Abstract
The aryl hydrocarbon receptor is a ligand-activated transcription factor responsive to both natural and synthetic environmental compounds, with the most potent agonist being 2,3,7,8-tetrachlotrodibenzo-p-dioxin. The aim of this work was to develop a categorical COmmon REactivity PAttern (COREPA)-based structure–activity relationship model for predicting aryl hydrocarbon receptor ligands within different binding ranges. The COREPA analysis suggested two different binding mechanisms called dioxin- and biphenyl-like, respectively. The dioxin-like model predicts a mechanism that requires a favourable interaction with a receptor nucleophilic site in the central part of the ligand and with electrophilic sites at both sides of the principal molecular axis, whereas the biphenyl-like model predicted a stacking-type interaction with the aryl hydrocarbon receptor allowing electron charge transfer from the receptor to the ligand. The current model was also adjusted to predict agonistic/antagonistic properties of chemicals. The mechanism of antagonistic properties was related to the possibility that these chemicals have a localized negative charge at the molecule's axis and ultimately bind with the receptor surface through the electron-donating properties of electron-rich groups. The categorization of chemicals as agonists/antagonists was found to correlate with their gene expression. The highest increase in gene expression was elicited by strong agonists, followed by weak agonists producing lower increases in gene expression, whereas all antagonists (and non-aryl hydrocarbon receptor binders) were found to have no effect on gene expression. However, this relationship was found to be quantitative for the chemicals populating the areas with extreme gene expression values only, leaving a wide fuzzy area where the quantitative relationship was unclear. The total concordance of the derived aryl hydrocarbon receptor binding categorical structure–activity relationship model was 82% whereas the Pearson's coefficient was 0.88.
Keywords: aryl hydrocarbon receptor, gene expression, modelling, SAR, COREPA, TCDD
1. Introduction
The aryl hydrocarbon receptor (AhR) is a ligand-activated cytosolic transcription factor that belongs to the basic helix-loop-helix Per-ARNT-Sim (bHLH/PAS) protein superfamily and regulates expression of diverse target genes in multiple species and tissues [1-5]. The AhR modulates the biochemical and toxic effects of a wide variety of environmental compounds and plays a role in adaptation to environmental stress [6]. Downstream effects of ligand-activated AhR are mediated by a multiprotein complex containing hsp90, XAP2 and p23. This complex undergoes nuclear translocation, after which the AhR is released from the complex by binding to ARNT (Ah Receptor Nuclear Translocator). The ligand:AhR:ARNT complex then binds to dioxin responsive elements (DREs) in target genes and induces transcription.
AhR-mediated responses are well characterized and include transcriptional induction of phase I and phase II metabolism genes such as CYP1A1 [7]. Typical high-affinity AhR agonists include chlorinated dioxin/furans, biphenyls and polycyclic aromatic hydrocarbons [8]. AhR antagonists have also been studied and their structural properties analysed. For example, ellipticine analogues, some which have been used by the European medical community to treat breast cancer, have been identified as AhR antagonists [9,10]. These ellipticine derivatives can competitively bind to AhR and inhibit AhR activation of gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, the most potent AhR agonist) [11-13]. Not only have numerous flavonoids been identified as AhR antagonists [14], but the AhR has been shown to bind and be activated and/or inhibited by structurally diverse natural and synthetic chemicals [8,15,16].
Although it is not yet possible to predict the pharmacokinetic, pharmacodynamic and toxicological properties of AhR ligands [14], the ligand:receptor interaction can be modelled using computation tools. These computational methods fall into two broad categories [17]. The first class of methods dock putative ligands onto previously determined structural models. Docking approaches based on existing structures of the specific protein receptor utilize either the molecular mechanism or an empirical scoring function to estimate the affinity of the ligand:receptor complex. Alternatively, when the structure of the protein receptor has not been determined, a homology model is generated using structures of a closely related protein or proteins. This process involves threading the sequence of the target receptor through an experimental template and mutating relevant amino acid residues in the template to match those of the target receptor. The second class of methods involve receptor mapping. In this approach, a model of the receptor is built based on the structural analysis of its ligands. 3D-QSAR (three-dimensional quantitative structure–activity relationship) is a receptor mapping approach, in which a series of ligands with known affinity are aligned. The strengths of the electrostatic and steric potentials of each ligand are then mapped onto a grid surrounding the molecule, and these data are correlated with the affinity of the ligand:receptor complex [18]. While a homology model of the AhR ligand binding pocket has been recently developed [19,20], the structure of the model still needs further refinement, validation and analysis of ligand binding using docking studies before the model can be used for these purposes. Thus, the latter approach is currently the optimal one for analysis of ligand binding computations.
Over the past 30 years, a great number of relatively simple QSARs, as well as many other in silico methods [21,22], have been used to model interactions between polychlorinated dibenzo dioxins/furans (PCDD/Fs) and the AhR. Some of these modelling studies have generated successful qualitative or quantitative predictions regarding ligand:receptor interactions. For example, a box model of 3 × 10 Å [23] was originally proposed, in which the planar skeleton plus halogen substituents in the lateral positions formed a rectangular shape. A stacking model [24] was also proposed, in which molecular polarizability and the separation distance between ligand and receptor were identified as critical determinants of ligand affinity. Furthermore, comparative molecular field analysis (CoMFA) provided a more detailed characterization of the ligand-binding domain of AhR [25-27], with the maximum dimensions of 14.0 Å in length, 12.0 Å along the medial axis and 5.0 Å along the perpendicular direction to the plane. Kafafi et al. [28] reported a structure–activity model of AhR interaction with PCDD/Fs, which considered molecular lipophilicities (Ls) quantified by octanol-water partition coefficient, electro affinities (EA), entropies (Ss) and the electronic energy gap (Eg). The relationship between AhR binding, enzyme induction and PCDD toxicity was evaluated, based on the equilibrium dissociation constant and the difference in the ionisation potential (IP) and electro affinity (EA). AhR binding was also analysed as a function of the energy of the lowest unoccupied molecular orbital (LUMO), hydrophobic factors and global steric indices [29]. Polarizabilities, as indices of effectiveness of the medium- and short-range interactions, were also computed for PCDDs with different basis sets: although no basis set effect was observed, the polarizability anisotropy was closely related to the position of chlorine substituents [30]. Recently, chemical softness, electronegativity [31] and electrophilicity index [32-34] (parameters derived from density functional theory (DFT)) were examined as potential determinants of AhR affinity and potency of PCDFs for AhR; however, only moderate relationships were observed [35].
The goal of this study was to define structural and parametric boundaries for AhR ligands within different AhR ranges of affinity/potency. SAR models were derived for different classes of chemicals grouped according to binding mechanism. The parametric boundaries were elucidated using the COmmon REactivity PAttern (COREPA) approach. The structural and parametric boundaries for antagonism and agonism were differentiated from each other. Furthermore, the categorical models for agonism and antagonism were correlated with the effects of agonists and antagonists on gene expression (GE), with GE measured using an AhR- and -DRE-responsive luciferase reporter gene assay. Based on this analysis, a system was developed for predicting GE outcomes from AhR binding affinity ranges, or conversely, binding affinity ranges from GE data. The system was developed on a training set composed of 142 chemicals, 23 of which had associated GE data, and tested on an external data set of 51 chemicals. The results demonstrated successful classification of non-binders, weak and strong agonists and antagonists of AhR, as well as successful verification based on experimental GE data.
2. Materials and methods
2.1 AhR binding affinity data
The categorical SAR model was developed using a training dataset composed of AhR binding affinities for 142 compounds falling into four chemical classes: polychlorinated biphenyls (PCBs), PCDFs, PCDDs, and ellipticines and flavones [22-25]. Binding affinities (Kd) for these compounds were converted into relative equivalent potency values (REP = Kd50TCDD/Kd50TEST CHEMICAL). The training set structures and their activity values are listed in Appendix 1. The synthetic ligands in the training set were grouped into three binding activity ranges: (1) strong binders with REP≥0.1 (30 chemicals), (2) weak binders with 0<REP<0.1 (52 chemicals), and (3) non-binders with REP=0 (60 chemicals). GE results for 51 chemicals were mostly collected from the literature [36-40] and used as an external set for model validation. Both AhR binding and GE data were available for 23 of these chemicals and they were used to establish a relation between binding and GE. The external validation set is listed in Appendix 2.
