Abstract
By identifying gene products whose knockdown is associated with phenotypic changes, large-scale RNA-mediated interference screens have demonstrated previously unknown components of biological pathways. This commentary provides general guidelines for using such screens to answer questions of immunological interest.
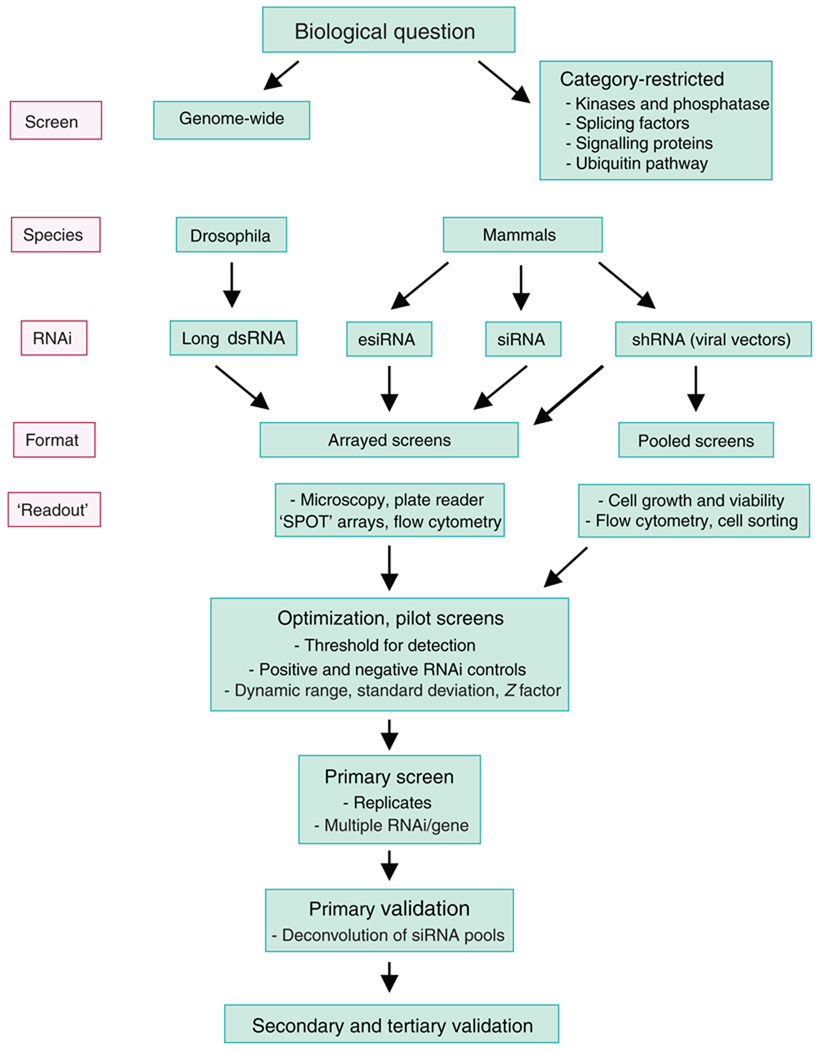
In barely a decade, RNA-mediated interference (RNAi) has evolved from a fascinating biological phenomenon into a powerful experimental tool. Progress in genome sequencing and annotation, as well as technological advances that permit biological assays to be executed and analyzed in high throughput, has sparked increasing interest in genome-wide RNAi screens through which the effects of gene silencing on biological phenotypes can be systematically explored. The prospective screener faces many challenges (Fig. 1). These include identifying the best experimental system with which to answer the biological question of interest, choosing whether to screen genome wide or to focus on subsets of already annotated genes, deciding whether to screen for targets individually or do pooled RNAi screens, and developing robust and reproducible assays for the identification and further characterization of interesting candidate genes. Technical considerations include the method of RNAi delivery, the inclusion of appropriate positive and negative controls, the elimination of false positives due to the inherent potential of RNAi-based technologies to produce off-target effects, the choice of statistical methods to define the initial ‘hit’ list, and the design of relevant secondary analyses to narrow the ‘hits’ and assess biological mechanism. We hope that this Commentary will be useful as a practical guide to students, postdoctoral fellows and other investigators trying to evaluate whether RNAi screens could be feasible and useful in their own research program.
Figure 1.
Steps involved in RNAi screening. esiRNA, endonuclease-prepared siRNA.
