periplasmic chaperone and the OM usher are colored yellow and blue, respectively; subunits are represented as in (A). Blowout topology diagrams of donor-strand complemented (lower right) and donor-strand exchanged (upper right) subunits are shown. The subunit’s N-terminal extension is labeled (Nte).
(C) Predicted domain organization in OM ushers. The OM translocation and middle domain crystallized in this study are represented in blue and magenta, respectively.
Type 1 and P pili have served as model systems for the elucidation of the CU biosynthetic pathway. All uropathogenic Escherichia coli produce type 1 pili that are responsible for attachment, invasion, and establishment of biofilms in the bladder (Wright et al., 2007, Justice et al., 2006). P pili are produced by pyelonephritic strains of E. coli and are required for colonization of the kidney (Roberts et al., 1994). P and type 1 pili are complex extended fibers composed of two distinct subassemblies: (1) a distal tip fibrillum measuring 2 nm in diameter that is connected to a (2) rigid helical rod of 6.8 nm diameter (Jones et al., 1995; Kuehn et al., 1992). At their distal end, type 1 and P pilus tips contain the FimH and PapG adhesin, respectively, targeting the bacteria to mannose-containing receptors on the bladder epithelium (FimH) or to Gal-α (1–4)-Gal-containing glycolipid receptors on the kidney epithelium (PapG) (Choudhury et al., 1999; Dodson et al., 2001). In type 1 pilus tips, FimH is followed by one copy each of the FimG and FimF subunits (Figure 1A, right) (Hahn et al., 2002; Jones et al., 1995; Saulino et al., 2000). In P pili, PapG is attached via the PapF adaptor subunit to a tip fibrillum formed by a PapE homopolymer, itself attached to the pilus rod via the PapK adapter subunit (Figure 1A, left) (Kuehn et al., 1992). The pilus rod consists of a homopolymer of over 1000 copies of the FimA or PapA subunits in type 1 and P pili, respectively (Hahn et al., 2002; Kuehn et al., 1992).
Fiber assembly requires a periplasmic chaperone (FimC and PapD for type 1 and P pili, respectively) that interacts with each pilus subunit, providing a platform onto which the subunits fold (Barnhart et al., 2000; Bann et al., 2004; Vetsch et al., 2004). Previous crystal structures of chaperone-subunit and subunit-subunit complexes have established a unifying mechanism for subunit stabilization and polymerization by the CU pathway (Choudhury et al., 1999; Sauer et al., 1999, 2002; Zavialov et al., 2003). Pilus subunits share a common noncanonical immunoglobulin (Ig)-like fold characterized by the absence of its C-terminal β strand (Figure 1B). After translocation of the newly synthesized pilus subunit across the inner membrane into the periplasm, the cognate periplasmic chaperone “donates” part of a β strand for the intermolecular complementation of the incomplete Ig fold of the pilus subunit, a process termed donor-strand complementation (Figure 1B, topology diagram at bottom) (Choudhury et al., 1999; Sauer et al., 1999). Polymerization of the subunits at the bacterial OM occurs through a similar fold complementation mechanism, which involves the donation of the N-terminal extension (Nte) peptide of an incoming pilus subunit to complement the incomplete Ig fold of the previously assembled subunit in the growing pilus fiber (Figure 1B, topology diagram at top) (Sauer et al., 2002; Zavialov et al., 2003). This process of donor-strand exchange, in which the chaperone donor strand is exchanged for another subunit’s Nte, occurs at the OM usher (FimD and PapC for type 1 and P pilus subunits, respectively) (Figure 1B).
In sharp contrast to the wealth of information available on chaperone-subunit and subunit-subunit interactions, little is known of the structure or mechanism of action of the usher (Nishiyama et al., 2005). OM ushers selectively recruit chaperone-bound subunits to the OM, facilitate their ordered polymerization, and translocate the nascent fiber across the OM. They comprise four predicted domains: an N-terminal periplasmic domain (~125 residues), a central β-barrel domain (predicted residues ~135 to ~640) interrupted by a middle domain (predicted residues ~220 to ~325), and a C-terminal periplasmic domain (~170 residues) (Figure 1C) (Capitani et al., 2006; Ng et al., 2004; Nishiyama et al., 2003; Saulino et al., 1998; Thanassi et al., 2002). Chaperone-subunit recruitment occurs via the usher’s N-terminal domain (Ng et al., 2004; Nishiyama et al., 2005). This domain discriminately recognizes different chaperone-subunit complexes by directly binding to the N-terminal domain of the periplasmic chaperone and part of the chaperone-bound subunit (Nishiyama et al., 2005). Subunit polymerization has been shown to be a concerted process, with a new subunit’s Nte binding at an initiation site (called the P5 pocket) on chaperone-bound subunits and gradually displacing the chaperone donor strand (Remaut et al., 2006; Vetsch et al., 2006). How the usher facilitates this process or what parts of the usher are involved is unknown. Cryo-electron microscopy (cryo-EM) images of the PapC usher reconstituted in E. coli lipids reveal that ushers form dimeric channels in the OM, compatible in size with the secretion of folded subunits (Li et al., 2004). Complementation studies in PapC loss-of-function mutants confirm OM ushers to be functional as dimers (So and Thanassi, 2006). The predicted central β-barrel domain is likely to form the translocation channel, but the roles of the predicted middle domain or the periplasmically located C-terminal domain in usher function are unclear. The latter is believed to participate in activating the usher for translocation in a process that is initiated by the chaperone-adhesin complex (So and Thanassi, 2006). Understanding the complex multistep processing of CU pilus subunits into a functional, membrane-anchored filamentous organelle has been hampered by the lack of high-resolution structural information on the usher and complexes thereof.
Here, we present the crystal structure of the PapC translocation channel and the cryo-EM structure of the closely related FimD usher bound to the translocating FimC:F:G:H quaternary tip complex. These structures capture a twinned-pores usher transporter in action and provide an integrated model of the CU-mediated subunit assembly mechanism.