2.2 Conformational analysis by genetic algorithm
A method for coverage of the conformational space by a limited number of conformers was developed [41]. Time-complexity for a systematic conformational analysis search increases exponentially with degrees of freedom, making it computationally intractable. Therefore, a genetic algorithm (GA) was employed instead of a systematic search, because it minimizes 3D similarity among generated conformers. This makes the problem computationally feasible even for large and flexible molecules. To minimize the effects of the non-deterministic character of GA on the reproducibility of generated conformers and their distribution in the structural space, a procedure was developed for saturating the conformation space [42]. This allows the conformational space of chemicals to be populated with an optimal number of conformers. Stable conformational distributions are then achieved across selected molecular descriptors, and the distributions are not perturbed by addition of new conformers. The conformer distributions associated with saturated conformation space are expected to provide reliable reactivity patterns (see the section on COREPA approach below).
Each of the generated conformers is submitted to a geometry optimization procedure by quantum-chemical methods. Usually, MOPAC 93 [43,44] is employed by making use of the AM1 Hamiltonian. Next, the conformers are screened to eliminate those whose heat of formation, DHf°, is greater than the DHf° associated with the lowest energy conformer by a user-defined threshold (20 kcal/mol). Subsequently, conformational degeneracy, due to molecular symmetry and geometry convergence, is detected within a user-defined torsion angle resolution.
2.3 The COREPA method
The COREPA is a probabilistic classification scheme identifying criteria which will classify an unknown object into predefined classes using a training set composed of objects from multiple classes [45,46]. The COREPA formalism uses a Bayesian probabilistic method to identify common structural characteristics among chemicals that elicit similar biological activity. Instead of single parameter values for each chemical corresponding to individual conformers, their probabilistic conformational distributions in the molecular descriptor space are analysed and compared, thus accounting for molecular flexibility. The common reactivity pattern is developed by seeking overlap between the conformer distributions of biologically similar chemicals in the specific structural space. The parameters discriminating common reactivity patterns of biologically dissimilar chemicals were considered to be related to the endpoint under investigation (Figure 1).
Figure 1.
Illustration of: (a) discrete conformer distributions of two chemicals across E(HOMO) parameters; (b) conformer distributions of both chemicals.
Thus, the problem of structure alignment typically used for similarity assessments is circumvented in COREPA by overlapping and comparing conformational distributions of chemicals across the descriptor axis. Deriving reactivity pattern by the COREPA approach does not require aligning of structures, and allows identifying the common reactivity pattern populated mainly by the conformers of biologically similar chemicals.
In the original formulation of the COREPA method, the common reactivity patterns were determined across a single parameter axis in terms of parameter ranges [47] (see Figure 1). While easy to interpret, the one-dimensional formulation significantly limits the discriminative power of the COREPA approach. In the current formulation of the COREPA method, multi-dimensional reactivity patterns (COREPA-M) are developed. To provide mechanistic transparency of the reactivity pattern, the number of parameters (molecular descriptors) was limited to three.
2.4 The model applicability domain
The reliability of the predictions made by the AhR model was evaluated by a stepwise approach and the model applicability domain was determined [48]. In this approach, four stages are applied to account for diversity and complexity of the QSAR models, reflecting their mechanistic rational and transparency. Three of the four steps are described in this work. General parametric requirements are imposed in the first stage. Here, the domain is only specified for those chemicals in the training set for which selected physico-chemical parameters fall in a specified range of variation.
The second stage analyses and defines the structural similarity between chemicals which are correctly predicted by the model. The structural neighbourhood of atom-cantered fragments is used to determine this similarity. Atom-cantered fragments are extracted from training set chemicals for which the QSAR model provides correct predictions (within user defined accuracy thresholds); thus, a list of ‘good fragments’ is compiled, which is then used to assess non-training set chemicals. If the atom-cantered fragments for each atom constituting an external chemical are elements of this list, then the chemical belongs to the structural domain of the model. If the atom-cantered fragments of any atoms constituting an external chemical are not elements of this list, it does not belong to this domain.
The third stage in defining the domain is based on a mechanistic understanding of the modelled phenomenon, i.e., domain of the mechanistic hypothesis. Here, the model domain combines reliability of specific reactive groups, hypothesized to cause the effect and the domain of explanatory variables, and determines the parametric requirements that elicit functional group reactivity.
3. Results and discussion
3.1 Basic modelling assumption
This study uses the following criteria/definitions to build categorical SARs for AhR binding states: (1) if a chemical binds AhR, but fails to trigger a cellular response (i.e., altered GE), the chemical is classified as an antagonist; (2) if a chemical binds AhR and elicits a GE response, the chemical is classified as an agonist. Chemicals that bind to the receptor but do not meet the structural boundaries for antagonism were categorized as agonists (chemicals that mimic the action of a naturally occurring substance). These chemicals were further categorized as strong or weak agonists.
3.2 AhR binding model
The training set includes three classes of AhR ligands: AhR dioxin-like compounds, PAHs and biphenyls. Dioxin-like compounds include chlorinated dibenzo-p-dioxins and chlorinated dibenzofurans, flavones and carbazoles. The assumption that dioxin-like compounds bind to the AhR in a similar manner is based on published studies (Procopio et al. [49]). According to these studies, the AhR binding affinity of polychlorinated dioxins is related to the negative molecular electrostatic potential at the extreme ends of the ligand's long axis and a depleted charge above and below the aromatic rings. This mechanism applies to chlorinated dibenzofurans, flavones and carbazoles, all of which interact with AhR by a so-called ‘dioxin-like mechanism’.
The concentration of nucleophilic sites at the central part of a molecule and electrophilic sites at the extreme ends of the molecule was a common structural feature for high and low-affinity dioxin-like AhR ligands (Figure 2). Oxygen, sulphur or nitrogen provides nucleophilic sites in the central part of the molecule, and halogen substituents provide electrophilic sites at the extreme ends. High-affinity AhR binding requires a nucleophilic centre surrounded by ≥ two bromines or four chlorines (at positions 2,3,7,8) per benzene ring. Low-affinity AhR binding is observed for dioxins with two or three chlorine atom substituents or furans with at least three chlorines.
Figure 2.
Structural requirements for dioxins and furanes distinguishing high (a) from low (b) AhR binding activity chemicals, where Rx=O or N or S; Ry=Cl or F and Ry1=Br or I.
Figures 3 and 4 summarize structural features required for binding of flavones, carbazoles and ellipticines to AhR by a ‘dioxin-like mechanism AhR’. High-activity flavones (Figure 3a) require strong electron accepting groups at the 4′ position (N3 or I or NO2), combined with strong electron donor groups at the 3′ position (OCH3 or OC2H5 or OCOCH3). Flavone activity significantly decreases (Figure 3b) in compounds with weak electron accepting (CN or NCS or CON3) and weak electron donor groups (CH3 or C2H5 or C3H7).
Figure 3.
Structural requirements for flavones distinguishing high (a) from low (b) AhR binding activity chemicals, where Rx=Rx1−C; Rx1=O or N or S; Rz=OCH3 or OC2H5 or OCOCH3 (electron donor groups); Rδ=N3 or I or NO2 (electron acceptor groups); Rz1=CH3 or C2H5 or C3H7 or OCH3 or OC2H5 or OC3H7 or OH or OCOCH3; Rδ1=CN or NCS or CON3.
Figure 4.
Structural requirements for carbazoles observed in the high activity range (a) and ellipticines observed in low activity range (b), where: Rx=O or N or S; Rz1=CH3 or C2H5 or C3H7 or OCH3 or OC2H5 or OC3H7 or OH or OCOCH3.
High-activity carbazoles (Figure 4a) require a keto group next to the heteroatom (N or O or S); low-activity carbazoles tend to have weak electron donor groups (CH3 or C2H5 or C3H7 or OCH3) near the heteroatom (Figure 4b)
Structural features of PAH-like and biphenyl-like compounds that bind AhR are presented in Figure 5. It is assumed that these compounds interact with AhR via a stacking-type mechanism [29]. According to this mechanism, the PCB phenyl ring with the greatest degree of chlorination is assumed to be parallel with the receptor, while the other phenyl ring is rotated to a minimum energy conformation consistent with quantum-mechanical calculations.
