Abstract
The eight-residue surface loop, 45–52 (Ser, Ala, Val, Gly, Asn, Ala, Glu, Ser), of the homotetrameric protein streptavidin has a “closed” conformation in the streptavidin-biotin complex, where the corresponding binding affinity is one of the strongest found in nature (ΔG ∼ –18 kcal∕mol). However, in most of the crystal structures of apo (unbound) streptavidin, the loop conformation is “open” and typically exhibits partial disorder and high B-factors. Thus, it is plausible to assume that the loop structure is changed from open to closed upon binding of biotin, and the corresponding difference in free energy, ΔF = Fopen – Fclosedin the unbound protein, should therefore be considered in the total absolute free energy of binding. ΔF (which has generally been neglected) is calculated here using our “hypothetical scanning molecular-dynamics” (HSMD) method. We use a protein model in which only the atoms closest to the loop are considered (the “template”) and they are fixed in the x-ray coordinates of the free protein; the x-ray conformation of the closed loop is attached to the same (unbound) template and both systems are capped with the same sphere of TIP3P water. Using the force field of the assisted model building with energy refinement (AMBER), we carry out two separate MD simulations (at temperature T = 300 K), starting from the open and closed conformations, where only the atoms of the loop and water are allowed to move (the template-water and template-loop interactions are considered). The absolute Fopen and Fclosed (of loop + water) are calculated from these trajectories, where the loop and water contributions are obtained by HSMD and a thermodynamic integration (TI) process, respectively. The combined HSMD-TI procedure leads to total (loop + water) ΔF = −27.1 ± 2.0 kcal∕mol, where the entropy TΔS constitutes 34% of ΔF, meaning that the effect of S is significant and should not be ignored. Also, ΔS is positive, in accord with the high flexibility of the open loop observed in crystal structures, while the energy ΔE is unexpectedly negative, thus also adding to the stability of the open loop. The loop and the 250 capped water molecules are the largest system studied thus far, which constitutes a test for the efficiency of HSMD-TI; this efficiency and technical issues related to the implementation of the method are also discussed. Finally, the result for ΔF is a prediction that will be considered in the calculation of the absolute free energy of binding of biotin to streptavidin, which constitutes our next project.
INTRODUCTION
An important objective of this paper is to further develop our method, the hypothetical scanning molecular dynamics (HSMD) for calculating the absolute entropy, S, and the absolute Helmholtz free energy, F (F = E-TS, where E is the energy and T is the absolute temperature). Calculation of these fundamental thermodynamic quantities is extremely difficult, in spite of the significant progress that has been made in the last 50 years.1, 2, 3, 4, 5, 6, 7, 8, 9, 10 Calculation of F is in particular challenging in structural biology due to the flexibility and strong long-range interactions characterizing bio-macromolecules such as proteins. Thus, the potential energy surface of a protein, E(x), is rugged (x is the 3N-dimensional vector of the Cartesian coordinates of the molecule’s N atoms), i.e., it is “decorated” by a tremendous number of localized wells and “wider” wells (called microstates) defined over regions, Ωm, with each wider well consisting of many localized ones. A microstate Ωm (e.g., the α-helical region of a peptide), which typically constitutes only a tiny part of the entire conformational space Ω, can be represented by a sample (trajectory) generated by a local molecular-dynamics (MD)11, 12 simulation (see further discussions in Refs. 13 and 14). A molecule will visit a localized well for a very short time [several femtoseconds (fs)] while staying much longer within a microstate,15, 16 which is therefore of a greater physical significance. A central aim in protein folding is the daunting task of finding the most stable microstate, i.e., that with the lowest free energy, Fm = –kBT ln Zm, where kB is the Boltzmann constant, and the partition function Zm is integrated over Ωm (rather than over the entire space).
In addition to the difficult problem of protein folding (where interest is typically in a single microstate), more manageable problems are commonly studied, where smaller systems are involved (e.g., cyclic peptides), or the focus is on specific protein regions, such as flexible surface loops, side chains, or bound ligands. These limited systems can populate significantly several microstates, Ωm in thermodynamic equilibrium, which should be identified, and their populations, pm = exp(–Fm∕kBT), calculated. It is of interest to know whether the conformational change adopted by a loop (a side chain, ligand, etc.) upon ligand binding has been induced by the ligand (induced fit17, 18) or alternatively whether the free loop interconverts among different microstates, one of which is selected upon binding (selected fit19). (Notice again that not only is the calculation of pm difficult, but defining a microstate in the high-dimensional conformational space is also not straightforward.) The topic of the present paper is related to this category of problems, as described later in the paper.
Thus, our central aim is to study the relative stability of the “open” and “closed” conformations (microstates) of a mobile loop existing in each chain of the homotetrameric protein streptavidin (see Fig. 1); this loop is found in the “closed” conformation in the streptavidin-biotin complex. To study the loop flexibility and stability, we carry out MD simulations of the protein in water and calculate the differences in free energy and entropy (ΔFmn and ΔSmn, respectively) between these microstates (denoted m and n). Notice, however, that carrying out such calculations with thermodynamic integration (TI) would be practically unfeasible if the structural variance between m and n were significant. Therefore, alternatively (and as in previous work) we use our HSMD method mentioned earlier,20, 21, 22, 23, 24, 25 which enables one to calculate the absolute S and F. Thus, only two (separate) local MD simulations of the loop in m and n are carried out, from which Fm, Fn, and ΔFmn = Fm – Fn (and ΔSmn = Sm – Sn) are obtained directly and the integration process is avoided. (Other methods for calculating the absolute F and S are reviewed, for example, in Ref. 7.)
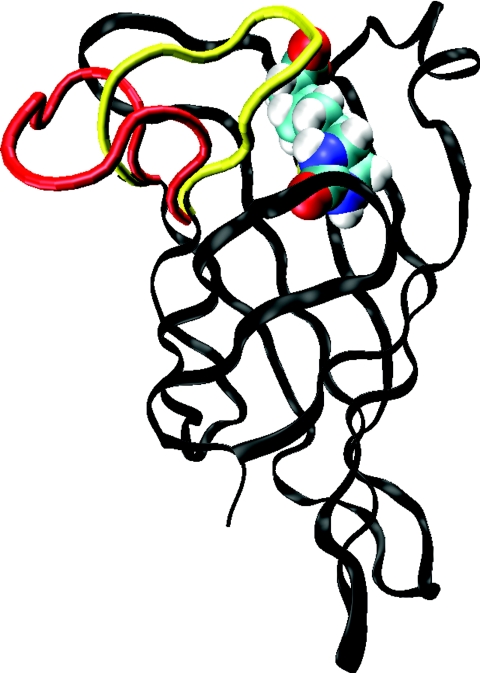
Figure 1.
The conformations of the closed loop (yellow tube) and the open loop (red tube) attached to a single monomer of streptavidin (black ribbon) complexed with biotin. The significant difference between these structures is evident.
With HSMD (or HSMC when Monte Carlo replaces MD), each conformation of a sample is reconstructed step-by-step (from nothing) using transition probabilities (TPs); the product of these TPs leads to an approximation Pi for the correct Boltzmann probability , where from Pi various free-energy functionals can be defined. While the TPs of HSMC(D) are stochastic in nature (calculated by MD or MC simulations), all the system interactions are taken into account (from now on, for simplicity, we shall omit in most cases the letters MC); in this respect, HSMD can be viewed as exact,20 where the only approximation involved is due to insufficient MD sampling for calculating the TPs. HSMD has unique features: it provides rigorous lower and upper bounds for F, which enable one to determine the accuracy from HSMD results alone without the need to know the correct answer. Furthermore, F can be obtained from a very small sample and in principle even from any single conformation (e.g., see the results for argon in Ref. 20). The HSMC(D) methodology has been developed systematically, as applied to systems of increasing complexity. The initial (HSMC) calculations of liquid argon, TIP3P water,20 self-avoiding walks,22 and polyglycine molecules23 have verified the validity of the theoretical predictions stated above by comparisons with accurate results obtained by other well-established techniques; a subsequent application of HSMD to peptides has led to an ∼100 times reduction in computer time.25
HSMD has also been applied to mobile loops which change their structure due to ligand binding. We studied initially the seven-residue surface loop, 304–310 (Gly-His-Gly-Ala-Gly-Gly-Ser), of the enzyme porcine pancreatic α-amylase in vacuum and in the generalized Born surface area (GB∕SA) implicit solvent.13 In a subsequent paper, the loop was capped with 70 TIP3P water molecules and an HSMD-TI procedure was developed in which the contribution of water to F is calculated by a TI process, which is more efficient than HSMD.14 Subsequently, HSMD-TI was applied to the loop 287–290 with the bulky residues, Ile, Phe, Arg, and Phe of acetylcholinesterase (AChE) from Torpedo California;26 reaction of AChE with the inhibitor diisopropylphosphorofluoridate (DFP) leads to a displacement of the loop’s backbone by roughly 4 Å, and experimental evidence suggests that the free-energy penalty for the loop displacement is on the order of 4 kcal∕mol (i.e., Ffree – Fbound ∼ −4 kcal∕mol). Therefore, AChE has been an ideal system for checking the performance of HSMD-TI, and in particular for examining the minimal number of water molecules needed to cap the loop. We have found that to recover the experimental free-energy difference, the water density should be close to that of bulk water, and our results, Ffree – Fbound = −3.1 ± 2.5 and −3.6 ± 4 kcal∕mol for a sphere containing 160 and 180 waters, are equal within error bars to the experimental value.26
The present study constitutes an additional step in the application of HSMD-TI to mobile loops. Streptavidin (isolated from the bacterium Streptomyces avidinni) is a tetrameric protein consisting of 159 residues per chain, where each subunit can bind noncovalently one biotin molecule (C10H16N2O3S). This binding (characterized first by Chaiet and Wolf27) is of extraordinarily high affinity (Ka ∼ 1013 M−1, ΔG ∼ –18 kcal∕mol)—one of the strongest found in nature.28 It is a property that has been exploited to devise powerful tools for affinity chromatography, biochemical assays, and many other application.29, 30, 31, 32 Biotin binds to the open end of the β barrel of each subunit of streptavidin, and an eight-residue surface loop (45–52) (Ser, Ala, Val, Gly, Asn, Ala, Glu, Ser) folds so as to cap the barrel (Fig. 1). This loop, which usually has an “open” structure in apo streptavidin, has been identified before as a major factor in the affinity of streptavidin to biotin.31 Deletion of the loop via circular mutation decreases the binding affinity by approximately 10 kcal∕mol.33
The x-ray structures of streptavidin complexed with biotin were first determined in 1989 by Hendrickson et al.34 and Weber et al.,31 where the latter study also provided the structure of apo streptavidin and its comparison with the complexed one. Today the protein data bank (PDB) contains 134 crystal structures of wild-type and mutated streptavidin, complexed with biotin and other ligands. To calculate the free-energy difference between the closed and open microstates of the loop, one needs to know the corresponding crystal structures. However, the open structure in apo streptavidin is (in most cases) partially disordered and the closed structure appears with a bound biotin. We were able to overcome these hurdles by suitably matching structures of the loop and protein taken from a set of crystal structures of the free and complexed streptavidin obtained by Freitag et al.,30 as described in detail in the next section.
Finally, it should be noted that the eight-residue loop and the 250 water molecules capping it constitute the largest system treated by HSMD-TI thus far. Testing the performance of HSMC-TI for systems of increasing size (N) is important due to the N1∕2 increase in the fluctuation of the absolute entropy and energy. In this paper, we suggest and test new reconstruction procedures for a loop. Our results shed more light on the somewhat unusual structural behavior of the loop of streptavidin exhibited in crystal structures. Also, the free-energy difference between the closed- and open-loop microstates (in the apo protein) should be taken into account in the calculation of the absolute free energy of binding of biotin to streptavidin. Thus, the present study is the first step in our long-range goal of developing HSMD-TI as a tool for calculating the absolute free energy of binding, where we intend to apply HSMD-TI initially to the streptavidin-biotin and avidin-biotin complexes.