RESULTS AND DISCUSSION
Structure of the Usher Translocation Channel
To gain a structural understanding of pilus biogenesis at the OM, we solved the structure of the translocation channel of the PapC usher. The presence of the flexible periplasmic domains in the mature 809-residue PapC prevented growth of crystals of sufficient quality for structure determination of the full-length protein. Through limited proteolysis, we identified a stable 55 kDa fragment (residues 130–640) corresponding to the predicted OM translocation domain and the recently identified middle domain (Capitani et al., 2006). As expected, PapC130–640 localized to the OM and the integrity of the β-barrel domain was confirmed by a heat-modifiable mobility shift on SDS-PAGE, typical of folded OM β-barrel proteins (results not shown). Two crystal forms were obtained. The structure of PapC130–640 was determined at 3.4 Å resolution using the multiwavelength anomalous dispersion phasing method on F222 crystals and refined against 3.2Å resolution data from crystals in the C2 space group to a final R and free R factor of 25.9% and 29.6%, respectively (see Experimental Procedures, Figure S1, and Table S1). It encompasses the full translocation pore consisting of a kidney-shaped, 24-stranded β-barrel (residues 146–635), ~45 Å in height and with outer and inner dimensions of 65 Å by 45 Å and 45 Å by 25 Å, respectively (Figures 2A, 2B, and 2C). OM ushers therefore correspond to the largest single-protein β-pores observed to date (previously determined β-pore structures contain 22 β strands at most). The β-barrel closes in an end-to-end fashion and positions the N and C termini on the periplasmic side of the membrane. The N- and C-terminal globular domains will thus be juxtaposed and reside in the periplasm, consistent with their role in chaperone-subunit recruitment and adhesin-induced pore activation (Ng et al., 2004; Nishiyama et al., 2003; Saulino et al., 1998; Thanassi et al., 2002). The predicted middle domain (residues 257–332) is formed by a long sequence between strands β6 and β7 and consists of a six-stranded, βsandwich fold (strands βA–βF; Figure 2C). The domain is positioned laterally inside the β-barrel pore (Figure 2B). As a result, the middle domain, hereafter referred to as the plug domain, completely occludes the luminal volume of the translocation pore, preventing passage of solutes or periplasmic proteins across the channel in its nonactivated form (Figures 2A and 2B). Globular plug or cork domains have also been observed in bacterial siderophore transporters (Ferguson et al., 1998). Unlike these, however, the PapC plug domain is inserted into the loop connecting two β strands (strands 6 and 7) rather than positioned at the N-terminal extremity of the β-barrel. The plug domain is held in place by a β-hairpin (strands β5 and β6, hereafter referred to as the β5-6 hairpin, Figures 2A, 2B, and 2C) that folds in from the barrel wall into the channel lumen. The inward curvature of the β5-6 hairpin creates a large gap in the side of the β-barrel extending well into the part submerged in the OM bilayer (Figure 2B), a feature unprecedented in previously known OM β-barrel structures. The point where the β5-6 hairpin breaks out of the barrel coincides with a bulge (Ser209-Arg210) in the adjacent strand β4 (Figure 3A). This bulge disrupts the local secondary structure interaction with the hairpin strand β5 and, in combination with a charged residue (Glu228) in β5 that is pointing into the nonpolar bilayer, pushes the β5-6 hairpin into the barrel lumen, leaving the unusual gap in the lining of the transmembrane barrel (Figure 2B). In addition, the luminal part of the β5-6 hairpin is capped from the extracellular side by the only helix in the structure, the α1 helix (residues 448–465). Inside the β-barrel, the β5-6 hairpin interacts with the inner surface of the channel and helix α1 through a patch of hydrophobic interactions formed by Ile231, Phe236 on the hairpin, Trp239, Phe357, and Val377 on the barrel wall, and Met451 on the helix (Figure 3B). In addition, the β5-6 hairpin forms two electrostatic interaction networks that bridge the plug domain with the channel wall (Glu361-Lys339-Asp234-Arg303) and α1 helix (Arg237-Glu467-Arg305-Asp323) and help position the plug domain laterally inside the translocation channel (Figure 3B). Given the conservation in sequence and predicted topologies in usher proteins, the translocator pore structure of the PapC usher can be considered a prototype for all bacterial ushers (sequence alignment; Figure S2).
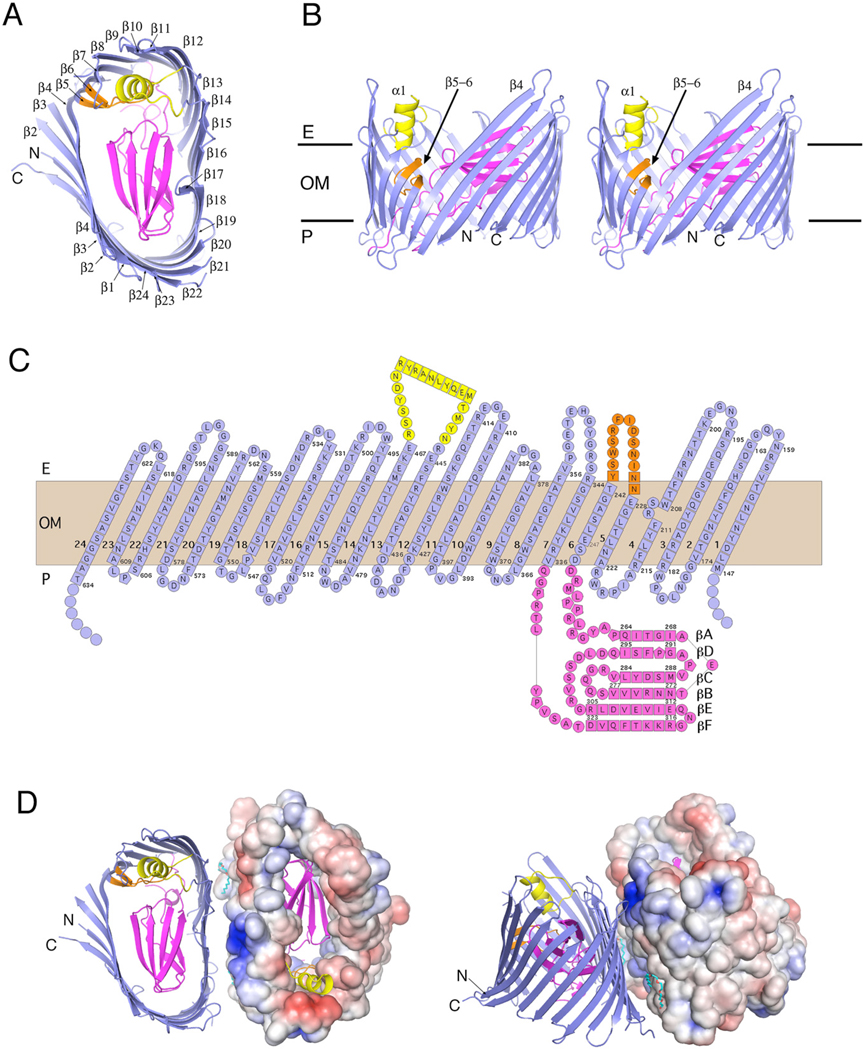
Figure 2. Crystal Structure of PapC130–640.