Figure 5.
Structural requirements for PAH-like (a) and biphenyl-like mechanisms (b).
The above structural data are not sufficient to discriminate between dioxin-, PAH- and biphenyl-like chemicals with different AhR binding affinities. Hence, to improve discrimination for high- and low-affinity AhR ligands, structural data were combined with parametric boundaries. For instance, high-activity dioxin-like chemicals (REP≥0.1) were separated from low-activity AhR binders (0<REP<0.1) by using the maximal donor delocalizability (D_max) as a discriminating electronic descriptor (Figure 6).
Figure 6.
The COREPA reactivity patterns of low active (left pattern) and highly active (right pattern) dioxin-like AhR binders across maximal donor delocalizability.
This analysis showed that binding affinity correlates positively with electron donation capacity of dioxin-like ligands, with the most active ligands (26 chemicals) characterized by higher D_max values than low activity ligands (13 chemicals). The high D_max value for the most active ligands indicates that electron charge transfer occurs from ligand to receptor. The positive correlation between AhR binding and electron donation capacity of dioxin-like ligands is illustrated by related electronic indices, such as orbital energies (Figure 7).
Figure 7.
Separation of the COREPA reactivity patterns of inactive (most left pattern) and low active (right pattern) dioxin-like chemicals across EHOMO.
Figure 7 shows that the energy of the highest occupied molecular orbital (EHOMO) separated reactivity patterns of 19 binders and five non-binders. The parametric boundary discriminating active dioxin-like chemicals is EHOMO>−9.39 eV. Active dioxins are characterized by higher EHOMO, which supports the hypothesis that electron charge transfer from ligand to AhR is facilitated. Three outliers (false positives) were identified in the COREPA discrimination scheme based on EHOMO; these chemicals meet the prefiltering requirement for active dioxins with respect to the number of attached halogens.
Only two chemicals acting by a PAH-like mechanism were strong AhR binders. Their reactivity pattern was compared with weak binders across the electronegativity descriptor (Figure 8).
Figure 8.
Comparison between reactivity patterns of highly active binders (left pattern) and low active binders (right pattern) acting by PAH-like mechanism across electronegativity.
Electronegativity values were lower (i.e., <−4.53 eV) for high-affinity PAHs than for low-affinity PAHs, which supports the hypothesis that electron charge transfer from ligand to AhR is facilitated, as described above for AhR dioxin-like compounds. Seven low-affinity PAHs were also distinguished from 10 non-binders (Figure 9) based on electronegativity. High-affinity PAHs are also characterized by high donor delocalizability (D_max>0.21 (a.u.)2/eV), which supports the same hypothesis.
Figure 9.
The COREPA reactivity patterns of inactive (left pattern) and low active PAHs (right pattern) across maximal donor delocalizability.
According to observed REP values, biphenyls populated the bins of low AhR-binding (five ligands) and non-AhR-binding ligands (26 ligands). Because one parameter was not sufficient to discriminate biphenyls with different activity, the multiparametric COREPA approach was used. Not surprisingly, the chemical hydrophobicity of the ligand binding cavity appeared to be an important parameter. Consequently, log(Kow) values were higher for active chemicals than for inactive chemicals. The second most important parameter was the energy of the lowest unoccupied molecular orbital (ELUMO). Active biphenyls were characterized by lower energetically exposed molecular orbitals, which facilitates electron charge transfer from receptor to ligand. This contrasts with the mechanism for dioxins and PAHs, which involves charge transfer from ligand to receptor. For biphenyls, higher activity correlated positively with electrophilicity (e.g., lower ELUMO), and charge transfer is from the AhR receptor to the ligand.
These data suggest that AhR ligands have two distinct mechanisms of AhR binding: one mechanism is characteristic of dioxin- and PAH-like chemicals, and the second mechanism is characteristic of biphenyl-like chemicals. For the former, the ligand donates charge to the receptor; for the latter, the receptor donates charge to the ligand.
The parameters used for deriving the categorical models for each mechanism and each activity bin are summarized in Table 1.
Table 1.
Binding mechanisms, activity bins and discriminating molecular parameters used for deriving the categorical models for AhR binding affinity.
| Mechanism | |||
|---|---|---|---|
| Activity | Dioxin - like | PAH - like | Biphenyl - like |
| Active High |

|

|
None |
| Low Active |

|

|

|
| Non- Active |

|

|

|
Models for each mechanism and activity bin were organized as a battery, which could be used for screening and model testing. The battery of AhR models is illustrated in Figure 10.
Figure 10.
The battery of categorical models associated with different binding mechanisms and activity bins.
The SAR battery was applied to the training set of 142 chemicals. The resulting statistics are summarized in Table 2 (omitting discrimination between strong and weak binders).
Table 2.
Statistics for the model implementation on training set with 142 chemicals, summarized for each mechanism.
| Training set |
||||
|---|---|---|---|---|
| Binders | Non-binders | Number of chemicals | Concord | |
| Pred/Obs | Pred/Obs | % | ||
| Dioxin-like | 52/60 (87%) | 1/3 (33%) | 63 | 84% |
| PAH-like | 9/9 (100%) | 5/10 (50%) | 19 | 74% |
| Biphenyl | 5/5 (100%) | 15b/15 (100%) | 29 | 100% |
| Non-bindersa | 0/8 | 23/23 (100%) | 31 | 74% |
Non-binder chemicals not meeting any model structural requirements.
Undefined chemicals not reaching the probabilistic threshold in the COREPA model (by default 0.7).
Thirty-one chemicals (21%) were predicted to be non-binders. The mechanism of these chemicals was not categorized, because they did not meet the structural requirement for binding. For nine chemicals (7%) the model failed to reach the probabilistic threshold in the COREPA model (by default 0.7). The model predicts binding mechanism with a concordance of 82% and Pearson's coefficient of 0.88.
3.3 Modelling AhR antagonism
A broad range of substituted dioxins, flavones and ellipticines have been studied as potential antagonists of AhR. The results suggest that antagonists of AhR are characterized by an electron-rich centre near or along a lateral position of the molecule [50]. Furthermore, Henry et al. [51] showed that when the AhR is bound by the most potent flavone antagonists (by formation of external H-bond), it remains in the cytosol, associated with hsp90, and consequently fails to initiate AhR-dependent signal transduction. Thus, distinct structural features are required for ligand binding and for ligand-induced activation of AhR and its downstream functions (i.e., signal transduction and altered GE).
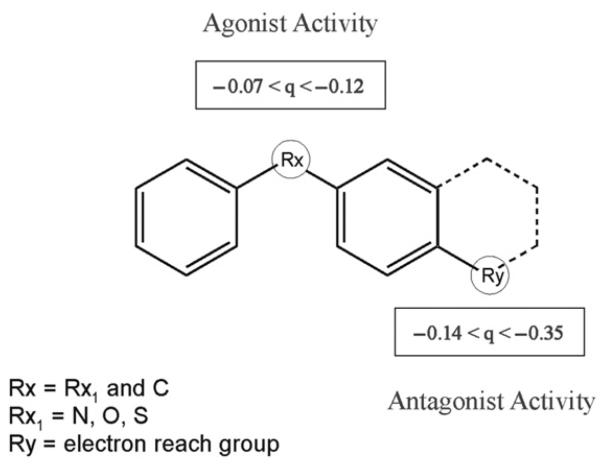
Here, a set of antagonists (flavonoids and ellipticines) and agonists were compared using the COREPA approach. The results revealed that negative charge (q) tends to be localized to the extremes of AhR antagonists (Figure 11), and that charge localization differentiates antagonists and agonists, as shown by comparing integral reactivity patterns across the dipole moment (Figure 12).
Figure 11.
Localization of negative charges in agonists and antagonists.
Figure 12.
Comparison of COREPA reactivity patterns of agonists (left pattern) and antagonists (right pattern) across dipole moment.