The biological question
What types of questions can be answered by RNAi screens? One approach is to do genome-wide screens that seek to identify all possible regulators of a general biological process: cell proliferation, cell size and shape, mitosis or the efficiency of viral infection. More specific ‘readouts’ yield more focused answers: regulators of signaling by Jak kinase–STAT transcription factors, Hedgehog and the transcription factor NF-κB have been identified by RNAi screens done in conjunction with reporter assays, and regulators of apoptosis or DNA-damage pathways have been sought with similar screens1–3. The second approach is narrower and aims to identify a ‘holy grail’: an unknown protein in a specific pathway that is presumed to exist but whose molecular identity is not yet known. This approach is especially fruitful if the desired protein falls into a known category, as the screen can then be limited to a defined subset of target genes; however, it becomes crucial that every candidate in the category is classified correctly through bioinformatics analyses and tested in the screen. Limited RNAi screens have been used to identify the deubiquitinating enzyme CYLD as a key regulator of the NF-κB pathway4 and the heterogeneous ribonucleoprotein hnRNPLL as an inducible regulator of CD45 pre-mRNA splicing5,6.
Genome-wide RNAi screens are most successful when they incorporate a focused search for predicted regulatory proteins. Drosophila RNAi screens have been used to identify regulators of store-operated Ca2+ entry, a process necessary for sustained nuclear translocation of the transcription factor NFAT7–9. Although calcium-activated NFAT proteins are present only in vertebrates, the pathways for NFAT activation and deactivation are conserved through evolution; thus, the screens yielded, as predicted, key proteins in the calcium entry pathway, the endoplasmic reticulum calcium sensor Stim and the channel pore subunit Orai, whose existence had been inferred from electrophysiological experiments. Conversely, a drosophila screen for negative regulators of NFAT nuclear translocation identified three distinct kinases10: CK1 and GSK3 were already known to phosphorylate two separate serine-rich motifs of NFAT1 and to promote its nuclear export, but a third kinase, DYRK, that was predicted to exist on the basis of phosphorylation of a third serine-rich motif, was discovered through the RNAi screen followed by targeted phosphorylation assays. Similar screens could be applied to the identification of regulators of other transcription factors that translocate to the nucleus.
Questions that can potentially be answered with RNAi screens abound. In principle, RNAi screens of cultured cells could identify the complete set of regulators (activators or inhibitors) of the differentiation of CD4+ and CD8+ T cells, the activation and differentiation of B cells and dendritic cells (DCs), cytokine production, the expression of cell surface or intracellular proteins, mast cell degranulation and the cytolytic activity of natural killer cells or cytolytic T cells. The fact that several regulators of each process are already known should not be a deterrent; these serve as invaluable positive controls in setting up the assay, and different family members may be discovered to have specific functions in the same pathway in different cell types. More complex assays could also be devised involving coculture of two or more cell types to investigate, for example, the mechanisms by which parasites elicit a T helper type 2 (TH2) response or regulatory T cells suppress effector T cell function. Screens for ‘synthetic lethality’ may be used to discover proteins that act in synergy with recognized factors (discussed below). And finally, limited RNAi screens could be used to identify ubiquitin-pathway enzymes linked to T cell and B cell signaling and anergy, specific small G proteins that participate in particular aspects of cellular signaling or intracellular trafficking and lipid transferases that modify transmembrane signaling proteins such as Lat.
Are whole-organism RNAi screens feasible and informative? In the nematode Caenorhabditis elegans, RNAi screens—done by arraying worms in 96-well plates and feeding them bacteria able to produce long double-stranded RNA (dsRNA) corresponding to individual annotated genes—have been critical in characterizing fundamental biological processes (such as growth, viability, post-embryonic morphology and motility)1–3. In mice, lentiviral short hairpin RNA (shRNA) directed against individual genes (such as the gene encoding the immunomodulatory receptor CTLA-4) has been used to compare hypomorphic phenotypes resulting from partial knockdown with the more severe phenotypes produced by total gene deletion11. However, whole-genome or even limited RNAi screens based on strictly in vivo ‘readouts’ are not practical in mice at present, mostly because of low efficiency and high expense. An alternative is pooled RNAi screens, in which populations of virus-transduced cells are reintroduced into mice and selected for loss or acquisition of a given phenotype (discussed below). Pooled RNAi screens can identify cell-intrinsic factors that modulate cancer growth and metastasis or control the activity (such as differentiation, function and homing) of cells in an organism.
Delivering small interfering RNA
RNAi is effected in cells by small interfering RNA (siRNA): 21- to 27-nucleotide double-stranded oligonucleotides with a characteristic 2-nucleotide 3′ overhang. These are produced from longer dsRNA by the cellular type III endoRNase Dicer. One strand or both strands of the siRNA become(s) incorporated into a large protein complex known as the ‘RNA-induced silencing complex’, which then targets and cleaves complementary messenger RNA (mRNA)12. Experimentally, RNAi can be induced in several different ways that are classified by the chemistry of the effector molecules (Fig. 2).