THEORY AND METHODOLOGY
Definition of the system
As pointed out earlier, we study the surface loop of Nres = 8 residues (45–52) (Ser, Ala, Val, Gly, Asn, Ala, Glu, Ser) of streptavidin. While the loop exists in each of the four subunits, HSMD-TI is applied only to a single loop of one subunit of the unbound protein. However, even such a limited treatment might be problematic because in the unbound structures of streptavidin which appear in the PDB, the (open) loop conformations are typically disordered, as can also be learned from the five x-ray structures (I–V) obtained by Freitag et al.30 under various crystallization conditions. Indeed, in structures I–IV there are 14 unbound subunits with 12 loops in the open conformation, where for 10 of these loops at least three residues could not be traced in the electron density maps and the other residues appear with elevated B factors. Only in structure III (PDB 1swc) were Freitag et al. able to resolve the free loop conformations of subunits 2 and 4 with average B factors of 33.8 and 48.6 Å2, respectively. It should also be noted that the loop of subunit 1 of the unbound structures I and II appears in the closed conformation, a fact that is attributed by Freitag et al. to crystal-packing interactions; however, the existence of closed conformations might suggest that the structural preference of the loop in unbound streptavidin is somewhat uncertain or that the loop responds to binding by a selected fit process.
We have a special interest in structure IV (1swd), in which two biotin molecules are bound to subunits 1 and 4; as expected, the corresponding loops are closed, and the loops of the unbound subunits 2 and 3 have partially disordered open conformations. We decided to use subunit 2 of structure IV as a basis to which initial open- and closed-loop structures will be attached for further optimization and MD simulations. First, we deleted the original loop coordinates of subunit 2 of IV and attached instead the closed-loop conformation of subunit 1 of IV by superimposing the structure of subunit 1 on subunit 2 (both of IV); similarly, the open-loop conformation of subunit 2 of III (1swc) (which has lower B-factors than the loop of subunit 4 of III) was attached to the same (subunit) basis to create an initial open-loop conformation; we denote these two tetramer structures as A and B for the closed and open loops, respectively. In both cases, the incomplete open loop in subunit 3 was also replaced by the open loop in subunit 2 of structure III. Notice that in the crystal structure of IV, only the coordinates of the (core) residues, 16–133, are provided.
Before discussing the various stages of system optimization and simulations, it should be pointed out that all calculations were performed with the AMBER99 force field35 where the amino acids Lys, Arg, Glu, Asp, and His are charged. The MD simulations were carried out in the NVT ensemble where some of the preliminary results of the tetramer were obtained with the AMBER 10 package;36 however, implementation of HSMD-TI is more convenient with the TINKER 5.0 program,37 therefore it was used in all of the free-energy calculations. The temperature was kept around 300 K with the Berendsen thermostat based on a time constant of 1.0 ps.38 The time step employed in the MD simulations was 2 fs, applying the RATTLE algorithm to constrain bonds involving hydrogen atoms.38 No periodic boundary conditions or cutoffs were used unless otherwise stated. The TIP3P model for water was used.39
Structural optimization for the free-energy calculations
We carried out two sets of MD simulations. In one set, which is aimed at testing the stability of the loop, the entire tetramer (soaked in water) was treated (see Sec. 3A). However, for calculating the free energy of the loop, only part of the protein was considered; below we define this partial system and describe its optimization, which constitutes the starting point for a set of simulations described in Sec. 3B.
The structural optimization started from structure B (1swd with the open-loop conformation attached to subunits 2 and 3). This structure was solvated with TIP3P water molecules in a sphere of radius Rwater = 15 Å around the center of the loop of subunit 2; to hold these water molecules around the loop, they were restrained with a flat-welled half-harmonic potential [a force constant of 10 kcal∕(mol Å2)] based on the distance from the “center” of the loop region. That is, the distance of each water molecule (in practice, the oxygen atom) is measured from the restraining center. If this distance is greater than 15 Å, a harmonic restoring force is applied; otherwise, the restraining force is zero. Also, harmonic restraints with a force constant of 5 kcal∕(mol Å2) were applied to the heavy atoms of the protein to eliminate bad atomic overlaps and strains in the original structure, while still keeping the atoms reasonably close to the PDB coordinates. The system was energy-minimized using 104 steps of steepest descent followed by 104 steps of conjugate gradient. The root-mean-square deviation (RMSD) values between the PDB coordinates of the structure without the loops before and after minimization are 0.15 Å for the backbone and 0.29 Å for the backbone and side chains, meaning that most of the change is due to the side-chain motion.
As in our previous HSMD-TI applications to loops, to reduce computer time we do not consider the entire protein but only a spherical part of it (the template) which is the closest to the loop and whose coordinates are held fixed during future simulations of the loop. To define this system, we first removed the water molecules from the minimized tetramer configuration obtained previously and defined a spherical template with a radius of 18 Å, centered at the middle point on the line connecting the α-carbon atoms of the first and last residues of the loop attached to subunit 2 of 1swd. Thus, if the distance of any atom of a residue from the middle point is less than 18 Å, the entire residue is included in the template; otherwise, the residue is eliminated. This “cutting” procedure reduced the number of protein atoms considered from 6805 to 957, representing a significant gain in simulation speed. We should note that most of the residues in the spherical cut belong to subunit 2, but some correspond to other subunits. Nevertheless, it is important to keep these extra residues because through visualization of the PDB files it became clear that they form a “wall” over the β barrel of subunit 2 which prevents the loop from moving to regions that are not available in the case of the whole tetramer.
Next, the loop was solvated with 250 TIP3P water molecules, distributed in a spherical cap with a radius of Rwater = 15 Å. Initially we used the same spherical center defined above for the template, but have found our template to be too “thin,” i.e., during MD simulations water molecules could “seep” through cavities in the template or around it to its “back side.” To avoid this undesired situation, we shifted the water center by 3.3 Å toward the “loop side” of the loop-template system, which was found to be the smallest distance necessary to prevent this effect. [Such a shift would not be needed for larger templates like that defined for the larger (single chain) protein AChE of 535 residues;26 see also Refs. 14 and 40]. The number of water molecules in the cap (250) was determined by the condition that their density is close to the bulk density of water in normal conditions of pressure and temperature (0.0350 Å−3); the actual density, 0.0355 Å−3, is indeed close to the target value. (Notice that the volume in which the waters move is not the full spherical volume, but a smaller one due to the presence of the loop and part of the template; it is about half the total volume.) The water molecules were restrained to stay in the spherical cap by a harmonic force with a force constant of 10 kcal∕(mol Å2) as described previously.
In this system, the template coordinates are always held fixed, i.e., only the loop and water atoms are allowed to move (no harmonic restraints were applied to the loop). Thus, the total potential energy Etotal is a sum of partial energies (the constant template-template energy is ignored),
| (1) |
where Eloop-loop is the intra loop energy and Eloop-tmpl is the energy due to the loop-template interactions; these energies define the total loop energy, Eloop. The interactions related to water are defined in a similar way, where their total is denoted by Ewater. This system was energy-minimized using the same procedure mentioned before, and equilibrated in a 500 ps MD simulation. This optimization procedure was also applied to structure A (1swd with its open subunit 2 loop replaced by its closed subunit 1 loop). Hence, both loops share exactly the same “frozen” template and the same 15 Å water sphere with 250 water molecules. The last loop∕water configurations of the 500 ps simulations for the open and closed loops become starting configurations for 2 ns “production” runs from which the free energy is calculated (see Secs. 3B, 3C).
One might consider our model to be limited as it is based on a partial frozen template (which reduces the system size, keeping the fluctuations of E and S manageable). However, this model is expected to be adequate, as a recent 250 ns MD simulation of the bound streptavidin tetramer in solution by Cerutti et al.41 has found the backbone of the 67 core residues of each subunit to be very stable, i.e., with RMSD values smaller than 0.7 Å; stronger fluctuations were observed there only in the loop regions. Also, as pointed out earlier, our free-energy results for the loop of AChE based on a frozen template are in a very good agreement with experiment.26 Since HSMD-TI can also treat a fluctuating template, we intend to test it in future studies, initially for a template whose heavy atoms are restrained by harmonic forces.
Statistical mechanics of a loop in internal coordinates
The theory of HSMD-TI has been developed in previous publications; therefore, we describe it briefly providing mainly equations that are related directly to the calculations. The reconstruction of the loop structure (of Nres = 8 residues) is carried out in internal coordinates; therefore, the loop conformations simulated by MD are transferred from Cartesians to the dihedral angles φi, ψi, and ωi (i = 1, Nres), the bond angles θi,l (i = 1, Nres, l = 1, 3), the side-chain angles χ, and the corresponding bond angles. For convenience, all these angles (ordered along the backbone) are denoted by αk, k = 1, K; as discussed in Sec. 2G, in this work we define two reconstruction procedures: one is based on K = 64 and the other on K = 104. We have argued in Refs. 13 and 25 that to a good approximation, bond stretching can be ignored, thus the bond lengths are considered to be constant.
The partition function of the loop∕water∕template system is
| (2) |
where E(xloop, xN) = Etotal is defined in Eq. 1, xloop are the Cartesian coordinates of the loop in microstate m, and xN are the 9Nwater Cartesian coordinates of the water molecules; Etotal also depends on the “frozen” template coordinates, which are omitted for simplicity. For the same reason, the letter m will be omitted in most of the equations and Nwater will be replaced in the theoretical section by N(N = Nwater). After changing the variables of integration from xloop to internal coordinates, the integral becomes a function of the K dihedral and bond angles, αk, k = 1, …, K, and a Jacobian cos(θi,l) that depends only on each of the bond angles θi,l,42, 43, 44
| (3) |
where [αk]=[α1 ,...,αK] and d[αk]=dα1 ...dαK . In Eq. 3, we have omitted a factor that depends on the bond lengths and is assumed to be the same (i.e., constant) for different microstates of the same loop and therefore does not affect entropy differences (e.g., see the discussion in Ref. 26). The Jacobian is also omitted for simplicity because we have shown13 that it cancels out (within the error bars) in entropy and free-energy differences (this conclusion was checked again and verified in the present study). The Boltzmann probability density corresponding to Z [Eq. 3] is
| (4) |
and the exact entropy S and exact free energy F (defined up to an additive constant) are
| (5) |
and
| (6) |
It should be noted that the fluctuation of the exact F is zero,45, 46 because by substituting ρB([αk ]) [Eq. 4] inside the curly brackets of Eq. 6, one obtains E([αk ])+kBTlnρB ([αk ])=−kTlnZ=F, i.e., the expression in the curly brackets is constant and equal to F for any set [αk ] within m. This means that the free energy can be obtained from any single conformation if its Boltzmann probability density is known. [Notice, however, that the calculation of ρB ([αk ],xN ) for a single conformation depends on the entire microstate, as is also evident from the HSMC(D) procedure discussed later.] Still, the fluctuation of an approximate free energy (i.e., one based on an approximate probability density) is finite and is expected to decrease as the approximation improves.45, 46, 20, 21, 22 Because HSMC(D) provides an approximation for ρB ([αk],xN ), one can, in principle, estimate the free energy of the system from any single structure.20, 21, 22 This is the reason why in practice reliable HSMC(D) results for F (but not necessarily for S and E) can be obtained from a relatively small sample.