The integral reactivity pattern for antagonists is shifted towards a higher dipole moment, which is consistent with the stronger localization of negative charges in these chemicals. The boundary providing best separation of antagonists and agonists is 2.72 Debye. Further, critical functional groups for AhR antagonists include a terminal electron reach group at the 4′ position (NO2, I, N3) and an electron donating group at the 3′ position (OCH3, OC2H5, OC3H7).
3.4 AhR binding and gene expression
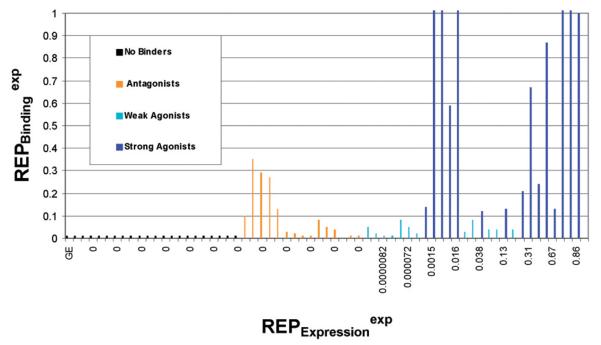
AhR modelling is often hampered by the limited availability of receptor binding data. In contrast, high-throughput GE data from ligand-treated cells is often available, and these data could potentially be used to support and extend AhR modelling studies. In the current work, the relationship between AhR binding and GE was explored using 23 training set chemicals for which GE data were available (Appendix 3). GE data for the 23 chemicals were plotted vs. binding data, as shown in Figure 13. Non-binders as well as strong and weak antagonists lacked GE effects (Figure 14a), and were clearly separated from agonists along the GE axis. Moreover (see also Figure 14b), the weak agonists (with REPGE between 0.0000 and 0.0004) have AhR binding in the low binding activity bin (0<REP<0.1), while the strong agonists (with REPGE higher than 0.14) appear to be strong binders (REP≥0.1). Based on this, it was hypothesized that the weak binders are weak agonists whereas the strong binders are strong agonists, respectively. This relationship was true for the extreme ranges of GE; however, in the range −0.0004≤REPGE≤0.1400, the relationship was poorly defined. This discrepancy could reflect inconsistency in the GE data in different test species.
Figure 13.
Observed binding versus expression data (REP) across different categories: non-binders, antagonists, weak agonists and strong agonists – ordered consecutively from left to the right.
Figure 14.
Binding versus expression based on the grouping of chemicals as agonist and antagonist.
3.5 AhR binding model validation
After including the GE intervals for agonists with high or low activity, the AhR model was used to make predictions for chemicals with known GE responses. The external test set with 51 chemicals with GE data was then used to validate the original binding model. The predictions for each chemical are presented in Appendix 2 and summarized in Table 3.
Table 3.
Summary of the prediction of the external set of 51 chemicals by the AhR model without (a) and with (b) counting the model domain.
| Prediction for the external set – number of chemicals and % from the external set |
|||
|---|---|---|---|
| Binders | Non-binders | Total chem | |
| (a) | |||
| Dioxin-like | 9 (56%) | 7 (44%) | 16 |
| PAH-like | 6 (100%) | 0 | 6 |
| Biphenyl | 2 (29%) | 1 (14%) | 7b |
| Non-bindersa | 22 (0%) | 0 | 22 |
| (b) | |||
| Dioxin-like | 7 (64%) | 4 (36%) | 11 |
| PAH-like | 5 (100%) | 0 | 5 |
| Biphenyl | 2 (67%) | 0 | 3b |
| Non-bindersa | 13 (0%) | 0 | 13 |
Non-binder chemicals not meeting any model structural requirements.
Undefined chemicals not reaching the probabilistic threshold in the COREPA model (by default 0.7).
Table 3a shows that of 16 dioxin-like chemicals, 56% are predicted binders and 44% are predicted non-binders. For PAH-like chemicals, 100% are predicted binders. For seven biphenyl-like chemicals, 29% are predicted binders, 14% are predicted non-binders. Four chemicals (57%) failed to reach the probability threshold of the multiparametric (COREPA) model. The most important result of this analysis is the prediction of 22 non-binders. However, if GE data is available for a chemical, the chemical (or a metabolite of the chemical) must be an agonist and must bind AhR. This suggests that the predictions include a large number of false negatives (i.e., 30 false negatives outside the model domain and 17 false negatives within the domain; Table 3b). This probably reflects the limited size of the training set for the AhR model. Therefore, the AhR model was re-derived using the following two steps:
The model structural domain was extended – the external set of 51 chemicals could not be added in entirety to the training set, because binding data was not available. However, incoming polychlorinated naphthalenes, which are associated with low GE, were included as low affinity binders, as shown in Figure 15.
- The model parametric domain was modified as follows:
- (2.1) ELUMO threshold used for discriminating dioxin-like mechanism at high activity range was shifted from −1.417 to −1.550 eV
- (2.2) The single multiparametric COREPA model derived for the biphenyl-like mechanism was replaced by two models applied subsequently: a single parametric model with requirement ELUMO<−0.890 eV and a new COREPA model with two discriminating parameters EHOMO and log (Kow).
Figure 16 shows the test battery of the re-derived AhR model.
Figure 15.
Extension of the structural requirements for PAH-like mechanism in low activity bin.
Figure 16.
Battery of the re-derived AhR model.
The re-derived model was re-tested with the original training set, and the results are shown in Table 4. Statistical analysis shows that the re-derived AhR binding model did not out-perform the original model with regard to the original training set. However, when re-tested on the external validation set, the re-derived model out-performed the original model (Table 5 versus Table 3). As shown in Table 5a, the number of chemicals that failed to meet any structural requirements of the model was reduced from 22 to eight chemicals (Table 5a, ‘non-bindersa’ row). The total number of predicted dioxin-like chemicals was 16 for both model versions; however, the number of non-binders was reduced from seven to three for the re-derived model. Similarly, the number of non-binding bi-phenyl-like chemicals was reduced to 0. Therefore, the total number of non-binders decreased from 30 to 11 (Table 5a). This includes non-binding chemicals that fail to meet any model structural requirements (column 1), as well as those that meet structural requirements but fail to meet parametric requirements of the model (column 2). In Table 5b, only one chemical was classified as a non-binder within the model domain. As above, this includes chemicals that fail to meet any model structural requirements (column 1), as well as those that meet structural requirements but fail to meet parametric requirements of the model (column 2).
Table 4.
Statistics from the prediction of 142 training set chemicals by the extended AhR model.
| Training set |
||||
|---|---|---|---|---|
| Binders | Non-binders | Number of chemicals | Concord | |
| Pred/Obs | Pred/Obs | % | ||
| Dioxin-like | 56/60 (93%) | 1/3 (33%) | 63 | 90% |
| PAH-like | 9/9 (100%) | 5/10 (50%) | 19 | 74% |
| Biphenyl | 5/5 (100%) | 9b/17 (53%) | 29 | 63% |
| Non-bindersa | 0/8 | 23/23 (100%) | 31 | 74% |
Non-binder chemicals not meeting any model structural requirements.
Undefined chemicals not reaching the probabilistic threshold in the COREPA model (by default 0.7).
Table 5.
Statistics of 51 external set chemicals predicted by the re-derived model without counting the domain (a) and with counting the domain (b).
| Prediction for the external set – number of chemicals and % from the external set |
|||
|---|---|---|---|
| Binders | Non-binders | Total chem | |
| (a) | |||
| Dioxin-like | 13 (81%) | 3 (19%) | 16 |
| PAH-like | 20 (100%) | 0 | 20 |
| Biphenyl | 7 (100%) | 0 | 7b |
| Non-bindersa | 8 (0%) | 0 | 8 |
| (b) | |||
| Dioxin-like | 11 (100%) | 0 | 11 |
| PAH-like | 5 (100%) | 0 | 5 |
| Biphenyl | 3 (100%) | 0 | 3b |
| Non-bindersa | 1 (0%) | 0 | 1 |
Non-binder chemicals not meeting any model structural requirements.
Undefined chemicals not reaching the probabilistic threshold in the COREPA model (by default 0.7).