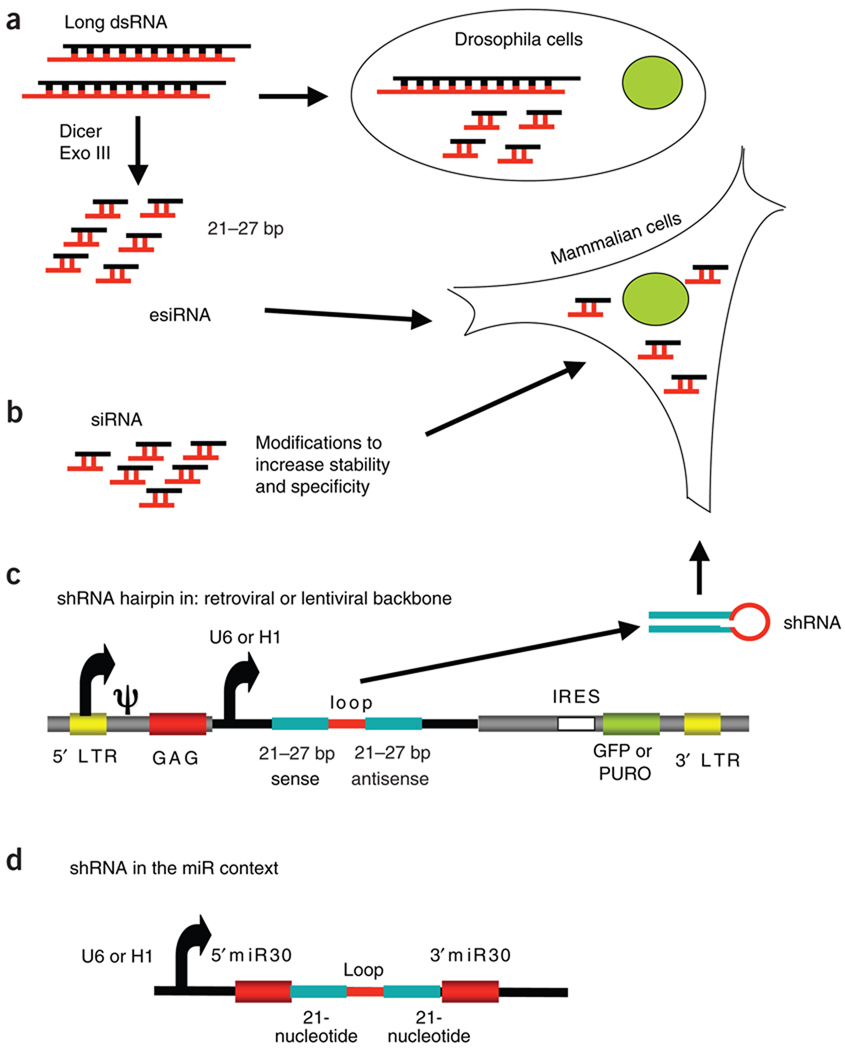
Figure 2.
Various types of RNAi chemistry. (a) Long strands of dsRNA (200–500 base pairs (bp)) are taken up by insect cells and are cleaved intracellularly into a pool of siRNA (21–27 base pairs); for mammalian cells, long strands of dsRNA are cleaved in vitro to generate endonuclease-prepared siRNA. (b) Synthetic siRNA molecules have chemically modified termini to increase stability and specificity of knockdown in cells. (c) Molecules of shRNA are ectopically expressed in cells from a U6 or H1 promoter; they can be delivered by a viral backbone with a selectable marker such as green fluorescent protein (GFP) or antibiotic resistance (for example, to puromycin (PURO)). LTR, long terminal repeat; GAG, group-associated antigen; IRES, internal ribosomal entry site. (d) The shRNA transcripts can also be expressed with additional micro-RNA (miR) sequences flanking the hairpin sequence.
Long dsRNA molecules are taken up and cleaved intracellularly by Dicer to yield a pool of overlapping siRNA molecules with high specificity for the target gene. This approach has been very useful in model organisms (C. elegans and drosophila) that lack the defensive type I interferon response of their mammalian counterparts. In mammalian cells, however, long dsRNA activates the antiviral type I interferon response, which can obfuscate phenotypic screening, in part through activation of the dsRNA-binding protein kinase R, which alters cellular transcription and translation13. The interferon response can be minimized through the use of pools of siRNA prepared by the digestion of long dsRNA templates in vitro with recombinant bacterial endoRNase III (ref. 14) or Dicer15, as these do not ‘trip’ cellular defenses when applied at low concentrations13.