(A) Ribbon representation of the PapC130–640 translocation channel viewed from the extracellular side. The β-barrel, plug domain, β5-6 hairpin, and helix α1 are colored blue, magenta, orange, and yellow, respectively. β strands are labeled β1 through β24, the labels N and C indicate the N and C termini of the translocation channel.
(B) Stereo representation of PapC130–640 viewed from the side. Structural elements are colored as in (A). The N and C termini, helix α1, strand β4, and the β5-6 hairpin are labeled.
(C) Topology diagram of PapC130–640. Residues in β strand, loop, or α-helical topology are indicated by squares, circles, or rectangles, respectively; they are colored blue, orange, and yellow for the β-barrel, β5-6 hairpin, and plug domain, respectively. The extracellular space, the expected OM spanning area, and periplasmic space are indicated as E, OM, and P, respectively. Strands are labeled 1–24 and βA–βF in the β-barrel and plug domain, respectively.
(D) Top (left) and side (right) views of the PapC130–640 dimer. The β-barrel in the right protomer is shown in surface representation colored by electrostatic potential, from−3 kT/e− (blue) to 8 kT/e− (red). The N and C termini are labeled. Two C8E4 molecules in the dimer interface are shown in stick representation (C and O atoms are colored cyan and red, respectively). Figures 2 and 3 were generated using PyMol and APBS (http://www.pymol.org/ and http://apbs.sourceforge.net/).
Figure 3. Stereo View of the β-Hairpin Interactions.
(A) Close-up of the site of inward orientation of the β5-6 hairpin. Residues in the β4 bulge and hairpin involved in the hairpin curvature are shown in stick representation. Color scheme is according to Figure 1.
(B) Close-up of the β5-6 hairpin interactions with the plug domain, helix α1, and the channel wall. Residues involved in the interaction are shown in stick representation.
(C) Charge interactions between the plug domain and the β-barrel, viewed from the periplasmic side of the membrane. Side chains of neighboring residues are labeled by residue number.
Available structural and biochemical evidence demonstrates OM ushers function as dimers organized in twinned pores (Li et al., 2004; So and Thanassi, 2006). Both the C2 and F222 crystal forms of PapC130–640 obtained in this study contain similar dimers formed by a crystallographic two-fold axis (Figure 2D). These dimers closely correspond in size and conformation to the twinned pores observed by negative stain EM of full-length PapC (Figure S3) and by cryo-EM imaging on single particles of detergent-solubilized full-length PapC and 2D crystals of PapC reconstituted in E. coli lipids (Li et al., 2004). The dimer interface in the 3D crystals is formed by the flat side of the kidney-shaped β-barrel, encompassing strands β11–β20 (residues 397–573; Figure 2D). However, contrary to what is seen in other oligomerizing OM proteins, the two PapC protomers in the dimer interface make little direct contacts with each other (side chains are typically between 4 and 6 Å apart; Figure 2D). Instead, the electron density at the dimer interface in the PapC130–640 crystals reveals that the interface between protomers is mediated by several detergent molecules. Likely, the extensive detergent wash steps (see Experimental Procedures) preceding PapC130–640 crystallization resulted in the exchange of interface lipids for detergent molecules. In this respect, it is interesting to note the two usher protomers in the PapC130–640 crystals (average Cα-Cα distance of 15 Å) are in somewhat closer proximity than seen in the lipid-reconstituted 2D crystals and in the FimD2:C:F:G:H complex (Cα-Cα distance of 27 Å see below). Presumably the loss of lipids in the former results in the two protomers being closer to one another. Finally, since the PapC130–640 construct used for crystallization lacks the N- and C-terminal periplasmic domains, it cannot be excluded that parts of these domains are involved in maintaining the usher dimer integrity.
Isolation of a Type 1 Pilus Assembly Intermediate
To understand the mechanism of pilus subunit translocation by the OM usher, we isolated pilus assembly intermediates by capturing type 1 pilus tip fibers during secretion through the FimD usher. FimD was expressed in bacteria together with the FimH adhesin, FimG and FimF tip subunits, and a His6-tagged FimC chaperone (FimCHis). By not providing the FimA helical rod subunit to the assembly system, pilus production stalls after incorporation of the last tip subunit FimF, resulting in production of quasihomogeneous FimD:tip complexes. Affinity and gel filtration chromatography of the detergent-solubilized OM fraction yielded the type 1 pilus tip complex as a single major peak in the chromatography profile (Figure 4A). Gel electrophoresis and Coomassie blue staining show all expected components (FimD:C:F:G:H) to be present in the complex (Figure 4B). Densitometry of the Coomassie-stained gel suggested a molar ratio for FimD:C:F:G:H of approximately 2:2:1:1:1, calculated by setting FimH equal to 1 mol (Figure 4B). However, since FimC adsorbs Coomassie blue dye twice as strongly as FimH, as demonstrated previously with dissolved 3D crystals of 1:1 FimC:FimH complex (Knight et al., 1997), we infer that the stoichiometry of the purified FimD:C:F:G:H complex is 2:1:1:1:1 rather than 2:2:1:1:1. Edman degradation analysis and cysteine labeling confirmed the 2:1:1:1:1 stoichiometry (see Supplemental Data).