3.6 Valuation of the GE/AhR binding relationship
The following analysis tested predictions of the re-derived AhR model using an external set of chemicals for which only GE data were available. For the external set of 51 chemicals, AhR binding affinity was predicted by the model and the results were compared with binding affinity inferred from GE values (see ‘AhR Binding and Gene Expression’, above). The results are listed in Appendix 2 and summarized in Table 6.
Table 6.
Valuation of the GE/AhR binding relationship.
| Predicted AhR binding | ||||
|---|---|---|---|---|
| GA/ binding relation |
||||
| Model prediction | Total chem number | REP<0.1 | 0<REP<1 | REP≥0.1 |
| REP<0.1 | 23 | 10 (43%) | 13 (57%) | 0 |
| REP≥0.1 | 17 | 2 (12%) | 9 (53%) | 6 (35%) |
| Non-binders | 11 | 2 (18%) | 7 (64%) | 2 (18%) |
Table 6 and Appendix 2 show that 23 chemicals were predicted to be weak binders (REP50.1). Of these, 43% had low binding activity based on GE (0<REPGA<0.0004). However, 57% fell in the ‘fuzzy area’ −0.0004≤REPGE≤0.14, such that the relationship between GE and AhR binding could not be determined. Nevertheless, no strong outliers or discrepancies were observed among predicted weak binders. The model predicted 17 strong binders, six (35%) of which had high binding activity based on GE, and nine of which (53%) fell in the fuzzy area. However, two chemicals (12%) were predicted to be weak binders based on GE, and were considered outliers. This could be due to experimental error. For this analysis, the model resulted in 11 (22%) incorrect predictions for non-binding chemicals, and failed to provide a definitive classification for seven chemicals AhR (Table 6, undefined chemicals).
In summary, the existence of a fuzzy area, namely GE (0.0004<REPGA<0.14), prohibited categorical predictions of AhR binding using GE data in this range. However, AhR binding and GE showed reasonable correlation outside of this range. This result could reflect use of GE data from different species and experimental systems in this analysis. Therefore, additional analysis is warranted to explain the lack of correlation in this activity range, to better understand the relationship between AhR binding and GE, and to improve the proposed AhR binding model.
4. Summary and conclusions
The AhR mediates the toxic and biological effects of a wide variety of environmental compounds. The goal of this study was to derive a COREPA-based categorical SAR model for AhR ligands within different ranges of affinity/potency.
The training set of 142 AhR ligands included dioxins, furans, biphenyls, PAHs, flavones/flavanones and carbazoles. The synthetic ligands in the training set were ranked into three reporter binding activity classes: (1) strong binders with REP≥0.1 (30 chemicals), (2) weak binders with 0<REP<0.1 (52 chemicals), and (3) non-binders with REP=0 (60 chemicals).
The conformational distributions of chemicals were analysed and compared to define the commonality between biologically similar chemicals within 2D and 3D structural space. The COREPA analysis suggested two different binding electronic mechanisms, which we refer to as the dioxin-/PAH-like binding mechanism and the biphenyl-like binding mechanism. In this investigation, the group of dioxin-like chemicals included dioxins, furans, flavonoids and carbazoles. These molecules appeared to donate electron density from the nucleophilic central part of the ligand to the receptor (these chemicals have higher energies of EHOMO and lower energies of ELUMO). PAHs formed a structurally different class of chemicals which interacted with the receptor by stacking-type electron charge transfer from ligand to the receptor. This assumption was confirmed by the positive correlation with donor delocalizability of those molecules used as a discriminating parameter. A stacking type of interaction with the AhR was identified for biphenyls; however, electron charge transfer was proposed to be from receptor to ligand. This mechanism was supported by the positive correlation with the electron acceptor capabilities of those chemicals (lower ELUMO energies). Based on these observations it could be concluded that three structural but only two electronic binding mechanisms are required to classify AhR ligands. The concordance of the COREPA-based categorical SAR model for AhR ligands was 82% and the Pearson's coefficient was 0.88.
The current model was also used to evaluate AhR ligands as agonists or antagonists. Antagonism correlated with stronger negative charge localization along the molecular axis of the ligand and electron donating properties of electron reach groups at the receptor surface. Therefore, it is predicted that AhR antagonists will form H-bonds with AhR which prevent its dissociation from hsp90.
The properties of AhR agonists and antagonists correlated with their GE effects. The highest increase in GE was elicited by strong agonists, lower increases in GE by weak agonists, and all antagonists (and non-AhR binders) had no effect on GE. This correlation was incorporated in the model, in order to predict AhR binding using only GE. The relationship was semi-quantitative for chemicals with extreme GE values. However, the quantitative relationship between AhR binding and GE was unclear in a large ‘fuzzy area’ along the GE axis, most likely due to variable characteristics of the GE data used in this study. Therefore, additional data and more comprehensive analyses are needed to refine the AhR binding model proposed in the present study.
Acknowledgements
M. Denison acknowledges support from the National Institutes of Environmental Health Science for studies on AhR ligands (ES012498 and ES07685). Funding provided by the Dow Chemical Company.
Appendices
Appendix 1.
142 AhR training set chemicals.
| CAS no | Chemical name | Observed REP value |
|---|---|---|
| 2422-79-9 | 12-Methylbenzanthracene | 0.01 |
| 57-97-6 | 9,10-Dimethyl-1,2-benzanthracene | 0 |
| 84761-86-4 | 1-Chlorodibenzofuran | 0.3 |
| 51230-49-0 | 2-Chlorodibenzofuran | 0 |
| 25074-67-3 | 3-Chlorodibenzofuran | 0 |
| N/A | 4-Chlorodibenzofuran | 0 |
| 64126-86-9 | 2,3-Dichlorodibenzofuran | 0 |
| 60390-27-4 | 2,6-Dichlorodibenzofuran | 0 |
| 5409-83-6 | 2,8-Dichlorodibenzofuran | 0 |
| 83704-39-6 | 1,3,6-Trichlorodibenzofuran | 0 |
| 76621-12-0 | 1,3,8-Trichlorodibenzofuran | 0 |
| 57117-34-7 | 2,3,4-Trichlorodibenzofuran | 0 |
| 58802-17-8 | 2,3,7-Trichlorodibenzofuran | 0.13 |
| 57117-32-5 | 2,3,8-Trichlorodibenzofuran | 0.01 |