Synthetic siRNA molecules are designed to mimic the end products of Dicer cleavage16 and may incorporate a range of chemical modifications designed to increase duplex stability and maximize the efficiency and specificity of depletion of the target mRNA17. Many siRNA libraries are commercially available with nearly complete genome coverage for several species; most libraries pool three or more target sequences per gene, with an estimated probability of silencing of over 95% (ref. 1). The siRNA molecules are readily delivered to established or transformed cell lines by transient transfection and can promote very efficient degradation of the target mRNA; however, they are ‘diluted out’ by cell division and multiple transfections may be needed. The extent of depletion of target proteins varies with their lifetime: short-lived proteins may be diminished to 10% or less of their normal abundance within 3–5 days, whereas long-lived proteins may show little or no depletion.
For primary cells, transient transfection is an inefficient method of siRNA delivery. Electroporation of primary T cells results in successful gene silencing only if very high siRNA doses are used (over tenfold higher than those needed for the established HeLa human cervical cancer cell line)18. Similarly, effective knockdown of the NF-κB subunit p50 in cultures of primary human DCs requires treatment with 500 nM siRNA19, compared with the 10–50 nM required for most established cell lines. The consequent high costs for siRNA oligonucleotides are likely to prove prohibitive for large-scale RNAi screening of primary cells.
Molecules of shRNA are ectopically expressed in cells as stem-loop (‘hairpin’) structures that resemble pre-microRNA, the endogenous substrate of Dicer20. Although U6 and H1 RNA polymerase III promoters were initially used and continue to be used to drive shRNA expression3,20,21, efficient delivery is obtained when effector hairpins are transcribed by RNA polymerase II in the context of the endogenous primary transcripts that encode microRNA22. Transient and stable transfection, as well as adenoviral, retroviral and lentiviral infection, have all been used1,3. Retroviral and adenoviral backbones are appropriate for most cultured lines and some primary cells, including hepatocytes, cardiomyocytes and activated lymphocytes, whereas delivery of shRNA to nondividing or post-mitotic cells, including neurons and immature DCs, is best achieved with lentiviral vectors21. Lentiviral delivery has also been successfully applied to gene silencing in primary human macrophages23, mouse osteoblasts24 and human T lymphocytes5. Retroviral and lentiviral methods are suited to primary cells in which transient transfection is inefficient; when delivered by viral transduction from a regulated promoter with a selectable marker gene, shRNA cassettes mediate stable and conditional gene silencing independently of transfection efficiency, an ideal approach for hematopoietic cells. The stable shRNA establishes an equilibrium between mRNA synthesis and degradation that in turn determines the extent of protein depletion. Several vector shRNA libraries are now available1,3, and backbones are continuously under refinement to improve long-term expression of the integrated shRNA cassette.
Transduction approaches place a burden on the screener in terms of reagent quality and screen optimization. High-titer virus stocks must be rigorously monitored for uniform and reproducible transduction and, ideally, uniform efficiency of target depletion. Also, researchers must strictly adhere to proper protocols for the handling infectious amphotropic virus, particularly in high-throughput analysis.
The experimental ‘readout’
The quality of an RNAi screen depends heavily on methods for detecting the desired phenotype robustly, reliably and in high-throughput format. Many traditional assays can be customized to a multiwell plate reader format (such as an enzyme-linked immunosorbent assay), to a microscopy-based format in which a robotic camera reads several positions in each well of a multiwell plate, or to a flow cytometry–based format in which an automated ‘head’ picks up and sequentially processes samples that have been incubated with antibodies in multiple wells of a multiwell plate. Flow cytometry is an ideal method for assessing the phenotypes of limited numbers of cells in suspension, in part because it controls for variations in cell number between samples. In ‘SPOT’ arrays, microchips are spotted with individual RNAi reagents and then overlaid with cells, and phenotypic changes are monitored directly by imaging of the cells overlying the spots25,26.
In an informative example of an siRNA screen, a classical assay for cell motility was adapted to a high-throughput screening format through the reproducible use of a robotic pin to ‘wound’ confluent monolayers of MCF-10A breast epithelial cells cultured in 96-well plates27. The objective was to identify genes involved in the regulation of epithelial cell migration. Cell migration into the wound area was visualized by fluorescence microscopy with vital dyes and results were normalized to control for changes in cell viability as a result of siRNA transfection. The authors were able to calculate wound-healing scores and to compile a high-confidence data set for candidate genes involved in cell migration and cell motility.