Figure 4. Isolation of the Type 1 Pilus Tip Assembly Intermediate.
(A) Gel filtration profile of FimD2:CHis:F:G:H complexes.
(B) Coomassie blue-stained gel of the isolated tip complex. Lanes 1 and 2 represent the peak fractions from the gel filtration column as indicated in (A). The identity and molecular weight of each Fim subunit is indicated on the right, and the molar ratio of each subunit relative to FimH—estimated by densitometry of lane 1—is indicated on the left.
(C) The purified FimD:tip complexes were incubated in SDS sample buffer at 25° or 95°C, separated by gel electrophoresis, and immunoblotted with anti-FimC:F, anti-FimC:G, or anti-FimC:H polyclonal antibodies, as indicated. The FimF, G, and H subunits are engaged in donor-strand exchange, as revealed by the disappearance of bands corresponding to the pilin monomers and the appearance of a band reacting with all three antibodies in samples incubated at 25°C.
(D) Negatively stained EM image of the purified tip complexes from the fraction shown in (B), lane 1, and a single class average (inset). Individual particles are indicated with white circles.
When polymerized, pilus subunits have undergone donor-strand exchange and form SDS-stable complexes at room temperature (Sauer et al., 2002). SDS-PAGE of the isolated tip complex both at room temperature and after boiling confirms that the complex contains the FimF:G:H subunits in donor-strand exchange rather than as separate, nonpolymerized chaperone-subunit complexes (Figure 4C). Thus, the isolated tip complex corresponds to a FimD usher dimer bound to the FimF:G:H tip polymer, with the last incorporated subunit (FimF) still in donor strand complementation with the FimC chaperone. The quality of the purified usher-chaperone-tip complex was assessed by EM characterization. Raw electron micrographs of the complexes stained with phosphotungstic acid revealed discrete and homogeneous particles (Figure 4D).
OM Ushers Function as Asymmetric Twinned Pores
The type 1 pilus tip complex described above was analyzed in higher detail by cryo-EM. A 3D map was computationally reconstructed from ~11,000 tip complex particles by the single particle method (Frank, 2006) (Figures 5A and 5B; see Experimental Procedures). The 3D map, rendered at a threshold of 2σ, has an estimated resolution of 23 Å (Figure S4) and corresponds in volume to the calculated molecular mass of the tip complex of 265 kDa (Figure 4B). The structure of the tip complex can be divided into three major regions (Figure 5C) so distinct that several major features are immediately recognizable. A bulky area of density with apparent twofold symmetry lies centrally located in the complex (Figure 5C; region 1). This density corresponds in size and symmetry to the usher twinned pores observed previously by cryo-EM imaging of 2D crystals of the homologous PapC usher reconstituted in E. coli lipids (Li et al., 2004). Indeed, the crystal structure coordinates of the PapC usher translocation domain, described above, fit well inside either lobe of region 1 of the 3D cryo-EM map (Figure 5C). Furthermore, the position of both protomers closely resembles that of the PapC130–640 dimer seen in both crystal forms (Figure 2D). Finally, FimD is the only molecular species in the FimD2:C:F:G:H complex to be present in two copies (Figure 4B). Thus, the density region 1 can confidently be assigned as the twinned translocation channels formed by the FimD usher dimer present in the purified tip complexes.
Figure 5. 3D Cryo-EM Reconstruction of the FimD2:C:F:G:H Type 1 Pilus Tip Assembly Intermediate.
(A) Cryo-EM image of the purified tip complexes from the fraction shown in lane 1 in Figure 4B. Three individual tip complex particles are marked by a white circle.
(B) A comparison of selected 2D class averages in rows 1 and 3 with their corresponding reprojections from the 3D map in rows 2 and 4.
(C) Front (top left), side (top right), periplasmic (bottom left), and extracellular (bottom right) views of the 3D reconstruction of the FimD:tip complex rendered at 2σ (white surface), and additionally at 4.5 σ (gray), and 5σ (green) in the upper left view in order to show the cryo-EM density’s center of mass. The density regions corresponding to the outer membrane, extracellular space and periplasm (OM, E and P) are labeled as 1, 2 and 3, respectively. Available structures or closely related models (see Experimental Procedures for details) were docked into the cryo-EM density as follows: FimH adhesin (green), FimD translocation pore 1 and 2 (blue and dark blue), FimDN:C:F (blue:yellow:red), and FimG (orange).
A second distinguishing density in the cryo-EM map lies on top of the assigned translocation pore (Figure 5C; region 2). This protruding density consists of two separate thin and elongated lobes, the volume and relative angle of which correspond closely to the subdomain arrangement seen in the crystal structure of the FimH adhesin (Choudhury et al., 1999). Labeling with a monoclonal anti-FimH antibody confirmed density region 2 indeed corresponds to FimH (Figure S5A). The FimH structure (PDB ID: 1QUN) (Choudhury et al., 1999) fits well as a single rigid body into the exposed density of the tip fiber (Figure 5C), with the linker region between the two domains docking nicely at the weakest middle section of the tip density. Haemagglutination experiments on bacteria expressing FimD2:C:F:G:H type 1 pilus assembly intermediates show that the FimH adhesin is exposed on the bacterial surface (Saulino et al., 2000). Thus, region 2 defines the extracellular side of the complex. Importantly, when contoured at a 4 or 5σ level, the electron density for FimH clearly shows it to be in continuous density with usher pore 1 (Figure 5C, upper left panel).