Exact stochastic future scanning procedure
It should first be pointed out that the Metropolis Monte Carlo (MC) and MD are exact dynamical methods that enable one to sample system configuration i correctly with its Boltzmann probability, PiB, while the value of PiB is not provided (due to the dynamical character of these methods). [To simplify the discussion, we use the probability PiB rather than the probability density ρB ([αk ],xN ) defined in Eq. 4.] Thus, properties such as the energy that are “written” on i can easily be calculated, while a direct calculation of the absolute S is difficult because lnPiB is unknown (it depends not only on i but on the entire ensemble through the partition function Z, which cannot be obtained from a finite sample).
Unlike the dynamical MC and MD, the exact future scanning method, which constitutes the basis of HSMC(D), is a growth procedure that enables one (at least in principle) to generate any system configuration (including fluids) from nothing by determining the atoms’ positions step-by-step with the help of TPs; the product of these TPs leads to the value of PiB, and hence to S and F. Practically, a loop∕water∕template configuration would be generated by initially building a loop structure (in the presence of moving water) followed by the construction of a configuration of the surrounding water molecules (in the presence of a fixed loop conformation). In this way, a sample of statistically independent system configurations can be obtained.47
For simplicity, this construction is described for a loop without side chains, consisting of M Gly residues, i.e., ordered along the chain are the 3M heavy atoms denoted k′ = 1, …, 3M and the 6M dihedral and bond angles denoted αk, 1 ≤ αk ≤ 6M = K, with values within microstate m; the loop is surrounded by Nwater water molecules moving within the volume defined by a sphere of radius, Rwater, the template, and the loop. We seek to generate a configuration of the entire system by first generating a loop conformation and then a configuration of the water molecules.
With the scanning procedure, the position of the heavy atoms (xloop) is determined step by step. Thus the position of the first atom k′ = 1 is defined by the simultaneous determination of the first pair of dihedral and bond angles α1, and α2. The maximum range Δα1Δα2 which will keep the loop within m is defined, and each of Δα1 and Δα2 is divided into nb small bins (of sizes Δα1∕nb and Δα2∕nb ) denoted j1 and j2, j1 = 1,...,nb , j2 = 1,...,nb , respectively. A long MD simulation of the whole system (loop + water) is carried out within microstate m, where a conformation is retained every l fs leading to a huge sample of size n; then, the number of conformations nj1j2 that visit simultaneously the (double) bin j1j2 is calculated from which the corresponding TP is obtained, pj1j2 = nj1j2 ∕n (or ρj1j2 = [nj1j2 ∕n]∕[Δα1Δα2∕]). A double bin is then selected by a random number according to the pj1j2 which defines the position of atom k′ = 1 (and its hydrogen or oxygen). The position of this atom is not changed in the next steps of the build-up process, i.e., it becomes part of the “past”. The position of the second atom (k′ = 2) is determined in the same manner from a long MD simulation of the future part of the system (i.e., atoms k′ = 2, …, 3M and water) where α3 and α4 are considered, bins Δα3∕nb and Δα4∕nb are defined, probabilities are calculated, and a “lottery” (like above) determines the values of α3 and α4 which define the position of atom k′ = 2; the process continues until the positions of all the loop’s atoms (and their hydrogens or oxygens) have been determined. A configuration of the Nwater molecules is then determined in a similar way step by step in the presence of the fixed loop structure previously constructed (for details, see Ref. 20). Obviously, the smaller the bins are, the higher is the accuracy of the construction process, provided that the statistics is adequate, i.e., that the (future) MD simulations are long enough; this stochastic scanning method becomes exact as the bin size →0 (nb → ∞) and n → ∞. Notice that in applications of the (deterministic) scanning method to lattice models, only part of the future has been considered (i.e., only f steps ahead), where this part has been scanned completely; therefore, the corresponding TPs are approximate but deterministic (rather than stochastic), and accurate results were obtained by using an additional importance sampling procedure.47
This procedure can be described more formally as follows: at step k′ (k = 2k′), the positions of k′ − 1 atoms have already been determined (from the values of the corresponding k − 2 angles α1, …, αk–2) and they are kept fixed (defining the “past”); αk–1 and αk (which will determine the position of atom k′) are defined with the exact TP density ρ(αk−1 αk |αk−2,...,α1)
| (7) |
where Zfuture (αk ,...,α1 ) is a future partition function. The term “future” indicates that the integration defining Zfuture is carried out over the positions of atoms k′ = k∕2 + 1, …, K∕2 (which affect angles, αk+1 ,...,αK ) and the 9N coordinates xN of the water molecules (which will be determined in future steps of the build-up process). Notice that this integration is carried out in a restrictive way where the corresponding conformations (of the loop) remain within microstate m. Also, in this integration the atoms treated in the past (1,...,k′ − 1) (which were determined by α1 ⋯αk–2) are held fixed in their coordinates. For simplicity, the integrations below are written over the angles rather than the Cartesian coordinates (xloop) of the loop atoms, k′ = k∕2 + 1, …, K∕2. Thus
| (8) |
where E [Eq. 1] is the total potential energy of the loop∕ template∕water system, which also imposes the loop closure condition. The product of the TPs [Eq. 7] leads to the (Boltzmann) probability density of the entire loop conformation, . After the loop structure has been constructed, a configuration of water molecules is generated step by step [in the presence of the constant loop structure where the product of the corresponding TPs leads to the probability density of the water configuration, . The probability density ρB ([αk ],xN ) of the loop∕water∕template configuration is the product of and ]. One can define for m “the loop entropy of mean force,” Sloop ,
| (9) |
where is defined up to an additive constant. Extending the exact scanning procedure to side chains is straightforward, where again the position of a side-chain atom is defined by two angles as described previously.
However, implementation of the exact scanning procedure [as described prior to Eq. 7] for generating Boltzmann-weighted configurations of a large loop∕water∕template system would be inefficient, due to the need to calculate (at each step) a large set of accurate TPs (i.e., for an extremely large number of small bins); this would require extremely long (future) MD simulations. Also, with long simulations it is difficult to guarantee that the loop will remain in microstate m (see the discussion in Ref. 13). However, the exact scanning procedure provides the theoretical basis for HSMC(D). Thus, the exact scanning method is equivalent to any other exact simulation technique (in particular, to MC or MD) in the sense that large samples generated by such methods lead to the same averages and fluctuations (the sample does not carry a memory of the simulation method with which it has been generated). Therefore, one can assume that a given MC or MD sample has rather been generated by the exact scanning method, which enables one to reconstruct each conformation i by calculating the TP densities that hypothetically were used to create it step by step. With HSMC(D), the efficiency problems discussed earlier for the exact scanning procedure are alleviated to a large extent since only a single TP is calculated at each step [rather than many TPs (pj) required with the scanning method], as described below; also, because we are mainly interested in entropy differences, approximations (e.g., ignoring the Jacobians and bond stretching) can be applied without compromising the accuracy of the results.
The HSMC(D) method
The theory of HSMC(D) is described again for a loop consisting of M Gly residues. Notice that while HSMD and the exact scanning method (described in the previous section) have some resemblance, they are different. With the scanning method a loop conformation is generated with whereas with HSMD an already generated structure (by MD) is reconstructed in order to obtain its probability. Thus, one starts by generating an MD sample of the loop in water in microstate m; the configurations of this sample will be reconstructed by HSMD (application of HSMC is similar). First, the conformations are represented in terms of the dihedral and bond angles αk,1 ≤ αk ≤ 6M = K, and the variability range Δαk is calculated,
| (10) |
where αk(max) and αk(min) are the maximum and minimum values of αk found in the sample, respectively. Δαk, αk(max), and αk(min) enable one to verify that the sample has not “escaped” from microstate m. Notice that in the discussion below, we define the loop conformation by the set of angles [αk] rather than by the positions of the loop atoms, xloop, which are determined by [αk].
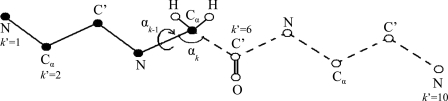
Each of the configurations (frames) ([αk ],xN ) (denoted i for brevity) of the sample is reconstructed in two stages, where the biotin structure is reconstructed first followed by the reconstruction of the water configuration xN. Because the position of atom k′ is defined by a dihedral and a bond angle, one has to calculate their TP simultaneously. Thus, at step k′ (k = 2k′) of stage 1, the k−2 angles αk−2 ⋯α1 have already been reconstructed and the TP density of αk−1αk , ρ(αk−1αk|αk−2 ,...,α1 ), is calculated from an MD run, where the entire future of the loop and water is moved (i.e., the loop’s atoms k′, k′+1, …, K∕2 and their connected hydrogens, and the water coordinates xN) while the past (loop’s atoms 1,2,...,k ′ −1 and their connected hydrogens) are held fixed at their values in conformation i. By considering a future conformation every 6 fs, a sample of size nf is generated. Two small segments (bins) δαk-1 and δαk are centered at αk-1(i) and αk(i), respectively, and the number of simultaneous visits, nvisit, of the future chain to these two bins during the simulation is calculated; one obtains (see Fig. 2)
| (11) |
where ρHS (αk−1 αk|αk−2 ,...,α1 ) becomes exact for very large nf (nf → ∞) and very small bins (δαk-1, δαk → 0). This means that in practice ρHS (αk−1αk|αk−2 ,...,α1 ) will be somewhat approximate due to insufficient future sampling (finite nf) and relatively large bin sizes. In Ref. 13, we have shown that δαk and δαk+1 can be optimized. Notice that with HSMD the future loop conformations generated by MD at each step k‘ remain in general within the limits of m, which is represented by the analyzed MD sample. The corresponding probability density related to the loop is
| (12) |
ρHS ([αk]) defines an approximate entropy functional, denoted , which can be shown (using Jensen’s inequality) to constitute a rigorous upper bound for [Eq. 9],20, 48
| (13) |
[Eq. 9] is the Boltzmann probability density of [αK] in m. ( is also known as Gibbs’ inequality.) Being an upper bound suggests that will decrease as the approximation improves. In practice, however, this inequality is satisfied only if the probabilities are well defined (e.g., they are correctly normalized). This is pointed out because ρHS (αK ,...,α1 ) is calculated stochastically and its error [and the error in , the estimation of , see Eq. 14] increases with increasing system size (i.e., increasing the number of TPs; see later discussions in Secs. 2F, 3C).
Figure 2.
An illustration of the HSMD reconstruction process of conformation i of a peptide consisting of three glycine residues, where for simplicity the oxygens and most of the hydrogens are discarded. At step k′ = 6, the TPs related to the “past” atoms k′ = 1, …, 5 (depicted by full spheres) have already been determined and these atoms are kept fixed in their positions at conformation i. At this step (6) one calculates the TPs of bond angle αk (defined by C′-Cα-N) and dihedral angle αk-1 (defined by C′-Cα-N-C′) which are related to C′, where k = 2k′ and thus k = 12. The TPs are obtained from an MD simulation where the as yet unreconstructed atoms k′ = 6, …, 10 (the “future” atoms) are moved (depicted by empty spheres connected by dashed lines) while the past atoms k′ = 1, …, 5 are kept fixed; note that the future part should remain within the limits of the microstate and future-past interactions are taken into account. Small bins δαk-1 and δαk are centered at the values αk-1 and αk in i. The TP is calculated from the number of visits [nvisit, Eq. 11] of the future part to δαk-1 and δαk simultaneously during the simulation. After ρHS (α11 α12|α10 ,...,α1) [Eq. 12] has been determined, the coordinates of C′ (and O) are fixed at their positions in i, i.e., they become “past” atoms and the process continues.
can be estimated (for microstate m) from a Boltzmann sample (of size ns) generated by MD using the arithmetic average,
| (14) |
where ρHS (t,m) is the value of ρHS ([α k ]) obtained for configuration t of the sample of m. (with the bar) is an estimation of the ensemble average [Eq. 13]; correspondingly, the ensemble averages of the energy are estimated from a sample of size ns and should appear with a bar as well. However, from now on only estimations will be considered, and for simplicity, all of them will appear without the bar, like the energies defined in Eq. 1. [Eqs. 13, 14] constitutes a measure of a pure geometrical character for the loop flexibility, i.e., with no direct dependence on the interaction energy. When the converged or the best value of is considered, it will be denoted by Sloop; thus, Floop = Eloop− TSloop is defined as the loop’s contribution to the total free energy, where Eloop is defined in Eq. 1. In the same way, the difference in the loop entropies between the open (o) and closed (c) microstates obtained for a specific set of parameters is denoted by while the converged values are denoted without “A” (i.e., ) and their difference is denoted by ΔS loop ,
| (15a) |
where
| (15b) |
One can define a free-energy difference for the loop, ,
| (16) |
whereΔEloop is obtained from Eq. 1.