Subcellular localization is an ideal ‘readout’ if suitable high-throughput assays (flow cytometry or high-content cellular imaging) are available. Cytoplasmic-to-nuclear shuttling is particularly easy to quantify with automated microscopy and image-analysis programs27. Certain biological processes (for example, degranulation, cytolysis, T cell signaling and T cell anergy) have steps that involve endocytosis or exocytosis; these can be ‘read out’ in high-throughput format as loss of a protein component from the plasma membrane or insertion of an intracellular membrane component (for example, LAMP-1) into the plasma membrane. Other convenient assays include changes in protein-protein association monitored by fluorescence resonance energy transfer and changes in phosphorylation status monitored with phosphorylation-specific antibodies. These assays are especially useful if they monitor a major end point of a complex biological pathway1,3.
If the pathway of interest shows some degree of evolutionary conservation, a nonmammalian system has several advantages. The RNAi reagents (long dsRNA) are publicly available, and the absence of an interferon response facilitates greater depletion of target proteins in drosophila cells than in mammalian cells1–3. Moreover, as illustrated by the NFAT RNAi screens, the genetic redundancy in model organisms is low: drosophila has only a single ortholog of the two mammalian Stim proteins and three mammalian Orai proteins7–9, which increases the feasibility of identifying previously unknown loss-of-function phenotypes. Finally, as also illustrated by the NFAT screen, off-target effects in nonmammalian systems may actually be an advantage: the complex mixture of overlapping 21- to 22-nucletoide siRNA molecules that results from the action of Dicer on long dsRNA molecules contains several siRNA molecules that target mRNA molecules encoding related family members with high efficiency. As a result, dsRNA-mediated knockdown of single representatives of the calcineurin and the CK1, GSK3 and DYRK kinase families leads to substantial depletion of other family members, which results in robust effects on the nuclear translocation of NFAT10.
Statistical analysis
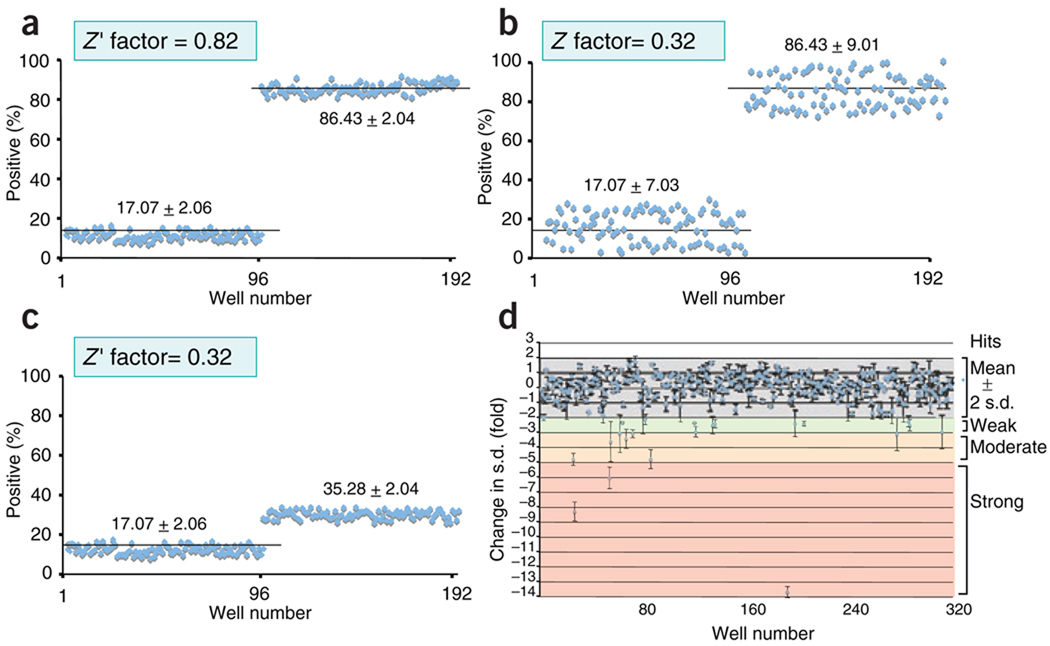
Whatever the format, a suitable primary assay for high-throughput screening must have a strong signal/background ratio and low variation28 (Fig. 3). Before primary screening, pilot experiments should be done in which RNAi doses, incubation times and other assay variables are ‘titrated’ to optimize assay responses to negative and positive control siRNA molecules; these control values determine baseline, signal and standard deviation (s.d.) values for the system. A parameter known as the ‘Z factor’ is now in general use: it reports assay quality regardless of how the assay is set up and thus can be used to determine which of a large number of possible assay formats is robust enough for a screen. The Z factor is calculated through the use of many replicates of the assay with positive and negative controls; a value of at least 0.5 (ideally, >0.7) indicates a suitable difference between signal and background values together with low variability (Fig. 3).