| N/A | 2,6,7-Trichlorodibenzofuran | 0.02 |
| 83704-30-7 | 2,3,4,6-Tetrachlorodibenzofuran | 0.03 |
| 83704-32-9 | 2,3,4,8-Tetrachlorodibenzofuran | 0.05 |
| 57117-37-0 | 2,3,6,8-Tetrachlorodibenzofuran | 0.05 |
| 51207-31-9 | 2,3,7,8-Tetrachlorodibenzofuran | 0.24 |
| 64126-87-0 | 1,2,4,8-Tetrachlorodibenzofuran | 0 |
| 83704-21-6 | 1,2,3,6-Tetrachlorodibenzofuran | 0.03 |
| 83704-22-7 | 1,2,3,7-Tetrachlorodibenzofuran | 0.09 |
| 58802-16-7 | 1,3,4,7,8-Pentachlorodibenzofuran | 0.05 |
| N/A | 2,3,4,7,9-Pentachlorodibenzofuran | 0.05 |
| 83704-53-4 | 1,2,3,7,9-Pentachlorodibenzofuran | 0.03 |
| 83704-50-1 | 1,2,4,6,7-Pentachlorodibenzofuran | 0.15 |
| 71998-74-8 | 1,2,4,7,9-Pentachlorodibenzofuran | 0 |
| 67517-48-0 | 1,2,3,4,8-Pentachlorodibenzofuran | 0.08 |
| 57117-41-6 | 1,2,3,7,8-Pentachlorodibenzofuran | 0.13 |
| 58802-15-6 | 1,2,4,7,8-Pentachlorodibenzofuran | 0.01 |
| 57117-31-4 | 2,3,4,7,8-Pentachlorodibenzofuran | 0.67 |
| 70648-26-9 | 1,2,3,4,7,8-Hexachlorodibenzofuran | 0.04 |
| 57117-44-9 | 1,2,3,6,7,8-Hexachlorodibenzofuran | 0.04 |
| 67562-40-7 | 1,2,4,6,7,8-Hexachlorodibenzofuran | 0 |
| 60851-34-5 | 2,3,4,6,7,8-Hexachlorodibenzofuran | 0.21 |
| 1746-01-6 | 2,3,7,8-Tetrachlorodibenzodioxin | 1 |
| 40321-76-4 | 1,2,3,7,8-Pentachlorodibenzodioxin | 0.13 |
| N/A | 2,3,6-Trichlorodibenzo-p-dioxin | 0.05 |
| 39227-28-6 | 1,2,3,4,7,8-Hexachlorodibenzodioxin | 0.04 |
| 50585-46-1 | 1,3,7,8-Tetrachlorodibenzodioxin | 0.01 |
| 58802-08-7 | 1,2,4,7,8-Pentachlorodibenzodioxin | 0.01 |
| 30746-58-8 | 1,2,3,4-Tetrachlorodibenzodioxin | 0.01 |
| 33857-28-2 | 2,3,7-Trichlorodibenzodioxin | 0.14 |
| 38964-22-6 | 2,8-Dichlorodibenzodioxin | 0 |
| 39227-61-7 | 1,2,3,4,7-Pentachlorodibenzodioxin | 0 |
| 39227-58-2 | 1,2,4-Trichlorodibenzodioxin | 0 |
| N/A | 1,2,3,4,6,7,8,9-Octachlorodibenzodioxin | 0 |
| 57465-28-8 | 3,4,5,3′,4′-Pentachlorobiphenyl | 0.08 |
| 32774-16-6 | 3,4,5,3′,4′,5′-Hexachlorobiphenyl | 0 |
| 32598-13-3 | 3,4,3′,4′-Tetrachlorobiphenyl | 0.02 |
| 32598-14-4 | 2,3,3′,4,4′-Pentachlorobiphenyl | 0 |
| 74472-37-0 | 2,3,4,4′,5-Pentachlorobiphenyl | 0 |
| 38380-08-4 | 2,3,3′,4,4′,5-Hexachlorobiphenyl | 0 |
| 65510-44-3 | 2′,3,4,4′,5-Pentachlorobiphenyl | 0 |
| 33025-41-1 | 2,3,4,4′-Tetrachlorobiphenyl | 0 |
| 31508-00-6 | 2,3′,4,4′,5-Pentachlorobiphenyl | 0 |
| N/A | 2,3′,4,4′,5,5′-Hexachlorobiphenyl | 0 |
| 35065-27-1 | 2,4,5,2′,4′,5′-Hexachlorobiphenyl | 0 |
| 2437-79-8 | 2,4,2′,4′-Tetrachlorobiphenyl | 0 |
| 88966-73-8 | 4′-Trifluoromethyl-2,3,4,5-tetrachlorobiphenyl | 0.03 |
| N/A | 4′-t-Propyl-2,3,4,5-tetrachlorobiphenyl | 0.08 |
| N/A | 4′-Iodo-2,3,4,5-tetrachlorobiphenyl | 0.01 |
| N/A | 4′-Bromo-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Ethyl-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Cyano-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Phenyl-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Acetyl-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-t-Butyl-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-n-Butyl-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-n-Acetylamino-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Nitro-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Methoxy-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Fluoro-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Methyl-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 4′-Hydroxy-2,3,4,5-tetrachlorobiphenyl | 0 |
| N/A | 2,3,4,5-Tetrachlorobiphenyl | 0 |
| 50585-41-6 | 2,3,7,8-Tetrabromodibenzodioxin | 6.67 |
| 50585-40-5 | 2,3-Dibromo-7,8-dichlorodibenzodioxin | 6.76 |
| 109333-32-6 | 2,8-Dibromo-3,7-dichlorodibenzodioxin | 22.37 |
| 109333-33-7 | 2-Bromo-3,7,8-trichlorodibenzodioxin | 0.87 |
| N/A | 1,3,7,8-Tetrabromodibenzodioxin | 5 |
| N/A | 1,2,4,7,8-Pentabromodibenzodioxin | 0.59 |
| N/A | 1,2,3,7,8-Pentabromodibenzodioxin | 1.51 |
| 51974-40-4 | 2,3,7-Tribromobenzodioxin | 8.55 |
| 39073-07-9 | 2,7-Dibromodibenzodioxin | 0.65 |
| N/A | 2,4,6,8-Tetrabromodibenzofuran | 0.11 |
| 120-12-7 | Anthracene | 0 |
| 85-01-8 | Phenanthrene | 0 |
| 1730-37-6 | 1-Methylfluorene | 0 |
| 1430-97-3 | 2-Methylfluorene | 0 |
| 779-02-2 | 9-Methylanthracene | 0 |
| 1523-23-5 | 1,9-dimethyl-anthracene | 0 |
| 50-32-8 | Benzo(a)pyrene | 0.03 |
| 238-84-6 | Benzo(a)fluorene | 0.01 |
| 243-17-4 | Benzo(b)fluorene | 0.01 |
| 129-00-0 | Pyrene | 0 |
| 92-24-0 | Naphthacene | 0 |
| 218-01-9 | Chrysene | 0 |
| 2498-76-2 | 2-Methylbenz(a)anthracene | 0 |
| 2319-96-2 | 5-Methylbenzanthracene | 0 |
| 316-14-3 | 6-Methylbenz(a)anthracene | 0 |
| 2541-69-7 | 7-Methylbenzanthracene | 0.01 |
| 2381-31-9 | 8-Methylbenzanthracene | 0 |
| 6111-78-0 | 11-Methylbenz(a)anthracene | 0.01 |
| 198-55-0 | Perylene | 0 |
| 56-49-5 | Methylcholanthrene | 0.02 |
| 191-24-2 | 1,12-Benzoperylene | 0 |
| 215-58-7 | 1,2:3,4-Dibenz[a]anthracene | 0.06 |
| 53-70-3 | 1,2:5,6-Dibenzanthracene | 0.12 |
| 213-46-7 | Picene | 0.18 |
| 56-55-3 | 1,2-Benz[a]anthracene | 0.03 |
| N/A | 3′-Nitro-7,8-benzoflavanone | 0 |
| N/A | 4′-Nitro-7,8-benzoflavanone | 0.01 |
| N/A | 3′-Amino-7,8-benzoflavanone | 0 |
| N/A | 4′-Amino-7,8-benzoflavanone | 0 |
| N/A | 3′-Methoxy-4′-nitroflavanone | 0.1 |
| N/A | 4′-Nitroflavanone | 0 |
| N/A | 3′-Methoxy-4′-nitro-7,8-benzoflavanone | 0.35 |
| N/A | 2,3,6,7-Tetrachlorodibenzodioxin | 0.06 |
| N/A | 4′-Iodo-3′-methoxyflavone | 0.29 |
| N/A | 4′-Triazido-3′-methoxyflavone | 0.27 |
| N/A | 4′-Nitro-3′-methoxyflavone | 0.13 |
| N/A | 4′-Thiocyanate-3′-methoxyflavone | 0.08 |
| N/A | 3′-Methoxyflavone | 0.05 |
| N/A | 4′-Nitro-3′-propyloxyflavone | 0.05 |
| N/A | 4′-Cyano-3′-methoxyflavone | 0.04 |
| N/A | 4′-Nitro-3′-hydroxyflavone | 0.015 |
| N/A | 5′-Iodo-4′-amino-3′-methoxyflavone | 0.013 |
| N/A | 4′-Amino-3′-methoxyflavone | 0.009 |
| N/A | 4′-Acetylamino-3′-methoxyflavone | 0.007 |
| 55786-24-8 | 7,8-Dehydrorutaecarpine | 0.71 |
| 84-26-4 | Rutecarpine | 0.14 |
| 77251-57-1 | 13-Oxoellipticine | 0.01 |
| 10371-86-5 | 9-Methoxyellipticine | 0.01 |
| 5263-05-8 | 8-Methoxy-5,11-dimethyl-6H-Pyridocarbazole | 0.003 |
| N/A | 5H-benzo[b]carbazole-6,11-dione | 0.88 |
| N/A | 6,11-dimethyl-5H-benzo[b]carbazole | 0.04 |
| N/A | 9-methoxy-5,11-dimethyl-6H-pyrido[4,3-b]carbazol-1-amine | 0.03 |
| N/A | 8-methoxy-5,11-dimethyl-10H-pyrido[2,3-b]carbazole | 0.02 |
| N/A | 5,11-dimethyl-6H-pyrido[3,4-b]carbazole | 0.01 |
| N/A | 5,11-dimethyl-10H-pyrido[3,4-b]carbazole | 0.01 |
| N/A | 10H-pyrido[3,4-b]carbazole-5,11-dione | 0.003 |
Appendix 2.