To reconstruct the water configuration, one can use in principle the HSMC(D) procedure for fluids mentioned previously,20 which would lead to and then to the contribution of the water configuration to the free energy, . However, this procedure for fluids has not been optimized yet and it is relatively time consuming.
Alternatively, as in Refs. 14 and 26, one can obtain Fwater ([αk ],xN ) by a TI procedure based on the same reference state for the open and closed structures. In this state, the water-water and water-template interactions are preserved but the (fixed) loop structure [αk ] does not “see” the surrounding waters, i.e., the loop-water interactions [electrostatic and Lennard-Jones (LJ)] are switched off. These interactions are gradually increased (from zero) during an MD simulation of water (while the loop structure remains fixed at [αk ]). For [αk ] of microstate m the integration leads to , which is then averaged over the ns sample configurations [as in Eq. 14]. The integration is performed in two stages but in an opposite direction to that described above, i.e., first the charges are gradually decreased to zero (by decreasing the parameter λ from 1 to zero; see Sec. 3D), followed by a similar decrease in the LJ potential, leading to and , respectively. Denoting the set of [αk ] in the sample by t and omitting m, one obtains
| (17) |
The difference in the free energy of water between the open and closed microstates is
| (18) |
and the difference in the total free energy between the open and closed microstates is
| (19) |
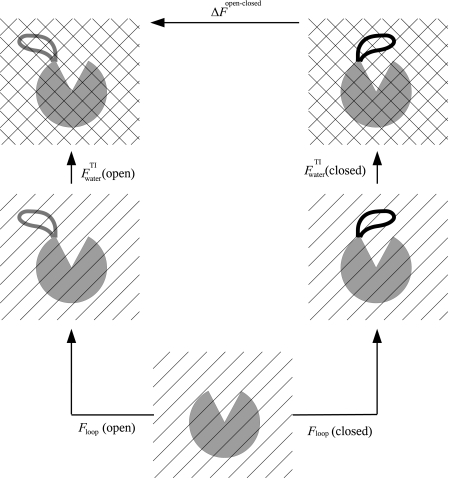
The corresponding thermodynamic cycle is described in Fig. 3, which shows that the free-energy calculation for both loops starts from the same reference state (0) of a (fixed) template surrounded by water, i.e., the equilibrium state is defined by water-water and water-template interactions. In the first step, ns loop structures are reconstructed independently in water and the corresponding free energies, Floop(o) and Floop(c), are obtained for the open and closed loops. The hatched background in the reference state and the first stage demonstrates that the free energy of water is not considered. In the second step, the loop-water interaction is gradually decreased to zero by the TI procedure described above for the ns configurations, where the (negative) average results are and ; the crossed-hatched background here demonstrates that the free energy of water is considered. The sums of the loop and water contributions are Ftotal(o) and Ftotal(c), and their difference is ΔFtotal = Ftotal(o) − Ftotal(c) [Eq. 19].
Figure 3.
A schematic illustration of the thermodynamic cycle used to calculate the difference in free energy between the open and closed microstates. For both microstates, the calculations start from the same reference state: a fixed template (gray partial circle) soaked in water (parallel lines) which appears in the bottom. In step 1, a set of 40 loop conformations selected (for each microstate) from a 2 ns MD trajectory are reconstructed in water by HSMD leading to the loop contribution to the free energy, Floop = Eloop − TSloop, where Eloop [Eq. 1] consists of the loop-loop and loop-template energy and Sloop is the converged results of the entropy defined in Eqs. 13, 14; the parallel lines indicate that the loop is soaked in water, which affects its behavior, while the free energy of water is not considered directly. In step 2, the contribution of water to the free energy, [Eq. 17], is calculated for each of the 40 configurations by a thermodynamic integration procedure where the loop-water interactions are increased gradually from zero (the reference state) to their full value (in practice these interactions were decreased to zero). The squared background means that the free energy of water is calculated. The total free-energy difference is [Eq. 19].
Analysis of the reconstruction results
First, notice that the heavy atoms are ordered along the loop and denoted k′ = 1, …, K∕2. Because in the reconstruction of step k′ the whole future is simulated (i.e., atoms k′, k′+1, …, K∕2), one can order the atoms in different ways, e.g., residue by residue, or the entire backbone first and the side-chain atoms later. In the present work, the atoms are considered residue by residue where for each residue the backbone is treated before the side chain. Also notice that the position of atom k′ is defined by the bond and dihedral angle, αk−1 and αk, where k = 2k′.
The MD simulation (in water) of the future chain at step k′ starts from conformation i, which we want to reconstruct, and every g = 6 fs the current conformation is retained; the ninit initial retained conformations are discarded for equilibration. For each of the next nf (retained) future conformations (i.e., based on the positions of the atoms k′, k′ + 1, …, K∕2) the dihedral and bond angles, αk−1 and αk, are calculated and if both are found to be within their corresponding bins, nvisit [Eq. 11] is increased by 1, which leads to the TP, ρloop (αk−1 αk|αk−2 ,...,α1 ) [Eq. 11]; when the entire loop structure has been reconstructed (i.e., the TPs have been calculated for the K∕2 pairs of angles), one obtains an estimation for the probability density of this structure, ρHS (αK ,...,α1 ) [Eq. 12]. Notice that in practice, one might encounter two opposite scenarios of over- or undercoverage of the future part of m.
Thus, if nf is too large, the loop might “overflow” to a neighbor microstate, leading to a number of counts (nvisit) which is too small, hence to too small TPs and probabilities, ρHS (αK ,...,α1 ); the larger nf is, the smaller are these probabilities. Therefore, the corresponding values of [Eq. 14] will increase with increasing nf rather than decrease as expected by theory for improving approximations [see the discussion following Eq. 13]. [Note that even at step k′, where the “past” of the loop (atoms 1,...,k′ −1) is kept fixed, the (future) unfixed part (k′ ,...,K∕2) can leave the microstate during long MD simulations; such an “overflow” is more likely to happen for small residues such as Gly and for small k.] To control an overflow, we have suggested carrying out the reconstruction in j shorter repetitive “units,”13, 14, 25 each based on < nf conformations where nf = jn′f. Using units of increasing length () and larger values of nf (i.e., larger j) enables one to gain control on the extent of coverage of a microstate by the future simulations (again, very small values will lead to undercoverage of the microstate while large might lead to an overflow). Obviously, each unit should start from the reconstructed structure i with a different set of velocities followed by an adequate equilibration of size ninit; an important test for an adequate coverage is verifying that decreases with increasing j [i.e., improving the estimation of ρHS (αK ,...,α1 )] which indeed has been found in several previous studies.13, 14, 23, 25
In the second scenario, the future conformation is located within m but nf is too small for adequately calculating a TP [Eq. 11], i.e., the ratio nvisit∕nf as yet has not been stabilized. As an example, consider a χ angle which visits (in m) more than one rotamer or all of them (i.e., Δαk = 360°); the corresponding ratio nvisit∕nf will decrease systematically as nf is increased, since most of the sampled angles will fall out of the considered bin, meaning that stabilization will occur only for very large nf. Thus, for practical nf values, TP(nf) will also decrease with increasing nf and if the number of such angles is significant they might lead to a systematic increase of .
In a normal situation, however, increasing nf will lead to an improved estimation of the TPs, and hence of ρHS , and the corresponding results for are expected to decrease as required by theory, provided that the probabilities are well defined [see the discussion following Eq. 13]. Indeed, in Sec. 3C we discuss a case where insufficient equilibration (i.e., too small ninit) leads to unnormalized ρHS which is followed by a systematic increase in as nf is increased.
Therefore, the analysis of the results for requires caution. It should first be noted that our main interest is in the differences between microstates m and n rather than in the absolute values themselves. For any practical set of nf and bin sizes, δαk, and will be approximate and thus their difference, , might be approximate as well. However, if is found to be stable for significantly improving sets of parameters, the stable value can be considered as the correct difference (within the statistical errors). Indeed, in applications of HSMD to peptides25 and loops,13, 14, 26 relatively small values of nf have already led to stable differences, meaning that the systematic errors in both and are comparable and thus are canceled in [we define the deviation, −Sloop as the systematic error]. In Ref. 13, we have provided theoretical arguments supporting this error cancellation, which, however, should be verified for each system studied.
Thus, using HSMC(D)-TI, the objective is not to obtain the most accurate free energies, Fm and Fn, but to minimize computer time by finding the worst HSMC(D)-TI approximations for Fm and Fn for which their difference is still correct within a required statistical error. It should be pointed out that the difficulties involved in the definition of a microstate discussed above are not characteristic only for HSMD-TI but are common to all methods for calculating entropy.
Various implementations of the reconstruction procedure
A somewhat technical issue is how to program the reconstruction procedure in the most general way that will be applicable to loops of different sequence and length. We describe two such procedures, where the reconstruction is performed atom by atom, and hence an order of the atoms along the chain should initially be determined, as discussed earlier.
Thus, at step k′ atom k′ is treated and the related dihedral and bond angles, αk-1, αk (k = 2k′), are obtained in the usual way from the positions of atoms k′, k′−1, k′−2, and k′−3, and k′, k′−1, and k′−2, respectively (see Fig. 2). In general, all heavy atoms and polar hydrogens are taken into account, but one can consider approximations based on a smaller number of atoms or include methyl hydrogens if, for example, the rotational entropy of the methyl groups of valine is of interest. In this way, if C′ (k′ = 4) is treated and the previous atoms are Cα (k′ = 3), N (k′ = 2), and H(N) (k′ = 1), these last three atoms are held fixed, while C′ and the atoms numbered k′ > 4 move in the MD reconstruction simulation; nvisit is calculated for the dihedral ψ and the bond angle defined by the positions of C′, Cα, and N. After the reconstruction simulation for k′ = 4 has been completed, C′ becomes fixed in its position at conformation i and O(C′) (k′ = 5) should be treated next (where C′, Cα, and N are its previous atoms to be considered); thus, the dihedral angle is ψ and the bond angle is defined by the positions of O, C′ and Cα. Notice, however, that after O has been fixed, the position of the next N (k′ = 6) is determined to a large extent as well. Here we define two procedures, the “conventional” procedure I and procedure II.
With procedure I, the atom O(C′) is not considered (i.e., O is not numbered) and ψ is defined by N(k′ = 5), C′ (4), Cα(3), and N(2) and the corresponding bond angle by N(5)-C′ (4)-Cα(3), where O [like N(5)] is allowed to move in the simulation because these angles also define the position of O to a large extent. With procedure II, on the other hand, O(C′) is considered as described above followed by the treatment of N (k′ = 6) for which the pair of angles are defined now by the positions of N (k′ = 6), O (k′ = 5), C′ (k′ = 4), and Cα (k′ = 3); however, these angles are not the conventional ones since the dihedral angle is defined around C′O and the bond angle N-O-C′ is based on the nonstandard N-O bond.
While procedure II is somewhat more accurate than procedure I, its implementation is more time consuming (more atoms and angles) and the contribution of the additional reconstructed angles to the entropy is expected to be comparable in different microstates. Therefore, entropy differences (our main interest) obtained with procedure I are not expected (in general) to change significantly by procedure II.