Figure 3.
Z-factor calculation and ranking of screen ‘hits’. (a) A good assay for high-throughput screening has a low s.d. and a high signal response in wells 1–96 (baseline average) relative to that in wells 97–192 (stimulated average). (b,c) In a poor assay, higher s.d. in the baseline and stimulated averages (b) or lower baseline/signal values (c) decrease the Z-factor calculated. (d) ‘Hits’ from a sample screen plate (n = 384) ranked by change in s.d. from the mean (fold): weak ‘hits’ are two to three s.d. from the mean; moderate hits are three to five s.d. from the mean; and strong hits are over five s.d. from the mean.
In the large-scale screen, negative and positive control siRNA molecules should ideally be included on every plate with experimental siRNA molecules to monitor quality control during screening. ‘Hits’ are typically identified as replicates that are beyond two to three s.d. values from the mean or beyond a predetermined phenotypic threshold (for screens that assess morphology or other parameters that cannot be readily quantified). Although mean and s.d. values may be calculated with control samples (not treated with siRNA), we recommend that they be calculated with the entire set of experimental siRNA molecules to control for general complications due to RNAi (discussed below); strong ‘hits’ (typically >5 s.d. from mean in replicate) should be excluded so as not to skew the calculated mean values. Arrayed screens should be done at least in duplicate, and preferably in triplicate, to minimize false discovery; for pooled screens, more replicates are preferable29–31.
Off-target effects and other caveats
Off-target effects are a major problem of RNAi-based screening and may contribute substantially to the false discovery of candidate proteins32. Three types of off-target effects can be distinguished. First, if an siRNA sequence is identical or nearly identical to a sequence present in an unrelated mRNA, the resulting degradation of the unrelated mRNA may produce a confusing phenotype or produce a false-positive result. This problem can be avoided by careful choice of the siRNA sequence used, and in fact such problem sequences are removed from commercial siRNA and shRNA libraries on a continuing basis. Second, if the ‘seed region’ of an siRNA pairs with a weakly complementary sequence in the 3′ untranslated region of an unrelated mRNA, the siRNA can act like a microRNA and cause depletion of non-target proteins through mRNA degradation or translational block32. This problem is intrinsic to RNAi screens and is much more difficult to avoid. Third, in mammalian cells, even short siRNA may activate the antiviral type I interferon response in a sequence-independent way, especially if saturating doses of siRNA or shRNA are delivered13. Additional unintended effects may arise through the activation of Toll-like receptor or other cellular signaling pathways; in some cases, this has been traced to the presence of GU-rich sequences in the siRNA or shRNA33
Off-target effects can be minimized by effective screen design. Instead of mock manipulations or buffer controls, nontargeting or scrambled RNAi sequences should be used, and interferon responses and cell activation and/or differentiation that might be induced by electroporation, nucleofection, viral infection or RNAi should be monitored so that nonspecific effects due to general cellular responses to siRNA delivery methods or to RNAi itself may be assessed. Changing the method of siRNA delivery and ‘titrating down’ the amount of siRNA delivered may decrease unwanted effects. Multiple siRNA species should be used for each target gene; these are often applied in pools of three to five to maximize knockdown and throughput but should be rescreened individually in the first step of ‘hit’ validation. At least two siRNA molecules in the pool should recapitulate the phenotype, and there should be a clear correlation between protein depletion and phenotypic severity. Whenever possible, parallel loss-of-function assays with null strains, pharmacological inhibitors or gene mutants should be used to confirm data from siRNA-mediated knockdown. The final word in target validation is to reintroduce an RNAi-resistant version of the target gene product into depleted cells and show recovery of function. This can be challenging for large sets of candidate genes but is essential for validating candidates of particular interest.