The assignment of region 1 and 2 as the usher density and the extracellularly protruding adhesin, respectively, locates the remaining density region 3 to the periplasmic side of the complex (Figure 5C). It is well established that the periplasmic side of the usher contains the N- and C-terminal domains as well as the last incorporated chaperone-subunit complex, which, in this case, is FimC:F (Hahn et al., 2002; Ng et al., 2004; Nishiyama et al., 2003, 2005; Saulino et al., 2000; Thanassi et al., 2002). This interpretation is further confirmed by the observation that binding of anti-His6 antibodies followed by visualization by cryo-EM clearly locates FimCHis in region 3 (Figure S5B). Noticeably, density region 3 is positioned asymmetrically, under just one of the usher pores, suggesting that only one pore is used for secretion (see below).
The previously determined crystal structure of the ternary complex of the FimD N-terminal usher domain (FimDN) bound to the FimC chaperone and the pilin domain of FimH (FimHP) (PDB ID: 1ZE3 [Nishiyama et al., 2005]) can be unambiguously docked as a single rigid body in region 3 (Figure 5C; Movie S1) with either manual or automated docking using Chimera (Pettersen et al., 2004). This complex is a good model for the FimDN:C:F complex as all pilin domains have similar folds and structures. The docked FimDN:C:Hp complex not only fits the electron density very well, but would also position FimF under usher pore 1, in a region of the electron density that is continuous with that of usher pore 1. FimF is in donor strand exchange with FimG, itself in donor strand exchange with FimH (Figure 4C). As FimH is in continuous electron density with usher pore 1 on the extracellular side and FimF locates under the same usher pore on the periplasmic side, the bridging subunit FimG must reside inside the translocating usher pore 1. Indeed, when modeled within usher pore 1 (see Experimental Procedures), FimG satisfies all known positional restraints for it to accept the FimF Nte and donate its own Nte to FimH (Figure 6A). Moreover, the low electron density between the two pores excludes the possibility that FimG could reside between the two pores. Our type 1 tip complex model fits the experimental cryo-EM density with an excellent global correlation coefficient of 0.70 as calculated by the Mod-EM density fitting method (Topf et al., 2005).
Figure 6. Model of Pilus Assembly at the OM Usher.
(A) Integrated, hypothetical model of pilus subunit incorporation at the OM usher. The model shows the FimD2:C:F:G:H complex as derived by cryo-EM (Figure 5C and main text), colored blue, dark blue, yellow, red, orange, and green for FimD usher pore 1, 2, the FimC chaperone, FimF, FimG, and the FimH adhesin, respectively. In addition, the model shows the tentative position (shaded in light gray) of an incoming FimC:A complex bound to usher 2’s N-terminal domain (FimDN2:C:A, colored dark blue:yellow:cyan and labeled N2, FimC, and FimA, respectively). N1 indicates the N-terminal domain of usher 1.
(B) Schematic diagram of pilus assembly integrating available biological and biochemical data with the new PapC130–640 structure and FimD2:C:F:G:H model. The diagram shows the PapC twinned pores (in blue and dark blue for usher 1 and 2, respectively), the ushers’ N-terminal domains (N1 and N2 for usher 1 and 2 in blue and dark blue, respectively), the plug domain (P in usher 2 and P′ and P″ in usher 1 for the two alternative models of gating [see text]; in magenta), the FimH adhesin (H; in green), FimG (G; in orange), FimC:F (C1:F; in yellow:red), and an incoming FimC:A complex (C2:A; in yellow:cyan). For clarity, the C-terminal domains of ushers 1 and 2 are not shown. At left, the FimD2:C:F:G:H complex recruits an incoming FimC:A complex through binding to the N-terminal domain of usher 2 (N2; step 1). The complex is brought within donor-strand exchange of FimF, resulting in the release of the FimF-bound chaperone (C1) and the dissociation of the N-terminal domain of usher 1 (N1) (middle panel; steps 2 and 3). N1 is now free to recruit another FimC:A complex (labeled C:A′; right panel, step 4), and bring the complex within proximity of the N2-bound FimC:A complex (step 5). Donor-strand exchange then releases N2 for recruitment of the next chaperone-subunit complex (step 6). Iteration of alternating binding to released usher N-terminal domains, followed by donor-strand exchange with the penultimate chaperone-subunit complex leads to stepwise growth of the pilus fiber (steps 1 through 6).
The position of the N-terminal usher domain in the docked FimDN:C:F complex lies approximately 62 and 75 Å from the N termini of the docked pore domain of usher 1 and 2, respectively, distances that are consistent with the pore domain and the N-terminal domain being connected by a 20-residue linker sequence. The resolution in our cryo-EM map does not allow the docked N-terminal domain to be allocated to either usher with certainty, although the shorter distance to pore 1 and the relative positions of the N and C termini of the pore suggest it to belong to usher 1. The cryo-EM map lacks density corresponding to the second FimDN domain, indicating that this domain is disordered in the periplasm when not bound to a chaperone-subunit complex. Similarly, the absence of density in our 3D reconstruction large enough to correspond to the C-terminal domains of ushers 1 and 2 argues for these domains to be disordered in the purified tip complex. Limited proteolysis also suggests these domains to be disordered in the unbound state (this study). Finally, the plug domain within the usher channel would not be resolved as distinct from the usher density at the present resolution of the cryo-EM density (23 Å).
Our structural and biochemical data on the purified type 1 tip complex and PapC firmly establish the usher dimer as the functional unit for CU pilus biogenesis. Our 3D cryo-EM reconstruction reveals translocation of the polymerized subunits occurs asymmetrically and through a single pore only (usher 1; Figure 5C).