Validation set of 51 chemicals having gene expression data only.
| Predicted AhR |
|||||
|---|---|---|---|---|---|
| CAS RN | Chemical name | GE observe |
by model | by GE analysis |
Discrepancy |
| 58863-14-2 | 1,2,3,4,5,6,7-Heptachloro naphthalene | 0.00052 | REP<0.1 | 0<REP<1 | (a) |
| N/A | 2,3,3′,4,4′,5,6-Heptachlorobiphenyl | 6.7E-06 | REP<0.1 | REP<0.1 | + |
| N/A | 1-Bromo-2,3,7,8-tetrachlorodibenzodioxin | 0.28 | REP>0.1 | REP>0.1 | + |
| N/A | 2-Bromo-1,3,7,8-tetrachlorodibenzodioxin | 0.37 | REP>0.1 | REP>0.1 | + |
| N/A | 2-Bromo-3,6,7,8,9-pentachlorodibenzodioxin | 0.19 | REP>0.1 | REP>0.1 | + |
| N/A | 2,2′,4,5′,6-Pentabromobiphenyl | 0.0028 | REP<0.1 | 0<REP<1 | |
| N/A | 3,3′,4,4′,5-Pentabromobiphenyl | 0.016 | REP<0.1 | 0<REP<1 | |
| N/A | 3,3′,4,4′,5,5′-Hexabromobiphenyl | 0.0047 | REP<0.1 | 0<REP<1 | |
| N/A | 1,2,3,6,7,8-Hexachloro naphthalene | 0.0028 | REP<0.1 | 0<REP<1 | |
| 82-05-3 | Benzanthrone | 1.6E-06 | REP<0.1 | REP<0.1 | + |
| 90-13-1 | 1-Chloronaphthalene | 0.000017 | REP<0.1 | REP<0.1 | + |
| 91-58-7 | 2-Chloronaphthalene | 0.000018 | REP<0.1 | REP<0.1 | + |
| 119-47-1 | Advastab 405 | 0.28 | Non-binder | REP>0.1 | − |
| 189-55-9 | Dibenzo(a,i)pyrene | 0.0429 | REP<0.1 | 0<REP<1 | |
| 189-64-0 | Dibenzo(a,h)pyrene | 0.0265 | REP<0.1 | 0<REP<1 | |
| 191-30-0 | Dibenzo(a,l)pyrene | 2.52E-05 | REP>0.1 | REP<0.1 | − |
| 192-65-4 | Dibenzo(a,e)pyrene | 0.00108 | REP>0.1 | 0<REP<1 | |
| 192-97-2 | Benzo(e)pyrene | 3.71E-05 | REP>0.1 | REP<0.1 | − |
| 205-82-3 | Benzo(j)fluoranthene | 0.0405 | Non-binder | 0<REP<1 | − |
| 205-99-2 | Benzo(b)fluoranthene | 0.049 | REP>0.1 | 0<REP<1 | |
| 224-42-0 | Dibenzacridine | 0.027 | Non-binder | 0<REP<1 | − |
| 225-11-6 | Benz(a)acridine | 0.004 | Non-binder | 0<REP<1 | − |
| 225-51-4 | Benz(c)acridine | 0.0026 | Non-binder | 0<REP<1 | − |
| 226-36-8 | Dibenz(a,h)acridine | 2.45 | Non-binder | REP>0.1 | − |
| 1825-31-6 | 1,4-Dichloro naphthalene | 0.000035 | REP<0.1 | REP<0.1 | + |
| 2050-75-1 | 2,3-Dichloro naphthalene | 0.000027 | REP<0.1 | REP<0.1 | + |
| 2498-66-0 | 7,12-Benz(a)anthraquinone | 0.000036 | Non-binder | REP<0.1 | − |
| 6640-24-0 | 1-(3-Chlorophenyl)piperazine | 6.2E-06 | Non-binder | REP<0.1 | − |
| 19408-74-3 | 1,2,3,7,8,9-Hexachlorodibenzodioxin | 0.061 | REP>0.1 | 0<REP<1 | |
| 34588-40-4 | 2,3,6,7-Tetrachloro naphthalene | 0.000041 | REP<0.1 | REP<0.1 | + |
| 35822-46-9 | 1,2,3,4,6,7,8-Heptachlorodibenzodioxin | 0.031 | REP>0.1 | 0<REP<1 | |
| 39001-02-0 | Octachlorodibenzofuran | 0.0016 | Non-binder | 0<REP<1 | − |
| 55673-89-7 | 1,2,3,4,7,8,9-heptachlorodibenzofuran | 0.044 | REP>0.1 | 0<REP<1 | |
| 57653-85-7 | 1,2,3,6,7,8-Hexachlorodibenzodioxin | 0.098 | REP>0.1 | 0<REP<1 | |
| 67562-39-4 | 1,2,3,4,6,7,8-Heptachlorodibenzofuran | 0.024 | REP>0.1 | 0<REP<1 | |
| 67733-57-7 | 2,3,7,8-Tetrabromodibenzofuran | 0.6 | REP>0.1 | REP>0.1 | + |
| 67922-26-3 | 1,2,3,4,6-Pentachloro naphthalene | 0.000068 | REP<0.1 | REP<0.1 | + |
| 70362-50-4 | 3,4,4′,5-Tetrachloro-1,1′-Biphenyl | 0.0045 | REP<0.1 | 0<REP<1 | |
| 72918-21-9 | 1,2,3,7,8,9-Hexachlorodibenzofuran | 0.11 | REP>0.1 | REP>0.1 | + |
| 77102-82-0 | 3,3′,4,4′-Tetrabromobiphenyl | 0.08 | REP<0.1 | 0<REP<1 | |
| 103426-92-2 | 1,2,4,5,7,8-Hexachloro naphthalene | 0.00006 | REP<0.1 | REP<0.1 | + |
| 103426-94-4 | 1,2,3,5,7,8-Hexachloro naphthalene | 0.00011 | REP<0.1 | REP<0.1 | + |
| 103426-95-5 | 1,2,3,5,6,8-Hexachloro naphthalene | 0.00049 | REP<0.1 | 0<REP<1 | |
| 103426-96-6 | 1,2,3,4,6,7-Hexachloro naphthalene | 0.0012 | REP<0.1 | 0<REP<1 | |
| 103426-97-7 | 1,2,3,5,6,7-Hexachloro naphthalene | 0.00048 | REP<0.1 | 0<REP<1 | |
| 107555-93-1 | 1,2,3,7,8-Pentabromodibenzofuran | 0.14 | REP>0.1 | REP>0.1 | + |
| 107555-95-3 | 1,2,3,4,6,7,8-Heptabromodibenzofuran | 0.0027 | Non-binder | 0<REP<1 | − |
| 110999-46-7 | 1,2,3,7,8,9-Hexabromodibenzodioxin | 0.017 | REP>0.1 | 0<REP<1 | |
| 129880-08-6 | 1,2,3,4,7,8-Hexabromodibenzofuran | 0.017 | Non-binder | 0<REP<1 | − |
| 131166-92-2 | 2,3,4,7,8-Pentabromodibenzofuran | 0.094 | REP>0.1 | 0<REP<1 | |
| 150224-16-1 | 1,2,3,6,7-Pentachloro naphthalene | 0.00058 | REP<0.1 | 0<REP<1 | |
empty fields represent those chemicals for which a prediction cannot be given either by the AhR model or GE analysis.
Appendix 3.