The candidates identified in RNAi screens may depend to a large extent on the experimental system, as illustrated by a group of three genome-wide siRNA screens done to characterize host cellular proteins involved in the life cycle and replication of human immunodeficiency virus type 1 (HIV-1)34–36. In these experiments, human embryonic kidney (HEK) cells36 or HeLa cells expressing the HIV receptor CD4 (refs. 34,35) were transiently transfected with pools of siRNA and then infected with HIV-1. Virus replication was assessed by monitoring of expression of the HIV-1 core antigen p24 or activation of an HIV-1 reporter construct. Each of the three screens identified several hundred previously unknown proteins essential for the virus life cycle, but direct comparison of the three data sets showed an overlap of only 13–15 genes37. The limited overlap in what should have been very similar screens may have reflected off-target effects, differences in cell types and critical assay parameters such as incubation time for siRNA transfection and virus infection, as well as key differences in secondary and tertiary filters applied to the primary data sets37.
Individual versus pooled screens
It has now become feasible to do functional analyses of RNAi-treated cells in a pooled format. Pooled screens have the advantage that a large set of shRNA molecules, perhaps targeting an entire genome, can be delivered to bulk populations of cells, thereby allowing rapid evaluation of large gene sets and/or parallel analyses of multiple cell types. Cells are infected with pools of shRNA lentiviruses or retroviruses to yield on average one stably integrated shRNA per cell, then the cells are selected for loss or acquisition of a given phenotype, either ex vivo or (in principle) after introduction into mice. The ‘readout’ must involve a selectable change in cell phenotype–cell viability or cell proliferation or altered expression of a protein that can be detected by flow cytometry (for example, a surface marker expressed through activation of a signaling pathway, or a fluorescent reporter responsive to transcription, splicing, somatic hypermutation or class-switch recombination). Cells that have lost or acquired the desired phenotype are collected, RNA is prepared and enriched or depleted shRNA sequences are identified through quantification of half-hairpins or other ‘bar codes’ by PCR amplification and microarray29–31. Pilot screens are necessary to determine the optimal cell number, multiplicity of infection and selection time that favor reliable enrichment or depletion of shRNA.
In an elegant study, two clinically distinct diffuse large B cell lymphoma subtypes, germinal center and activated B cell–like, were analyzed to identify discrete gene sets associated with cell survival29. Diffuse large B cell lymphoma cell lines of the two subtypes were retrovirally transduced with doxycyline-inducible shRNA cassettes, and hairpins specifically eliminated from the populations expressing doxycyline-induced shRNA were identified. The lymphoid-specific scaffolding protein CARMA1 was shown to be a key mediator for survival of the activated B cell–like subtype but not the germinal center lymphoma subtype, through constitutive activation of NF-κB signaling29.
Validation: secondary analyses
Secondary assays that refine and characterize a functional gene set should involve assays that are complementary (orthogonal) to those used in the primary screen. If mice are available in which a candidate gene has been disrupted in the genome, the experiments should be repeated with cells from these mice; if kinases or other enzymes are identified in the primary screen, known specific inhibitors should be tested to see if they have the same effect; and if candidate transcription factors are identified, they should be tested to see if they affect transcription of relevant genes important in the biological pathway assessed in the screen. Epistasis experiments involving combined knockdown and/or overexpression should be done to determine whether two candidates identified by the screen are in the same signaling pathway; coimmunoprecipitation or tandem affinity purification may be used to establish physical interactions, if any, among candidates; and database mining and computational analyses may be used to establish network relationships among a set of candidates validated in the screen.
Some of the richest examples of orthogonal strategies are relevant to immunology. The discovery of the drosophila protein Orai in a functional genome-wide RNAi screen for NFAT regulators was validated through a second genome-wide screen for single-nucleotide polymorphisms in an extended family that included two homozygous patients with hereditary severe combined immunodeficiency, as well as heterozygous carriers who were not immunodeficient but could be identified because their T cells had 50% as much store-operated Ca2+ entry7. Linkage analysis identified a 9-megabase genomic region that contains ORAI1, one of the three human homologs of the drosophila gene encoding Orai. Similarly, on the basis of previously published findings that growth of the activated B cell–like subtype of diffuse large B cell lymphoma is associated with activating mutations in the sequence encoding the coiled-coiled motif of CARMA1 (refs. 29,38), proteins that interact with CARMA1 have been characterized by coimmunoprecipitation and mass spectrometry38. This strategy has identified CK1α as a CARMA1-specific kinase, and subsequent viability screening with pooled shRNA molecules has indicated that like CARMA1 expression, CK1α expression is essential for the survival of activated B cell–like lymphoma cells. When a similar pooled viability screen identified a requirement for the transcription factor IRF4 in multiple myeloma cell lines, the orthogonal approaches used to delineate an IRF4 signaling network in B cell malignancies were gene-expression profiling of IRF4-depleted B cell lymphoma subtypes, as well as genome-wide IRF4 chromatin immunoprecipitation and DNA microarray39.