Gating the Usher Translocation Pore
The usher mediates the translocation of folded, polymerized protein units across the OM (Sauer et al., 2002; Remaut et al., 2006; Vetsch et al., 2006). The width of the PapC translocation pore (inner diameter of 45 Å × 25 Å) corresponds well with the passage of P pilus subunits in an upright orientation, along the length of the polymer (P pilus subunit dimensions are 55 Å × 30 Å × 25 Å, 57 Å × 32 Å × 23 Å and 60 Å × 27 Å × 22 Å for PapE (PDB ID: 1N12), PapK (PDB ID: 1PDK), and PapA (PDB ID: 2UY6), respectively (Sauer et al., 1999, 2002; Verger et al., 2007). However, in its nonactivated form, the PapC channel is obstructed by the plug domain (Figures 2A and 2B). Therefore, the adhesin-induced activation of the usher (Saulino et al., 1998) must include the displacement of the plug domain from the translocation channel. A rotation of the plug domain out of the pore lumen and into the periplasm would create a channel of 37 Å × 25 Å (or 45 Å × 25 Å when the β5-6 hairpin and α1 helix are displaced from the channel as well) (Figure 6B, position P′). Alternatively, a conformational change in the β5-6 hairpin and α1 helix could allow an upward rotation of the plug domain inside the pore, thereby liberating a translocation channel of approximately 27 Å × 25 Å (Figure 6B, position P″). Substrate-induced gating of the transduction channel is also seen in siderophore importers like FhuA. In this family of importers, an inner membrane (IM) embedded partner, TonB, couples energy generated at the IM by the proton motive force to the active gating of a globular “cork domain” in the OM channel (Gumbart et al., 2007; Pawelek et al., 2006). In contrast, OM ushers function autonomously and independently of a hydrolyzable energy source or a proton gradient (Jacob-Dubuisson et al., 1994). Gating of the plug domain in the usher therefore relies solely on conformational changes induced by the binding of the chaperone-adhesin complex. Powering the large conformational rotation of the plug domain must therefore come from the energy emanating from the binding of the chaperone-adhesin complex and/or must be stored as structural strains in the nonactivated PapC channel. One such area of apparent structural strain is seen in the β5-6 hairpin that breaks out of the β-barrel lining. The exposed part of the adjacent strand β4 represents a large open-edged β sheet structure. Such exposed edges form highly aggregative surfaces for the edge-to-edge docking of β strands or β sheets (Richardson and Richardson, 2002). As part of the activation process, the β 5-6 hairpin could line up with β 4 into the barrel wall. This would break the interaction with the plug domain (Figures 3A and 3B) and allow its upward rotation or displacement out of the barrel lumen (Figure 6B). Another area of structural strain could reside in the interaction of the plug domain with the barrel lumen, which includes a number of like-charge residues (Figure 3C). It seems plausible that during activation, these repulsive forces will aid in expelling the plug domain from the barrel lumen.
An Integrated Model for Fiber Assembly at the Usher
Based on the cryo-EM-derived model (Figure 6A), we propose a mechanism where the two ushers in the twin pores cooperate in pilus polymerization by alternately recruiting new chaperone-subunit complexes through their N-terminal domains (Figure 6B). This mechanism provides a rationale for the known requirement of a dimeric usher complex: one usher provides the secretion channel, but two ushers are needed for successive rounds of subunit binding and fiber assembly.
The structural information presented here and the available biochemical background can be combined into an integrated model for subunit recruitment, subunit polymerization, and fiber translocation during pilus biogenesis at the OM usher (Figure 6B). Our structure of the FimD2:C:F:G:H tip complex captures the fiber assembly process after FimH, FimG, and FimF have assembled into a pilus tip. In this complex, the last incorporated chaperone-subunit complex (FimC:F) is bound to the N-terminal domain of usher pore 1. In our model, the next subunit to be incorporated into the fiber is recruited to the twinned pores through the N-terminal domain of usher 2 (step 1; Figure 6B). The N-terminal domain of the usher resides in the periplasm, tethered by a 20-residue flexible linker to the translocation pore. This spacer between the usher translocation pore and its N-terminal domain allows reorientation of the chaperone-subunit complex to position the Nte of the newly recruited subunit in proximity with the pilin domain of the previously recruited chaperone-subunit complex (bound to usher 1), as shown in Figure 6A. Donor-strand exchange is a concerted process, with the subunit’s Nte binding at an initiation site (called the P5 pocket) on chaperone-bound subunits and gradually displacing the chaperone donor strand (Figure 6A; step 2, Figure 6B) (Remaut et al., 2006). This strand exchange reaction results in the release of the chaperone from the previously recruited chaperone-subunit complex and its dissociation from the N-terminal domain of usher 1 (the usher lacks any detectable affinity for the chaperone when not in complex with the adhesin or a pilus subunit [Dodson et al., 1993; Nishiyama et al., 2003; Saulino et al., 1998]) (step 3; Figure 6B). Upon release of the chaperone, the subunit can now enter the translocating pore of usher 1 and the N-terminal domain of usher 1 is free to bind a new chaperone-subunit complex from the periplasmic pool and bring it within proximity of the previously recruited chaperone-subunit complex for donor-strand exchange (steps 4 and 5; Figure 6B). In this way, iterations of the alternating recruitments of new chaperone-subunit complexes at either usher’s N-terminal domain, followed by donor-strand exchange with the previously assembled subunit, allow the stepwise polymerization and translocation of the pilus fiber (steps 5 and 6; Figure 6B).
The model for CU pilus assembly presented above incorporates all known biochemical data into the new structural framework. It does however lack the usher C-terminal domain. Earlier studies in the P pilus system showed that the C-terminal domain of PapC is involved in the activation of the usher (So and Thanassi, 2006). From studies in type 1 pili, it is known that FimH recruitment triggers a conformational change in the usher required for its activation (Saulino et al., 1998). This activation step depends on the presence of the FimH adhesin domain (Munera et al., 2007). We propose that, in its inactive state, the C-terminal domain (not shown in Figure 6B) is positioned at least partially under the PapC or FimD channel and is in contact with the β5-6 hairpin and plug domain. This is consistent with cryo-EM data showing that in the absence of the C-terminal domain, the electron density within the channel is weaker (Li et al., 2004). When the chaperone-adhesin complex is recruited to the usher via the usher’s N-terminal domain, a putative additional interaction between the adhesin and the usher’s C-terminal domain may relay a conformational change to the plug domain and the β5-6 hairpin and result in opening of the channel. Such an interaction has been observed between the PapC C-terminal domain and the PapD:G chaperone-adhesin complex (So and Thanassi, 2006). As our FimD:tip complex captures a later stage of pilus biogenesis, a stage at which the twinned pores are already activated and the C-terminal domains may no longer be involved, it is not surprising that the C-terminal domains should not be seen. The C-terminal domains may lay idle, possibly tethered to a long linker (as for the N-terminal domain). However, further functional and structural work is needed to elucidate the role of the usher C-terminal domain and the mechanism of channel gating.