RESULTS AND DISCUSSION
Simulation of the entire tetramer
As discussed in the Introduction, an interesting question is whether the conformational transition of the loop upon binding demonstrates an induced fit, i.e., whether it moves due to interactions induced by the ligand, or alternatively the open loop interconverts among different microstates, one of which is selected upon binding (selected fit). In the x-ray structures of Freitag et al.30 and others, the loop in the bound protein is always in the closed conformation with well-resolved coordinates. Without biotin, the loop in most cases is partially disordered and exhibiting elevated B-factors (>40 Å2); however, in structures I and II of Freitag et al. where biotin is not bound, the loop in subunit 1 is closed suggesting that a selected fit is a possibility. To check the extent of flexibility of the loop in the closed and open microstates, we carried our two MD simulations as described below.
The first simulation is based on the x-ray structure of tetramer IV (1swd) of Freitag et al., where the disordered loop conformations of subunits 2 and 3 were replaced by the complete open-loop conformation of subunit 2 in crystal struc- ture III (1swc). Thus, the “retouched” 1swd structure consists of two open loops (subunits 2 and 3) and two closed loops (subunits 1 and 4) which cover the corresponding bound biotins. This tetramer of 6820 atoms (including hydrogens, not counting biotins) was solvated with a 10 Å buffer of TIP3P water (13688 water molecules) and 6 Na+ ions to neutralize the total charge. The system was treated with periodic boundary conditions, where the particle mesh Ewald (PME) summation method was used for electrostatics; the real-space terms were evaluated with a cutoff distance of 15 Å and the simulations were carried out in the NVT ensemble.
The system was minimized without restraints for 15000 steps of steepest descent and 15000 steps of conjugate gradient, followed by a 1 ns MD run where the system was heated gradually to 300 K. In the subsequent 2.5 ns production run, no significant structural changes were observed in the protein; in particular, the conformations of the two closed loops (with biotins) and the two open ones only exhibited localized fluctuations.
Next, our tetramer was changed further by moving the biotin of subunit 1 (below the closed loop) to the empty pocket of subunit 3 where the loop has an open conformation. Our objective has been to check whether the open loop of subunit 3 would move to a closed conformation due to its interaction with biotin, and whether the empty pocket in subunit 1 would cause the closed loop there to open.
The system was first energy minimized for 104 steps of steepest descent and another 104 steps of conjugate gradient, then was heated to 300 K during a 50 ps MD run, followed by additional 50 ps run at pressure of 1 atm and 300 K; however, the production simulation was carried out at fixed temperature and volume (NVT ensemble). During the first 2 ns, the loops underwent only small local conformational fluctuations. To check the stability further, the temperature was increased gradually to 500 K followed by a 5 ns MD run. The local fluctuations of the closed loop without biotin (subunit 1) increased significantly and the loop moved to a different microstate, but no transition from the closed to the open conformation has been observed. The fluctuations of the open loop in subunit 3 were also increased significantly, but in this case no transition to a different microstate was observed.
While these simulations are relatively short, they suggest that both the open and closed conformations (even without biotin) have considerable local stability; furthermore, according to our later results the free energy of the open loop is lower by ∼27 kcal∕mol than that of closed one. Therefore, it is plausible to assume that the conformations of the open loop (in apo streptavidin) do not cover the closed microstate, and the conformational response of the loop to ligand binding is probably not of a selected fit type (i.e., it is an induced fit). The fact that the open-loop structures in the presence of biotin did not change to the closed microstate and the closed-loop structures without biotin did not move to an open microstate is probably due to our relatively short simulation. This interpretation is in accord with a Gaussian network model (GNM) analysis of 1swd, where the three lowest-frequency modes have only a minor effect on the open loop, which undergoes relatively large fluctuations only in the fourth mode. The fluctuations of the closed-loop structures are not significant in any of the 20 lowest modes. In particular, no open-closed transition is observed.49
Simulations of the loop∕water∕template systems
In Sec. 2B we defined loop∕water∕template systems (for calculating the loop free energy) and described their optimization by a process that ended in 500 ps MD simulations applied to the open and closed loop. The last loop∕water∕template structures obtained in these optimization runs became the starting structures for (two) 2 ns MD production runs from which loop∕water∕template conformations were retained every 2 ps (i.e., 1000 configurations) for a future free-energy analysis. It should be pointed out that determining the exact limits of a microstate in conformational space is practically impossible and therefore it is commonly defined by the underlying MC or MD trajectories initiated from a microstate’s structure. Thus, the microstate’s size typically increases with simulation time, t, and for large enough t the loop might move to a significantly different region in conformational space; obviously, E, S, and F depend on t as well. In previous publications, we have developed procedures for checking that the system remains in its microstate during a simulation (see Ref. 13 and references cited therein). Thus, in the case of two microstates m and n one should verify that the differences, ΔEmn(t), ΔSmn (t), and ΔFmn(t), are stable during a long enough time, Δt, meaning that the conformational changes in both microstates are small and comparable. Notice that the present trajectories of 2 ns are significantly larger than the ∼0.5 ns trajectories studied in our previous papers.13, 14, 23, 24, 25, 26, 54
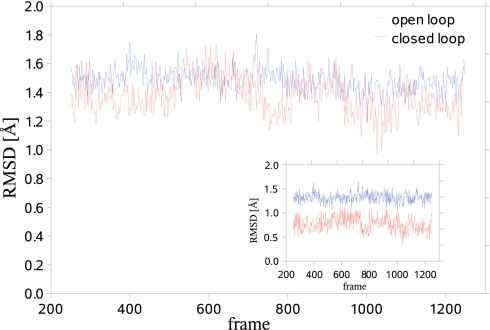
First, we calculated for each run (trajectory) the RMSD of the loop’s conformations from its initial one, and the results are presented in Fig. 4 as a function of simulation time. The figure reveals that the closed loop has moved from its initial conformation slightly more than the open loop, as is also demonstrated by the average RMSD values, 1.49 and 1.38 Å, respectively. This is expected as the closed loop is attached to the template of the open one. On the other hand, larger RMSD oscillations are observed for the open loop than for the closed one, which are expressed by the corresponding standard deviations of the RMSD values, 0.13 and 0.08 Å; this is in accord with the standard deviations observed for the potential energy, 9.8 and 9.5 kcal∕mol, respectively (data not shown). These larger fluctuations of the open loop suggest that its entropy is higher than that of the closed loop, which agrees with the experimental observation of elevated B-factors and partial disorder of the open loop in crystal structures of sreptavidin.30
Figure 4.
Root-mean-square deviation (RMSD) of the heavy atoms from the x-ray structure for the open and closed loops obtained from MD trajectories of 2 ns, where a frame is defined every 1 ps. While lower RMSD values are observed for the open loop, their fluctuations are the largest, suggesting that the entropy of the open loop is larger than that of the closed one, ΔSloop > 0 [Eq. 15b]. The inset shows the RMSD for backbones.
The higher flexibility of the open microstate is also demonstrated by results for αk(min), αk(max), and Δαk [Eq. 10] presented in Table 1 for the backbone dihedral angles φ, ψ, and ω and the side-chain angles χ. These values, obtained (for the open and closed loops) from the samples of 1000 conformations defined above, show that the Δαk values for the backbone are relatively small (in most cases smaller than 80°), but as expected are sometimes higher for Gly and the χ angles. A detailed comparison of the Δαk values reveals that altogether they are larger for the open loop than for the closed one, which again suggests that ΔSloop [Eq. 15b] is positive.
Table 1.
Minimum and maximum values of dihedral angles, αk(min) and αk(max), and their differences Δαk (in degrees) for the open and closed samples.a
| Open loop |
Closed loop |
||||||
|---|---|---|---|---|---|---|---|
| Residue | Dihedrals | Min | Max | Δ | Min | Max | Δ |
| SER | φ | –168 | –120 | 48 | –159 | –110 | 49 |
| ψ | –25 | 45 | 70 | 134 | 176 | 42 | |
| ω | 143 | 205 | 62 | 160 | 205 | 45 | |
| χ1 | –178 | 178 | 356 | –29 | –210 | 181 | |
| ALA | φ | 25 | 98 | 73 | –166 | –118 | 48 |
| ψ | 5 | 68 | 63 | –41 | 15 | 56 | |
| ω | 144 | 195 | 51 | 147 | 193 | 46 | |
| VAL | φ | 100 | 214 | 114 | 39 | 96 | 57 |
| ψ | –61 | 82 | 143 | 108 | 176 | 68 | |
| ω | 153 | 212 | 59 | 155 | 200 | 45 | |
| χ1 | –103 | 102 | 205 | –99 | –42 | 57 | |
| GLY | φ | –163 | –58 | 105 | –175 | –92 | 83 |
| ψ | –36 | 87 | 123 | –71 | 8 | 79 | |
| ω | 151 | 215 | 64 | 154 | 205 | 51 | |
| ASN | φ | –101 | –32 | 69 | –175 | –93 | 82 |
| ψ | 92 | 152 | 60 | –3 | 62 | 65 | |
| ω | 158 | 204 | 46 | 170 | 219 | 49 | |
| χ1 | 145 | 195 | 50 | –101 | –34 | 67 | |
| χ2 | 33 | 126 | 93 | 83 | 245 | 162 | |
| ALA | φ | –89 | –43 | 46 | 25 | 91 | 66 |
| ψ | –67 | –15 | 52 | 124 | 178 | 54 | |
| ω | 152 | 195 | 43 | 155 | 202 | 47 | |
| GLU | φ | –137 | –87 | 50 | 165 | 236 | 71 |
| ψ | –36 | 10 | 46 | 1 | 77 | 76 | |
| ω | 161 | 199 | 38 | 148 | 208 | 60 | |
| χ1 | –78 | –28 | 50 | –180 | –29 | 151 | |
| χ2 | 55 | 121 | 66 | 137 | 223 | 86 | |
| χ3 | –152 | 164 | 316 | –180 | 180 | 360 | |
| SER | φ | –137 | –87 | 50 | 165 | 235 | 70 |
| ψ | –10 | 29 | 39 | 144 | 190 | 46 | |
| ω | 164 | 186 | 22 | –175 | –150 | 25 | |
| χ1 | –83 | –29 | 54 | 172 | 220 | 48 | |
αk(min), αk(max), and Δαk are defined in Eq. 10; their values were calculated from samples of 1000 loop conformations generated for the open and closed microstates by retaining a conformation every 2 ps from the corresponding 2 ns trajectories.
Results for the loop entropy
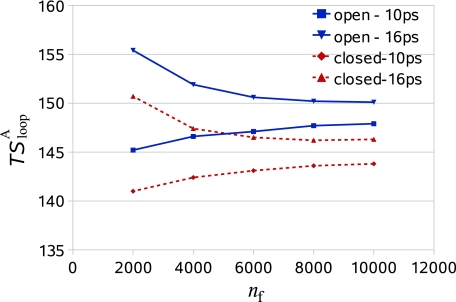
Results for the loop entropy, [Eq. 14], appear in Table 2 for the microstates of the open and closed loops and for their difference [see the discussion preceding Eq. 15a]. These results were obtained by independently reconstructing ns = 40 loop structures, distributed homogeneously along the entire sample of 1000 system configurations. At each reconstruction step of each configuration, the future part of the loop (and water) was simulated by MD, and a structure was retained every 6 fs for a later analysis. We carried out several rounds of analysis as described below.
Table 2.