‘Synthetic lethal’ screens
The idea of synthetic lethality derives from yeast genetics, in which two mutations are said to be ‘synthetically lethal’ if cells with single mutations are viable but cells with both mutations are unable to survive. Synthetic lethal phenotypes are a clear indication that there is a functional interaction between the gene products whose mutant versions confer lethality when combined. Modified screens for synthetic lethal mutations, done in cancer cells by combining low concentrations of a chemotherapeutic drug with whole-genome siRNA screens, can provide clues to combinations of therapies that might be particularly useful for specific cancer subtypes40: depletion of several core proteasome components enhances the sensitivity of a non-small-cell lung cancer cell line to paclitaxel, a microtubule-disrupting drug41. The idea can be extended to cellular phenotypes other than lethality; for example, subtle but important modifiers of TH1, TH2, TH-17 or inducible regulatory T cell differentiation may be identified by partial knockdown of key lineage-specific transcription factors (for example, T-bet, GATA-3, RORγt or Foxp3, respectively) in differentiating CD4+ T cells, in conjunction with genome-wide or subset-restricted depletion of candidate gene products.
Conclusion
Despite the fact that genome sequence information has been available for many years, most genes remain poorly annotated and their functions remain unknown. Genome-wide or subset-limited RNAi screens are an ideal method for discovering genes that encode molecules with previously unknown functions in a biological pathway of interest. We hope that this Commentary has provided readers with ideas for how to use RNAi screens effectively in their own research and has clarified critical points of screen design and the technical issues and limitations involved.
ACKNOWLEDGMENTS
Supported by the US National Institutes of Health (A.R.), the Juvenile Diabetes Research Foundation (A.R.), the Canadian Institutes for Health Research (S.S.) and the Leukemia and Lymphoma Society (S.S.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/natureimmunology/.
References
- 1.Echeverri CJ, Perrimon N. Nat. Rev. Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 2.Perrimon N, Mathey-Prevot B. Genetics. 2007;175:7–16. doi: 10.1534/genetics.106.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moffat J, Sabatini DM. Nat. Rev. Mol. Cell Biol. 2006;7:177–187. doi: 10.1038/nrm1860. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 5.Oberdoerffer S, et al. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, et al. Immunity. 2008;29:839–841. [Google Scholar]
- 7.Feske S, et al. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 8.Vig M, et al. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang SL, et al. Proc. Natl. Acad. Sci. USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gwack Y, et al. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Stockton J, Mathis D, Benoist C. Proc. Natl. Acad. Sci. USA. 2006;103:16400–16405. doi: 10.1073/pnas.0607854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannon GJ, Rossi JJ. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 13.Sledz CA, Williams BR. Biochem. Soc. Trans. 2004;32:952–956. doi: 10.1042/BST0320952. [DOI] [PubMed] [Google Scholar]
- 14.Kittler R, et al. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 15.Liou J, et al. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbashir SM, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bramsen JB, et al. Nucleic Acids Res. 2009;10:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McManus MT, et al. J. Immunol. 2002;169:5754–5760. doi: 10.4049/jimmunol.169.10.5754. [DOI] [PubMed] [Google Scholar]
- 19.Laderach D, Compagno D, Danos O, Vainchenker W, Galy A. J. Immunol. 2003;171:1750–1757. doi: 10.4049/jimmunol.171.4.1750. [DOI] [PubMed] [Google Scholar]
- 20.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 21.Stewart SA, et al. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva JM, et al. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto T, et al. Blood. 2006;108:3305–3312. doi: 10.1182/blood-2006-04-014829. [DOI] [PubMed] [Google Scholar]
- 24.Wein MN, Jones DC, Glimcher LH. Methods Mol. Biol. 2008;455:149–155. doi: 10.1007/978-1-59745-104-8_11. [DOI] [PubMed] [Google Scholar]
- 25.Ziauddin J, Sabatini DM. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 26.Neumann B, et al. Nat. Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 27.Simpson KJ, et al. Nat. Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 28.Zhang JH, Chung TD, Oldenburg KR. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 29.Ngo VN, et al. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 30.Luo B, et al. Proc. Natl. Acad. Sci. USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlabach MR, et al. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Echeverri CJ, et al. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 33.Judge AD, et al. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 34.Brass AL, et al. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 35.Zhou H, et al. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 36.König R, et al. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goff SP. Cell. 2008;135:417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bidère N, et al. Nature. 2009;458:92–96. doi: 10.1038/nature07613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaffer AL, et al. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaelin WG., Jr Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 41.Whitehurst AW, et al. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]