The structure of the translocating unit of the PapC usher provides a unique molecular view of a twinned-pore transporter in action. In combination with the cryo-EM reconstruction of the type 1 pilus usher:tip complex, it provides the first view of a translocase bound to a secretion intermediate. High-resolution structural information of a translocating unit that operates independent of an energized protein partner or cellular forms of stored energy (ATP or proton gradient) for pore gating and secretion of folded proteins has been lacking. The PapC130–640 structure suggests this gating could be powered by structural strain stored in the translocation pore. The cryo-EM model presented here reveals that usher twinned pores facilitate the asymmetrical translocation of a single polymer and function as a dimer during the polymerization step. It elucidates some of the most important molecular details of the mechanism of action of CU ushers and will inform future avenues of research in the field of protein transporters both in prokaryotes and eukaryotes.
EXPERIMENTAL PROCEDURES
PapC130–640 Expression and Purification
Details of this section of the experimental procedures can be found in the Supplemental Data.
PapC130–640 Structure Determination
PapC130–640 crystals were grown from detergent-solubilized protein by hanging drop vapor diffusion against a solution containing 5%–20% 3-methyl-1,5-pentanediol (MPD), 6%–12% polyethylene glycol 4000 (PEG4000), 50 mM Na citrate (pH 5.6), and 100 mM ammonium sulfate. Diffraction data were collected from loop-mounted crystals at 100K on the ESRF beamline ID29. Two crystal forms were obtained. C2 and F222 crystals have cell parameters of a = 166.5 Å, b = 101.9 Å, c = 113.7 Å, β = 128.2, and a = 170.9;Å, b = 178.4 Å, and c = 255.3 Å, containing 2 and 3 molecules per asymmetric unit, respectively (see Table S1 for data statistics). The structure was solved from a 2 wavelength MAD experiment (collected at λ = 0.98023 Å and λ = 0.98034 Å, corresponding to the measured selenium K-edge absorption peak and inflection point) on a F222 crystal diffracting to 3.4 Å. Data were integrated and scaled using the XDS package (Kabsch, 1993), and the 18 positions out of the expected 21 selenium sites were derived and refined using ShelxD-HKL2MAP (Pape and Schneider, 2004) and Sharp (de La Fortelle and Bricogne, 1997). Experimental electron density maps calculated from solvent-flipped phases (de La Fortelle and Bricogne, 1997; CCP4, 1994) allowed the manual building of the PapC130–640 main chain using COOT (CCP4, 1994) (Figure S1). Refinement of the initial model was completed in data collected on the C2 crystals to 3.2 Å, resulting in a final R and free R factor of 25.9% and 29.6%, respectively (CNS 1.2, [Brunger, 2007]). A two-fold noncrystallographic symmetry restrain was applied throughout refinement. The final model contains two copies of PapC130–640, encompassing main chain and side chain atoms for residues 146–637 (with gaps in 178–180, 248–252, 348–353, 430– 435, and 589–592), of which 83.7% and 15.7% are located in the most favored and additionally allowed regions of the Ramachandran plot, respectively. The modeled part of the solvent contains 43 water molecules, 2 LDAO molecules, and 2 C8E4 molecules.
Purification of FimD-Tip Complex and Electron Microscopy
Details of this section of the experimental procedures can be found in the Supplemental Data.
Image Analysis and 3D Reconstruction
No symmetry was assumed or imposed during image processing and 3D reconstructions. We used the software packages EMAN (Ludtke et al., 1999) and SPIDER (Frank et al., 1996) to process the cryoimages of the usher-chaperone-tip complex. A total 11,000 particles were manually picked with a square box 96 pixels in size. The parameters of the contrast transfer function (CTF) for each micrograph were calculated based on an averaged image of all selected particles from that micrograph, and these parameters were used for the phase correction of the raw particle images. The phase-corrected and contrast-reversed raw particles were centered and subjected to multivariate statistical analysis and reference free classification.
For 3D reconstruction, the starting models were calculated from representative class averages of the raw particle images, based on the cross-common line technique. These models were low-pass filtered and refined against the raw images until convergence. Refinements from slightly different starting models calculated from different selections of the 2D class averages resulted in similar final maps. The consistency in the refined structures is attributed to the refinement strategy designed for removing initial model bias (Ludtke et al., 1999) and to the distinct structural features of the tip complex. The resolution of the final map (23 Å) was estimated by Fourier shell correlation of two maps calculated separately from two halves of the data set (Figure S4). The map was then low-pass filtered to the estimated resolution. The resulting cryo-EM map had enough resolving features to be immediately interpretable (see main text). Antibody labeling (anti-FimH and anti-His tag) confirmed the composition and orientation of the complex (see main text; Figure S5). The monoclonal anti-His6-tag antibody (Covance in Berkeley, CA) or the monoclonal anti-FimH antibody (a gift from MedImmune in Gaithersburg, MD) was mixed with the FimD:tip complex at an estimated molar ratio of 1:1 and incubated at room temperature for 30 min before preparing the cryo-EM grids.
The handedness of the cryo-EM map was initially undetermined with the common-line-based reconstruction method. However, we found that the crystal structures of both the FimH adhesin and the FimDN:C:HP complex only fit the cryo-EM map well as a single rigid body if we mirrored the original map. This resulted in the presented handedness of the map.