HSMD results (in kcal∕mol) for the loop entropy, , and for entropy differences between the open and closed microstates at T = 300.a
|
|
|
|
||||||
|---|---|---|---|---|---|---|---|---|
| Equilbration | bin size, δ | nf | 10 ps | 16 ps | 10 ps | 16 ps | 10 ps | 16 ps |
| Procedure I | Δαk∕10 | 10000 (9) | 150.9 | 157.6 | 147.8 | 155.4 | 3.1 | 2.2 |
| Δαk∕20 | 10000 (9) | 148.7 | 151.8 | 144.8 | 148.4 | 3.9 | 3.4 | |
| Δαk∕40 | 10000 (9) | 148.1 | 150.4 | 144.1 | 146.6 | 4.0 | 3.8 | |
| Δαk∕60 | 500 | 141.9 | 138.4 | 3.5 | ||||
| 1000 | 143.6 | 139.3 | 4.3 | |||||
| 2000 (1) | 145.2 | 155.4 | 141.0 | 150.7 | 4.2 | 4.7 | ||
| 4000 (3) | 146.6 | 151.9 | 142.4 | 147.4 | 4.2 | 4.5 | ||
| 8000 (7) | 147.7 | 150.2 | 143.6 | 146.2 | 4.1 | 4.0 | ||
| 10000 (9) | 147.9 | 150.1 | 143.8 | 146.3 | 4.1 | 3.8 | ||
| Δαk∕80 | 500 | 141.9 | 138.4 | 3.5 | ||||
| 1000 | 143.6 | 139.3 | 4.3 | |||||
| 2000 (1) | 145.2 | 155.4 | 140.8 | 150.6 | 4.4 | 4.8 | ||
| 4000 (3) | 146.5 | 151.8 | 142.2 | 147.3 | 4.3 | 4.5 | ||
| 8000 (7) | 147.5 | 150.1 | 143.4 | 145.9 | 4.1 | 4.2 | ||
| 10000 (9) | 147.7 | 149.8 | 143.6 | 146.0 | 4.1 | 3.8 | ||
| Δαk∕100 | 500 | 141.9 | 138.5 | 3.4 | ||||
| 1000 | 143.6 | 139.4 | 4.2 | |||||
| 2000 (1) | 145.1 | 155.4 | 140.9 | 150.8 | 4.2 | 4.6 | ||
| 4000 (3) | 146.4 | 151.8 | 142.1 | 147.5 | 4.3 | 4.3 | ||
| 8000 (7) | 147.2 | 150.1 | 143.2 | 146.0 | 4.0 | 4.1 | ||
| 10000 (9) | 147.3 | 149.7 | 143.3 | 145.8 | 4.0 | 3.9 | ||
| Converged | 4.0 ± 1.0 | 3.9 ± 1.0 | ||||||
| Procedure II | 4.0° | 3333 | 5.3 ± 1.0 | 5.3 ± 1.0 | ||||
| QH I | 8000 | 198.3 | 198.3 | 192.6 | 192.6 | 5.7 ± 2.5 | ||
Results obtained with procedure I based on two equilibration times of 10 and 16 ps for the loop entropy, [Eqs. 13, 14], and the differences, [Eq. 15a]; they were obtained by reconstructing 40 loop structures selected homogeneously from larger MD samples (of 1000 water-loop configurations) of the open and closed microstates. The results are calculated as a function of the bin size δ = Δαk∕l [Eq. 10] and nf [Eq. 11], the sample size of the future chains used in the reconstruction process. For the 16 ps results, the nf values appear in parentheses, e.g., nf = 8000 (7) means that nf = 8000 and 7000 for the 10 and 16 ps results, respectively. is defined up to an additive constant that is expected to be the same for both microstates. The maximal statistical error of the results for is 0.7 kcal∕mol. The results for nf = 104 are bold-faced. For procedure II, the result is presented only for with a constant bin size of 4°. [Eq. 20] is the quasiharmonic entropy; these results were obtained from larger samples of 8000 loop conformations (see text).
As discussed earlier, the reconstruction simulation starts at each step from the structure to be reconstructed and the initial part of the simulation is used for equilibration and is thus discarded. To emphasize the important effect of equilibration, we have carried out two sets of analyses. The first set is based an equilibration of 10 ps (ninit = 1667) followed by a relatively long MD simulation of 60 ps (nf = 104 structures) where the probability ρloop (αk−1αk |αk−2 ,...,α1 ) [Eq. 11] and were calculated for nf = 500, 1000, 2000, 4000, 8000, and 104. In the second set, another 1000 loop conformations (6 ps) are ignored, i.e., the equilibration is increased to 16 ps (ninit = 2667) and results are thus calculated for nf = 1000, 3000, 7000, and 9000; these samples are denoted in the table by 1, 3, 7, 9, respectively. Results for appear in Table 2 for different bin sizes, δ = Δαk∕l, l = 10, 20, 40, 60, 80, and 100 centered at αk (i.e., αk ± δ∕2) [Eqs. 13, 14]. If the counts of the smallest bin are smaller than 50, the bin size is increased becoming δ1 = δ + 0.2δ and if necessary it is increased again to δ2 = δ1 + 0.2δ, etc. until the number of counts becomes 50; in the case of zero counts, nvisit is taken to be 1; however an event of zero counts is very rare.
Relying on the discussion following Eq. 13, one would expect the results for to decrease as the approximation improves, e.g., as the bin size is decreased for a given value of nf. The table always shows this expected behavior for the largest values of nf, nf = 104 9000, 8000, and 4000, while for the other nf values the results in some cases remain constant (rather than decrease) within the (larger) error bars. Similarly, for a given bin one would expect to decrease with increasing nf, which is always satisfied for the set based on the 16 ps equilibration but is never satisfied for the other set (of 10 ps equilibration), where the results always increase rather than decrease; this is an example where insufficient equilibration probably leads to unnormalized probabilities, which cause the unexpected behavior of [see the discussion following Eq. 13]. More specifically, because the reconstruction simulation starts from the bin, the bin will remain overoccupied after a short equilibration, and during the following short production run (small nf); thus, the number of counts nvisit [Eq. 11] will be too large leading to a too low ; as nf increases the entropy increases (due to the decrease in ninit), approaching its correct value (from below) for a large nf where the effect of the initial conditions is gradually eliminated. Note that the required equilibration time is system-dependent, where the present time (16 ps) is significantly larger than the 2.5 ps used in our previous studies,13, 14, 26 probably due to the larger loop studied here and its relatively high stability as discussed in Sec. 3A. Also, the reconstruction simulations (∼60 ps) are significantly larger than those used before for loops (e.g., 12.5 ps). Still, the 10 and 16 ps results for the open (closed) microstate are approaching each other from above and below, respectively, where for the largest nf they differ by ∼2.5 kcal∕mol (Extrapolating the results in the table suggests that for nf ∼ 16 000 the entropies obtained by the two sets will become equal within the error bars.) See also Fig. 5.
Figure 5.
The approximate loop entropy, (in kcal∕mol + const) [Eq. 14], is presented as a function of the number of future reconstruction steps, nf [Eq. 11] for the open and closed microstates. These results (taken from Table 2) are based on a bin size, Δαk∕l, l = 60 [Eq. 10], and are shown for two equilibrations of 10 and 16 ps. It is demonstrated that for 10 ps, the results for both the open and closed microstates increase while those for 16 ps decrease. Thus, for the open (closed) microstate the 10 and 16 ps results approach each other deviating by ∼2.5 kcal∕mol for nf = 104 (or 9000).
The results for (our main interest) for the 10 ps set converge nicely to ∼4 kcal∕mol, where is systematically too low (∼3.5 kcal∕mol). The convergence of the 16 ps set is less pronounced, probably due to lower statistics, i.e., the removal of the (first) 1000 structures that contribute most significantly to nvisit, which leads to a relatively high result, ∼4.6 kcal∕mol; therefore, for the 16 ps set the decrease of the [] results to the nf = 9000 value (3.9 kcal∕mol for l = 100) is stronger than that observed for the 10 ps set. Relying on the results for nf = 104 and 9000, we estimate kcal∕mol. Notice, however, that estimations for exceeding by ∼0.4 the 3.9 kcal∕mol value can be obtained from smaller (nf) samples. We have verified again13 that the Jacobian has no effect on results for within the statistical errors. For comparison, we have also calculated the entropy by the quasiharmonic (QH) approximation using the equation44
| (20) |
where σ is the covariance matrix and N = 64 is the number of internal coordinates. Clearly, SQH (obtained from two samples of 8000 configurations, where a configuration was retained every 1 ps from an 8 ns MD trajectory) constitutes an upper bound for S since correlations higher than quadratic are neglected. Indeed, the QH results are larger (by ∼34%) than the HSMD results (which do take higher-order correlations into account), in accord with our previous studies.13, 14, 23, 25, 26 Still, the QH result = 5.7 ± 2.5 kcal∕mol is equal to the HSMD value within relatively large error bars; this result required 88 h CPU time on our computers (see below), where most of the time was devoted to generating the two samples of 8000 configurations. It should be pointed out that in a detailed study, Chang et al.50 found QH to be unreliable when used in Cartesian coordinates or applied (in internal coordinates) to several microstates; on the other hand, it was found suitable for treating a single microstate, while the convergence of the results is slow and large samples are typically needed. Still, entropy differences are expected to be reliable (see also Ref. 51). We also provide in the table the value of obtained with procedure II using only one bin value of 4° for all angles; the result = 5.3 ± 1.0 kcal∕mol is close to that obtained with procedure I.
The error bars in Table 2 (and in Tables 3, 4) are SD∕(ns1∕2), where SD is standard deviation and ns is the number of structures. Using this formula is justified because every successive pair of the (40) reconstructed structures is separated by 50 ps along the MD trajectory, and thus the energy and entropy of these structures can be considered as uncorrelated. Notice that 40 structures were reconstructed also in Ref. 26; to verify further that this number is adequate, we have calculated results for ns = 20 and found them to agree within the error bars to those obtained by ns = 40. Reconstruction of a single loop conformation with 250 water molecules based on 104 future configurations (60 ps) with 2 ps equilibration requires ∼30 h on a single core of a Quad-Core AMD Opteron(tm) Processor 2380 (∼2500 MHz); however, we have shown that the correct result for is already obtained from a reconstruction based on 6 ps (1000 future configurations), which requires only 3 h CPU time. The longer calculations of nf = 104 were performed for validating convergence. In general, the required nf will increase with loop size; in Ref. 26, for example, where a smaller loop of acetylcholineesterase has been studied, the maximal value of nf is 2000.
Table 3.
Free energy of water (in kcal∕mol) calculated with thermodynamic integration (TI) for the open and closed loops.a
| (ch) | (LJ) | |||||||
|---|---|---|---|---|---|---|---|---|
| Window (ps) | Open | Closed | δ (Å2) | Open | Closed | Open | Closed | ΔFwater |
| 20 | –54.4 | –117.2 | 0.5 | ∞ | ∞ | ∞ | ∞ | – |
| 2.0 | 45.2 | 32.6 | –9.2 | –84.6 | 75.4 | |||
| 3.0 | 41.6 | 30.1 | –12.8 | –87.1 | 74.3 | |||
| 40 | −53.4 | −116.2 | 2.0 | 46.5 | 34.9 | −6.9 | −81.3 | 74.4 |
| Errors | ±1.0 | ±0.8 | ±0.6 | ±0.4 | ±1.0 | ±0.9 | ±1.3 | |
(ch) and (LJ) were obtained by eliminating the loop-water electrostatic (charge) and Lennard Jones (LJ) interactions, respectively; is their sum [Eq. 17]. Each integration is based on 22 windows, where the (best) results for windows of 40 ps are bold-faced. δ defines the soft-core LJ potentials [Eq. 21]. ΔFwater [Eq. 18] is the difference in the water free energy between the open and closed configurations. “Errors” stand for statistical errors [(standard deviation)∕401∕2].
Table 4.