Type 1 Tip Complex Model Generation
Details of this section of the Experimental Procedures can be found in the Supplemental Data.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Helen Saibil and Dr. Maya Topf for comments on the manuscript and suggestions for cryo-EM map fitting. We thank staff of beamline ID29 at the ESRF-Grenoble for technical assistance in data collection. This work is supported by MRC grant 58149 to G.W., by NIH grant GM062987 to D.G.T., by Brookhaven Laboratory LDRD grant 05-112 and NIH grant GM074985 to H.L., and by NIH grants AI029549, AI48689, and AI049950 to S.J.H.
Footnotes
ACCESSION NUMBERS
The PapC130–640 X-ray coordinates and the type 1 tip complex cryo-EM map have been deposited in the Protein Data Bank and Electron Microscopy Data Bank with accession codes 2vqi and EMD-5009, respectively.
SUPPLEMENTAL DATA
Supplemental Data include Supplementary Experimental Procedures, Supplemental References, five figures, and one movie and can be found online at http://www.cell.com/cgi/content/full/133/4/640/DC1/
REFERENCES
- Bann JG, Pinkner JS, Frieden C, Hultgren SJ. Catalysis of protein folding by chaperones in pathogenic bacteria. Proc. Natl. Acad. Sci. USA. 2004;101:17389–17393. doi: 10.1073/pnas.0408072101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Pinkner JS, Soto GE, Sauer FG, Langermann S, Waksman G, Frieden C, Hultgren SJ. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc. Natl. Acad. Sci. USA. 2000;97:7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protocols. 2007;2:2728–2730. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Capitani G, Eidam O, Grutter MG. Evidence for a novel domain of bacterial outer membrane ushers. Proteins. 2006;65:816–823. doi: 10.1002/prot.21147. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Dodson KW, Jacob-Dubuisson F, Striker RT, Hultgren SJ. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc. Natl. Acad. Sci. USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson KW, Pinkner JS, Rose T, Magnusson G, Hultgren SJ, Waksman G. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell. 2001;105:733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- Frank J. Their Native State, Second Edition. New York: Oxford University Press; 2006. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules. [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gumbart J, Wiener MC, Tajkhorshid E. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys. J. 2007;93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Haner M, Taschner N, Burkhard P, Aebi U, Muller SA. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 2002;323:845–857. doi: 10.1016/s0022-2836(02)01005-7. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Striker R, Hultgren SJ. Chaperone-assisted self-assembly of pili independent of cellular energy. J. Biol. Chem. 1994;269:12447–12455. [PubMed] [Google Scholar]
- Jones CH, Pinkner JS, Roth R, Heuser J, Nicholes AV, Abraham SN, Hultgren SJ. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Hultgren SJ. Filamentation by Escherichia coli subverts innate immunity during urinary tract infection. Proc. Natl. Acad. Sci. USA. 2006;103:19884–19889. doi: 10.1073/pnas.0606329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- Knight S, Mulvey M, Pinkner J. Crystallization and preliminary X-ray diffraction studies of the FimC-FimH chaperone-adhesin complex from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 1997;53:207–210. doi: 10.1107/S0907444996011511. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Heuser J, Normark S, Hultgren SJ. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature. 1992;356:252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- Li H, Qian L, Chen Z, Thibault D, Liu G, Liu T, Thanassi DG. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 2004;344:1397–1407. doi: 10.1016/j.jmb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Munera D, Hultgren S, Fernandez LA. Recognition of the N-terminal lectin domain of FimH adhesin by the usher FimD is required for type 1 pilus biogenesis. Mol. Microbiol. 2007;64:333–346. doi: 10.1111/j.1365-2958.2007.05657.x. [DOI] [PubMed] [Google Scholar]
- Ng TW, Akman L, Osisami M, Thanassi DG. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 2004;186:5321–5331. doi: 10.1128/JB.186.16.5321-5331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Vetsch M, Puorger C, Jelesarov I, Glockshuber R. Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD. J. Mol. Biol. 2003;330:513–525. doi: 10.1016/s0022-2836(03)00591-6. [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Horst R, Eidam O, Herrmann T, Ignatov O, Vetsch M, Bettendorff P, Jelesarov I, Grutter MG, Wuthrich K, et al. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 2005;24:2075–2086. doi: 10.1038/sj.emboj.7600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Cryst. 2004;37:843–844. [Google Scholar]
- Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. Donor strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol. Cell. 2006;22:831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Marklund BI, Ilver D, Haslam D, Kaack MB, Baskin G, Louis M, Mollby R, Winberg J, Normark S. The Gal(alpha 1–4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- Sauer FG, Remaut H, Hultgren SJ, Waksman G. Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Saulino ET, Thanassi DG, Pinkner JS, Hultgren SJ, Lombardo MJ, Roth R, Heuser J. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulino ET, Bullitt E, Hultgren SJ. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl. Acad. Sci. USA. 2000;97:9240–9245. doi: 10.1073/pnas.160070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So SS, Thanassi DG. Analysis of the requirements for pilus biogenesis at the outer membrane usher and the function of the usher C-terminus. Mol. Microbiol. 2006;60:364–375. doi: 10.1111/j.1365-2958.2006.05111.x. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Stathopoulos C, Dodson K, Geiger D, Hultgren SJ. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 2002;184:6260–6269. doi: 10.1128/JB.184.22.6260-6269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J. Struct. Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Verger D, Bullitt E, Hultgren SJ, Waksman G. Crystal structure of the P pilus rod subunit PapA. PLoS Pathog. 2007;3:674–682. doi: 10.1371/journal.ppat.0030073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsch M, Pourger C, Spirig T, Grauschopf U, Weber-Ban EU, Glockshuber R. Pilus chaperones represent a new type of protein-folding catalyst. Nature. 2004;431:329–333. doi: 10.1038/nature02891. [DOI] [PubMed] [Google Scholar]
- Vetsch M, Erilov D, Moliere N, Nishiyama M, Ignatov O, Glockshuber R. Mechanism of fibre assembly through the chaperone-usher pathway. EMBO Rep. 2006;7:734–738. doi: 10.1038/sj.embor.7400722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell. Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.