Contribution of the loop and water to the energy, entropy, and free energy (in kcal∕mol) of the open and closed microstates.a
| Ewater | TSwater | Eloop | TSloop | Floop | ||
|---|---|---|---|---|---|---|
| Open | –2609.3 ± 3.6 | –2602.4 ± 3.8 | –6.9 ± 1.0 | –200.8 ± 1.5 | 149.7 ± 0.6 | –350.5 ± 1.6 |
| Closed | –2689.0 ± 3.5 | –2607.7 ± 3.7 | –81.3 ± 0.9 | –103.2 ± 1.3 | 145.8 ± 0.7 | –249.0 ± 1.4 |
| Open-closed | ΔEwater | TΔSwater | ΔFwater | ΔEloop | TΔSloop | ΔFloop |
| 79.7 ± 4.9 | 5.3 ± 5.1 | 74.4 ± 1.3 | –97.6 ± 1.7 | 3.9 ± 1.0 | –101.5 ± 1.9 | |
|
Etotal |
TStotal |
Ftotal |
||||
| Open | –2810.1 ± 1.6 | –2452.7 ± 1.9 | –357.4 ± 1.8 | |||
| Closed | –2792.2 ± 1.6 | –2461.9 ± 1.7 | –330.3 ± 1.6 | |||
| Open-closed | ΔEtotal | TΔStotal | ΔFtotal | |||
| −17.9 ± 1.6 | 9.2 ± 2.1 | −27.1 ± 2.0 |
The water and loop energies, Ewater and Eloop, are defined in Eq. 1. [Eq. 17] is the water free energy obtained by a TI procedure. The loop entropy TSloop [Eqs. 13, 14] and its difference TΔSloop [Eq. 15b] are taken from Table 2; Floop = Eloop−TSloop. TΔSwater is obtained from ΔEwater − ΔFwater. Etotal [Eq. 1] and ΔEtotal are the total energy and its difference for the open and closed microstates. Ftotal is the sum of the loop and water free energies and its difference is ΔFtotal ]Eq. 19]; TΔStotal is obtained from ΔEtotal − ΔFtotal. The errors are defined in Table 3. Entropies and free energies are defined up to additive constants, which are expected to be equal for both microstates.
Contribution of water to the free energy
This contribution is obtained by calculating [Eq. 17 and Fig. 3], i.e., by eliminating the loop-water interactions in a TI process keeping the template and loop fixed, as described in Sec. 2E. Thus, starting from the complete loop∕template∕water system, the loop-water interactions were gradually annihilated using a parameter λ, where the electrostatic interactions were removed first followed by the removal of the LJ interactions (in the presence of zero electrostatic interactions). This TI was carried out independently for each of the 40 reconstructed loop structures using soft-core potentials defined by parameters λ and δ; thus, the LJ potential ϕ (based on the usual LJ parameters σ and ε) becomes52
| (21) |
The integration is based on 22 windows (λ = 0.95, 0.90, 0.85, …, 0.10, 0.05, 0.03, 0.01, 0.00), i.e., altogether 44 windows for both interactions. The higher density of λ points close to λ = 0 is necessary to accurately integrate the larger LJ fluctuations in this region. As in our previous study,26 integration step i starts by minimizing the last structure of the previous step, i−1, according to the potential energy of the current λ(t), followed by a 5 ps equilibration, which uses the set of velocities of the last i−1 structure (for λ = 0.95 we used a longer equilibration of 15 ps). After equilibration, a 20 ps production run is performed.
The average TI results (over 40 structures) for , , their sum, [Eq. 17], (for the open and closed microstates), and the difference, ΔFwater [Eq. 18], between (o) and (c) are presented in Table 3. (Note that while the interactions are eliminated by TI, the signs of the above free-energy functionals are reversed describing a TI procedure where the loop-water interactions are increased from zero.) The LJ results were calculated for several δ values, and those for δ = 2 also for windows of 20 and 40 ps, where the larger window time was also used in the electrostatic integrations. All these tests enable one to estimate more reliably the errors, in addition to the error estimation provided by the standard deviation.
It should be pointed out that in a typical application of TI (and free-energy perturbation), ligand a is transformed into ligand b in both the active site of a protein (P) and the solvent environment, which leads to the free-energy differences, ΔFPa,Pb and ΔFa,b, respectively. The success of this method lies in the fact that only the interactions of the mutated part of the ligand with the environment are directly considered and the fluctuations are therefore small. However, conformational changes in the entire protein (e.g., “jumps” of side chains among rotamers) occur constantly and the results might not converge for long simulation times; in other words, the microstate of Pb (and to some extent also of Pa) keeps changing as the simulation time increases. This is the main source of errors, which are commonly assessed by calculating ΔF also in the reverse direction, i.e., in going from b to a.
In our case, however, this problem does not exist because the conformations of both the protein (template) and the loop are kept fixed during TI (where the loop-water interactions are gradually eliminated). Furthermore, we carry out many such TI processes (40 in this paper) and average the results, where the fluctuations define the errors; therefore, performing reversed TI runs is not needed (they might also lead to further complexity; see below). Indeed, Table 3 shows that the errors of all the free-energy components are relatively small and for the open (closed) conformation the results for each component (based on different parameters) can differ by up to 6 kcal∕mol. However, the corresponding “open” and “closed” results are highly correlated as both change in the same direction and thus their differences, ΔFwater (which is our main interest), are very stable with much smaller deviations of ∼1 kcal∕mol. We obtain ΔF water = 74.4 ± 1.3 kcal∕mol, i.e., the free energy due to water is lower for the closed loop than for the open one.
Returning to the reversed TI, it should be pointed out that in the initial preparation of the systems, some water molecules typically become trapped within the template and they remain there during the generation of the MD trajectories, the reconstruction simulations, and the TI runs based on eliminating the loop-water interactions. Therefore, these waters can be considered as part of the fixed template. However, in the equilibration of water, which is the initial step of a reversed TI process (under zero water-loop interactions), a new set of trapped water molecules might be created which would lead to changes in the TI results. This undesirable effect can in principle be prevented but would require applying additional computational means.
The LJ results in Table 3 can intuitively be explained by considering an LJ integration where the loop-water interactions are increased (rather than decreased) from zero to their full value. In this process, the water molecules should create a void for the loop which requires investing energy. The results show that larger energy is required for the open loop (46.5), which is highly exposed to water, than for the closed loop (34.9 kcal∕mol), which tends to lie against the template. Explaining the different results for the electrostatic integrations is not straightforward. The integration time per frame is ∼140 h CPU on a single processor.
Combined results for the entire system
In the upper part of Table 4, we summarize the contributions of the loop and water (and implicitly also of the template) to the total energy, entropy, and free energy. The water contributions are the energy Ewater [Eq. 1] (including water-water, water-loop, and water-template interactions) and the free energy [Eq. 17 and Table 3], from which the water entropy is calculated, TSwater = Ewater − . The loop contributes its energy, Eloop [Eq. 1] (which includes the loop-loop and loop-template interactions), and the reconstructed entropy, TSloop [Eq. 14 and Table 2], which both lead to our definition of the loop’s free energy, Floop = Eloop – TSloop. Notice again that S and F are defined up to additive constants and thus only their differences ΔS and ΔF are physically meaningful; thus, the differences of all these parameters between the open- and closed-loop conformations are also provided in the table. Finally, in the lower part of the table, results are presented for the entire loop∕water∕template systems, i.e., for the total energy, entropy, and free energy, and their differences. It is seen again that the errors of the differences are smaller than the sum of errors of the corresponding open and closed properties, in some cases again due to systematic error cancelation.
It should be noted that the contributions of TΔSwater to ΔFwater and TΔSloop to ΔFloop are relatively small, being around 7% and 4%, respectively, i.e., most of the contribution is due to the corresponding energy differences, ΔEwater and ΔEloop. However, these entropy differences are positive, meaning that the total entropy of the open system is larger than that of the closed one, in accord with the elevated B factors and conformational disorder observed in crystal structures of unbound sreptavidin.30 Interestingly, TΔStotal = 9.2 kcal∕mol [the sum of 3.9 (loop) and 5.3 (water)] contributes significantly to ΔFtotal, about 34%, because ΔEwater and ΔEloop are of opposite signs and the absolute value of their sum (ΔEtotal) is relatively small. This demonstrates again that, in general, ΔE alone is not a reliable criterion of stability since (as in the present case) entropic effects could be significant.13, 14, 23, 25, 26
Notice that ΔEtotal is negative whereas TΔStotal is positive, while one would expect these quantities to share the same sign, as lower energy is generally correlated with lower entropy. However, as pointed out earlier, the higher entropy of the open-loop system agrees with the crystal structures of the unbound protein. Also, the fact that in most of these unbound structures the loop is open suggests that the open microstate is more stable than the closed one, and according to our calculations, this higher stability is gained from both a higher entropy and a lower energy and hence a lower total free energy, ΔFtotal = − 27.1 ± 2.0. The fact that the structure of the closed loop is attached to the template of an unbound protein might also contribute to the decrease of ΔFtotal.
SUMMARY AND CONCLUSIONS
In a previous publication,39 we capped loops with an increasing number of TIP3P water molecules and studied the effect of this number on the loop stability by monitoring the RMSD of the loop (from its x-ray structure) along MD trajectories. We have found that for several loops, a minimal number of ∼10 water molecules have already stabilized the structure. However, the general validity of this somewhat surprising result was tested further in a subsequent study26 with respect to a stricter free-energy criterion, where HSMD-TI was applied to a loop of the protein AChE. It has been found that the loop must be well soaked in water with the density of bulk water; thus, to cap the relatively large loop of streptavidin, at least 250 water molecules are needed, which makes the entire system the largest treated thus far with HSMD-TI. Testing HSMD-TI as applied to systems of increasing size is important because the fluctuation of the absolute entropy (and energy) increases as N1∕2 with the number of atoms, N.
Indeed, applying the loop reconstruction part of the method HSMD-TI required longer simulations. Thus, we have found that to obtain well defined (i.e., normalized or close to normalized) probabilities, which lead to the expected decrease of , one needs significantly longer equilibration (∼16 ps) than that applied in previous studies of smaller loops (2.5 ps);13, 14, 23, 25 the reconstruction simulations (∼60 ps) are also significantly larger than those used before (e.g., 12.5 ps). The TI component of the method also appears in methodologies for calculating the absolute free energy of binding.5, 6, 7, 8, 9, 10, 53 In this procedure, one should check, in particular, that the gradual elimination in the loop-water LJ interactions is adequately performed, which has been verified for the present larger loop∕template∕water system.
We initially carried out MD simulations of the entire tetramer in a box of water. While these runs were relatively short, they suggest that both the open and closed conformations have considerable local stability at room temperature even without biotin, and this in turn may suggest that the conformational response of the loop to ligand binding is of an induced- rather than a selected-fit type. However, longer simulations are needed to corroborate this point.
Our finding that the open loop is more stable than the closed one (by ΔFtotal = Fopen− Fclosed = –27.1 ± 2.0 kcal∕mol) is expected since experimentally the open conformation is found in most of the crystal structures of unbound streptavidin (the closed loops observed in two subunits are caused by crystal-packing interactions30). While this value seems large, it should be pointed out that ΔFloop = –25.2±7 kcal∕mol has been obtained recently (using a different method) for the mobile loop of avidin;53 also, Lazaridis et al.54 have estimated the effect of the conformational changes in avidin upon its binding of biotin (using implicit solvent) to find an energy of ∼ –21 kcal∕mol, where most of the conformational change is attributed to the loop (avidin and streptavidin are proteins with similar structures). As mentioned earlier, the fact that both loop conformations are attached to the same open template also contributes to the higher stability of the open loop. Therefore, one would seek to study each loop attached to its own template. However, a recent application of HSMD-TI to the loop of AChE using two templates has shown that such a comparison might be problematic partially due to the different accuracy of the corresponding x-ray structures.55
HSMD-TI provides the entropy of the loop and water as by-products of the simulation. In a recent paper, Singh and Warshel56 showed that various components of the entropy (e.g., solvation, hydrophobic) can be obtained by applying and releasing harmonic restraints. In the present paper, the contribution of TΔStotal (9.2 kcal∕mol) to ΔFtotal is significant at about 34%, which demonstrates that, in general, ΔE alone is not a reliable criterion of stability. Also, ΔEtotal is negative whereas TΔStotal is positive, while one would expect these quantities to share the same sign. Thus, the higher stability of the open loop is a contribution of both higher entropy and lower energy.
On the technical side, we have described in detail the implementation of two reconstruction procedures, and developed a set of programs for analyzing the reconstruction results. Thus, all the reconstructed structures were generated within TINKER and were saved on files for a later analysis based on a set of general programs written in C. This separation between the MD simulation and the analysis allows one to use any MM∕MD package and to carry out a flexible analysis (without the need for additional simulations) where parameters (e.g., bin size) can be changed and their effect on the results can be studied.
One advantage of HSMD-TI is the fact that the free energy can be obtained from a small number of structures, in principle even from any single structure. Indeed, the main result of this study, ΔFtotal = −27.1±2.0 kcal∕mol, is based on reconstructing only 40 structures [in agreement with our previous calculations of the loop of AChE (Ref. 26)]. The relatively high accuracy of this result stems also from cancellation of errors in entropy and energy differences, hence in free-energy differences. The fact that the number of water molecules is not large enables one to calculate their contribution to the total entropy, ΔStotal, which was found to be slightly larger than the contribution of the loop.
Finally, it should be pointed out again that while mobile loops appear in many enzymes, calculating ΔF = Fopen – Fclosed by the conventional methods (e.g., TI) is not straightforward, and very few such calculations are available.57 However, ΔF (without the ligand) should be considered in the calculation of the total absolute free energy of binding, which has not been done thus far. With HSMD-TI, calculation of ΔF is straightforward and reliable, as has been shown for a loop of AChE in our previous paper.26 In a subsequent project, we are calculating now the absolute free energy of binding of the biotin-streptavidin complex, where the present result ΔF = −27.1 kcal∕mol will be taken into account. In the next step in the development of HSMD-TI, we intend to extend it to a protein model where the template is not fixed.
ACKNOWLEDGMENTS
This work was supported by NIH Grant No. 2-R01 GM066090-4 A.
REFERENCES
- Beveridge D. L. and DiCapua F. M., Annu. Rev. Biophys. Biophys. Chem. 18, 431 (1989). 10.1146/annurev.bb.18.060189.002243 [DOI] [PubMed] [Google Scholar]
- Kollman P. A., Chem. Rev. 93, 2395 (1993). 10.1021/cr00023a004 [DOI] [Google Scholar]
- Jorgensen W. L., Acc. Chem. Res. 22, 184 (1989). 10.1021/ar00161a004 [DOI] [Google Scholar]
- Meirovitch H., in Reviews in Computational Chemistry, edited by Lipkowitz K. B. and Boyd D. B. (Wiley-VCH, New York, 1998), Vol. 12, p. 1. [Google Scholar]
- Gilson M. K., Given J. A., Bush B. L., and McCammon J. A., Biophys. J. 72, 1047 (1997). 10.1016/S0006-3495(97)78756-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boresch S., Tettinger F., Leitgeb M., and Karplus M., J. Phys. Chem. B 107, 9535 (2003). 10.1021/jp0217839 [DOI] [Google Scholar]
- Meirovitch H., Curr. Opin. Struct. Biol. 17, 181 (2007). 10.1016/j.sbi.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Zhou H.-X. and Gilson M. K., Chem. Rev. 109, 4092 (2009). 10.1021/cr800551w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foloppe N. and Hubbard R., Curr. Med. Chem. 13, 3583 (2007). 10.2174/092986706779026165 [DOI] [PubMed] [Google Scholar]
- Van Gunsteren W. F., Bakowies D., Baron R., Chandrasekhar I., Christen M., Daura X., Gee P. J., Geerke D. P., Glättli A., Hünenberger P. H., Kastenholz M. A., Oostenbrink C., Schenk M., Trzesniak D., Van der Vegt N. F. A., and Yu H. B., Angew. Chem. Int. Ed. 45, 4064 (2006). 10.1002/anie.200502655 [DOI] [PubMed] [Google Scholar]
- Alder B. J. and Wainwright T. E., J. Chem. Phys. 31, 459 (1959). 10.1063/1.1730376 [DOI] [Google Scholar]
- McCammon J. A., Gelin B. R., and Karplus M., Nature (London) 267, 585 (1977). 10.1038/267585a0 [DOI] [PubMed] [Google Scholar]
- Cheluvaraja S. and Meirovitch H., J. Chem. Theor. Comput. 4, 192 (2008). 10.1021/ct700116n [DOI] [PubMed] [Google Scholar]
- Cheluvaraja S., Mihailescu M., and Meirovitch H., J. Phys. Chem. B 112, 9512 (2008). 10.1021/jp801827f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elber R. and Karplus M., Science 235, 318 (1987). 10.1126/science.3798113 [DOI] [PubMed] [Google Scholar]
- Stillinger F. H. and Weber T. A., Science 225, 983 (1984). 10.1126/science.225.4666.983 [DOI] [PubMed] [Google Scholar]
- Getzoff E. D., Geysen H. M., Rodda S. J., Alexander H., Tainer J. A., and Lerner R. A., Science 235, 1191 (1987). 10.1126/science.3823879 [DOI] [PubMed] [Google Scholar]
- Rini J. M., Schulze-Gahmen U., and Wilson I. A., Science 255, 959 (1992). 10.1126/science.1546293 [DOI] [PubMed] [Google Scholar]
- Constantine K. L., Friedrichs M. S., Wittekind M., Jamil H., Chu C. H., Parker R. A., Goldfarb V., Mueller L., and Farmer B. T., Biochemistry 37, 7965 (1998). 10.1021/bi980203o [DOI] [PubMed] [Google Scholar]
- White R. P. and Meirovitch H., J. Chem. Phys. 121, 10889 (2004). 10.1063/1.1814355 [DOI] [PubMed] [Google Scholar]
- White R. P. and Meirovitch H., J. Chem. Phys. 124, 204108 (2006). 10.1063/1.2199529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. P. and Meirovitch H., J. Chem. Phys. 123, 214908 (2005). 10.1063/1.2132285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheluvaraja S. and Meirovitch H., J. Chem. Phys. 122, 054903 (2005). 10.1063/1.1835911 [DOI] [PubMed] [Google Scholar]
- Cheluvaraja S. and Meirovitch H., J. Phys. Chem. B 109, 21963 (2005). 10.1021/jp0529691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheluvaraja S. and Meirovitch H., J. Chem. Phys. 125, 024905 (2006). 10.1063/1.2208608 [DOI] [PubMed] [Google Scholar]
- Mihailescu M. and Meirovitch H., J. Phys. Chem. B 113, 7950 (2009). 10.1021/jp900308y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiet L. and Wolf F. J., Arch. Biochem. Biophys. 106, 1 (1964). 10.1016/0003-9861(64)90150-X [DOI] [PubMed] [Google Scholar]
- Green N. M., Adv. Protein Chem. 29, 85 (1975). 10.1016/S0065-3233(08)60411-8 [DOI] [PubMed] [Google Scholar]
- Bayer E. A. and Wilchek M., Methods Enzymol. 184, 49 (1990). [DOI] [PubMed] [Google Scholar]
- Freitag S., Trong I. L., Klumb L., Stayton P. S., and Stenkamp R. E., Protein Sci. 6, 1157 (1997). 10.1002/pro.5560060604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Ohlendorf D. H., Wendoloski J. J., and Salemme F. R., Science 243, 85 (1989). 10.1126/science.2911722 [DOI] [PubMed] [Google Scholar]
- Weber P. C., Pantoliano M. W., and Thompson L. D., Biochemistry 31, 9350 (1992). 10.1021/bi00154a004 [DOI] [PubMed] [Google Scholar]
- Chu V., Freitag S., Trong I. L., Stenkamp R. E., and Stayton P. S., Protein Sci. 7, 848 (1998). 10.1002/pro.5560070403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A., Pahler A., Smith J. L., Satow Y., Merritt E. A., and Phizackerley R. P., Proc. Natl. Acad. Sci. USA 86, 2190 (1989). 10.1073/pnas.86.7.2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., K. M.MerzJr., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., and Kollman P. A., J. Am. Chem. Soc. 117, 5179 (1995). 10.1021/ja00124a002 [DOI] [Google Scholar]
- Ponder J. W., TINKER—Software Tools for Molecular Design, Version 5.0, 2009, Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis.
- Case D. A., Darden T. A., T. E.CheathamIII, Simmerling C. L., Wang J., Duke R. E., Luo R., Crowley M., Walker R. C., Zhang W., Merz K. M., Wang B., Hayik S., Roitberg A., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Mathews D. H., Seetin M. G., Sagui C., Babin V., and Kollman P. A., AMBER 10, 2008, University of California, San Francisco.
- Allen M. P. and Tildesley D. J., Computer Simulation of Liquids (Clarenden, Oxford, 1987). [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L., J. Chem. Phys. 79, 926 (1983). 10.1063/1.445869 [DOI] [Google Scholar]
- White R. P. and Meirovitch H., J. Chem. Theory Comput. 2, 1135 (2006). 10.1021/ct0503217 [DOI] [PubMed] [Google Scholar]
- Cerutti D. S., Trong I. L., Stenkamp R. E., and Lybrand T. P., J. Phys. Chem. B 113, 6971 (2009). 10.1021/jp9010372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gō N. and Scheraga H. A., J. Chem. Phys. 51, 4751 (1969). 10.1063/1.1671863 [DOI] [Google Scholar]
- Gō N. and Scheraga H. A., Macromolecules 9, 535 (1976). 10.1021/ma60052a001 [DOI] [PubMed] [Google Scholar]
- Karplus M. and Kushick J. N., Macromolecules 14, 325 (1981). 10.1021/ma50003a019 [DOI] [Google Scholar]
- Meirovitch H. and Alexandrowicz Z., J. Stat. Phys. 15, 123 (1976). 10.1007/BF01012031 [DOI] [Google Scholar]
- Meirovitch H., J. Chem. Phys. 111, 7215 (1999). 10.1063/1.480050 [DOI] [Google Scholar]
- Meirovitch H., Int. J. Mod. Phys. 1, 119 (1990). 10.1142/S0129183190000062 [DOI] [Google Scholar]
- Meirovitch H., Phys. Rev. A 32, 3709 (1985). 10.1103/PhysRevA.32.3709 [DOI] [PubMed] [Google Scholar]
- Yang L. W., Rader A. J., Liu X., Jursa C. J., Chen S. C., Karimi H. A., and Bahar I., Nucleic Acids Res. 34, 24 (2006). 10.1093/nar/gkl084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. E., Chen W., and Gilson M. K., J. Chem. Theory. Comput. 1, 1017 (2005). 10.1021/ct0500904 [DOI] [PubMed] [Google Scholar]
- Singh N. and Warshel A., Proteins 78, 1724 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Straatsma T. P., and McCammon J. A., J. Chem. Phys. 100, 9025 (1994). 10.1063/1.466707 [DOI] [Google Scholar]
- General I. J., Dragomirova R., and Meirovitch H., J. Phys. Chem. B (in press). [DOI] [PMC free article] [PubMed]
- Lazaridis T., Masunov A., and Gandolfo F., Proteins 47, 194 (2002). 10.1002/prot.10086 [DOI] [PubMed] [Google Scholar]
- Mihailescu M. and Meirovitch H., Entropy 12, 1946 (2010). 10.3390/e12081946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. and Warshel A., Proteins 78, 1705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. A., Proteins 57, 645 (2004). 10.1002/prot.20294 [DOI] [PubMed] [Google Scholar]