23 chemicals possessing both AhR binding and gene expression data.
| CAS RN | Chemical name | AhR binding observe (REP) |
GE observe (REP) |
|---|---|---|---|
| N/A | 2,3′,4,4′,5,5′-Hexachlorobiphenyl | 0.0200 | 0.82 × 10−5 |
| N/A | 4′-t-Propyl-2,3,4,5-tetrachlorobiphenyl | 0.0500 | 0.0004 |
| N/A | 4′-Cyano-2,3,4,5-tetrachlorobiphenyl | 0.0100 | 0.89 × 10−5 |
| 53-70-3 | 1,2,5,6-Dibenzanthracene | 0.1200 | 0.0600 |
| 1746-01-6 | 2,3,7,8-Tetrachlorodibenzodioxin | 1.0000 | 1.0000 |
| 32598-13-3 | 3,4,3′,4′-Tetrachlorobiphenyl | 0.0200 | 0.0014 |
| 33857-28-2 | 2,3,7-Trichlorodibenzodioxin | 0.1400 | 0.0015 |
| 39227-28-6 | 1,2,3,4,7,8-Hexachlorodibenzodioxin | 0.0400 | 0.0750 |
| 40321-76-4 | 1,2,3,7,8-Pentachlorodibenzodioxin | 0.1300 | 0.7300 |
| 50585-40-5 | 2,3-Dibromo-7,8-dichlorodibenzodioxin | 6.7600 | 0.8600 |
| 50585-41-6 | 2,3,7,8-Tetrabromodibenzodioxin | 6.6700 | 0.7700 |
| 51207-31-9 | 2,3,7,8-Tetrachlorodibenzofuran | 0.2400 | 0.6700 |
| 51974-40-4 | 2,3,7-Tribromobenzodioxin | 8.5500 | 0.0330 |
| 57117-31-4 | 2,3,4,7,8-Pentachlorodibenzofuran | 0.6700 | 0.5800 |
| 57117-32-5 | 2,3,8-Trichlorodibenzofuran | 0.0100 | 0.0001 |
| 57117-41-6 | 1,2,3,7,8-Pentachlorodibenzofuran | 0.1300 | 0.1400 |
| 57117-44-9 | 1,2,3,6,7,8-hexachlorodibenzofuran | 0.0400 | 0.1400 |
| 57465-28-8 | 3,4,5,3′,4′-Pentachlorobiphenyl | 0.0800 | 0.0380 |
| 60851-34-5 | 2,3,4,6,7,8-Hexachlorodibenzofuran | 0.2100 | 0.3100 |
| 70648-26-9 | 1,2,3,4,7,8-Hexachlorodibenzofuran | 0.0400 | 0.1300 |
| 88966-73-8 | 4′-Trifluoromethyl-2,3,4,5-tetrachlorobiphenyl | 0.0300 | 0.0333 |
| 109333-33-7 | 2-Bromo-3,7,8-trichlorodibenzodioxin | 0.8700 | 0.6700 |
| 74472-37-0 | 2,3,4,4′,5-Pentachlorobiphenyl | 0.0024 | 0.0001 |
References
- 1.Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt JV, Bradfield CA. Ah Receptor Signaling Pathways. Annu. Rev. Cell. Dev. Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 3.Denison MS, Elferink CF, Phelan D. Toxicant-Receptor Interactions in the Modulation of Signal Transduction and Gene Expression. Taylor and Francis; Philadelphia, PA: 1998. pp. 3–33. [Google Scholar]
- 4.Ma Q. Induction of CYP1A1. The AhR/DRE paradigm: transcription, receptor regulation, and expanding biological roles. Curr. Drug Met. 2001;2:149–164. doi: 10.2174/1389200013338603. [DOI] [PubMed] [Google Scholar]
- 5.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu. Rev. Pharm. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 7.Denison MS, Fisher JM, Whitlock JP. The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J. Biol. Chem. 1988;263:17221–17224. [PubMed] [Google Scholar]
- 8.Denison MS, Seidel SD, Rogers WJ, Ziccardi M, Winter GM, Heath-Pagliuso S. In: Natural and synthetic ligands for the Ah receptor,in Molecular Biology Approaches to Toxicology. Puga A, Wallace KB, editors. Taylor and Francis; PA: 1998. pp. 393–419. [Google Scholar]
- 9.LePecq JB, Dat-Xuong N, Gosse C, Paoletti C. A New Antitumoral Agent: 9-Hydroxyellipticine. Possibility of a Rational Design of Anticancerous Drugs in the Series of DNA Intercalating Drugs. Proc. Nat. Acad. Sci. USA. 1974;71:5078–5082. doi: 10.1073/pnas.71.12.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Multon E, Riou JF, LaFevre D, Ahomadegbe JC, Riou G. Topoisomerase II-mediated DNA cleavage activity induced by ellipticines on the human tumor cell line N417. Biochem, Pharmacol. 1989;38:2077–2086. doi: 10.1016/0006-2952(89)90060-9. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez N, Roy M, Lesca P. Binding characteristics of Ah receptors from rats and mice before and after separation from hepatic cytosols. 7-Hydroxyellipticine as a competitive antagonist of cytochrome P-450 induction. Eur. J. Biochem. 1988;172:585–592. doi: 10.1111/j.1432-1033.1988.tb13929.x. [DOI] [PubMed] [Google Scholar]
- 12.Kurl RN, DePetrillo PB, Olnes MJ. Inhibition of Ah (dioxin) receptor transformation by 9-hydroxy ellipticine: involvement of protein kinase C? Biochem. Pharmacol. 1993;46:1425–1433. doi: 10.1016/0006-2952(93)90108-9. [DOI] [PubMed] [Google Scholar]
- 13.Gasiewicz TA, Kende AS, Rucci G, Whitney B, Willey JJ. Analysis of Structural Requirements for Ah Receptor Antagonist Activity: Ellipticines, Flavones, and Related Compound. Biochem. Pharmacol. 1996;52:1787–1803. doi: 10.1016/s0006-2952(96)00600-4. [DOI] [PubMed] [Google Scholar]
- 14.O'Prey J, Brown J, Fleming J, Harrison PR. Effects of dietary flavonoids on major signal transduction pathways in human epithelial cells. Biochem. Pharmacol. 2003;66:2075–2088. doi: 10.1016/j.bcp.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Denison MS, Pandini A, Nagy S, Baldwin E, Bonati L. Ligand binding and activation of the Ah receptor. Chem. Biol. Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 16.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 17.Stahura FL, Bajorath J. New methodologies for ligand-based virtual screening. Curr. Pharm. Des. 2005;11:1189–1202. doi: 10.2174/1381612053507549. [DOI] [PubMed] [Google Scholar]
- 18.Piparo EL, Koehler K, Chana A, Benfenati E. Virtual Screening for Aryl Hydrocarbon Receptor Binding Prediction. J. Med. Chem. 2006;49:5702–5709. doi: 10.1021/jm060526f. [DOI] [PubMed] [Google Scholar]
- 19.Pandini A, Denison MS, Song Y, Soshilov A, Bonati L. Structural and functional characterization of the AhR ligand binding domain by homology modeling and mutational analysis. Biochemistry. 2007;23:696–708. doi: 10.1021/bi061460t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandini A, Soshilov AA, Song Y, Zhao J, Bonati L, Denison MS. Detection of the TCDD binding fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis. Biochemistry. 2009;48:5972–5983. doi: 10.1021/bi900259z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner M, Sadowski J, Gasteiger J. Autocorrelation of molecular surface properties for modeling Corticosteroid Binding Globulin and cytosolic Ah receptor activity by neural networks. J. Am. Chem. Soc. 1995;117:7769–7775. [Google Scholar]
- 22.Tuppurainen K, Ruuskanen J. Electronic eigenvalue (EEVA): a new QSAR / QSPR descriptor for electronic substituent effects based on molecular orbital energies. A QSAR approach to the Ah receptor binding affinity of polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs) and dibenzofuranes (PCDFs) Chemosphere. 2000;41:843–848. doi: 10.1016/s0045-6535(99)00525-1. [DOI] [PubMed] [Google Scholar]
- 23.Poland A, Knutson JC. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 24.McKinney JD, Darden T, Lyerly MA, Pederson LG. PCB and related compound binding to the Ah receptor(s) Theoretical madel based on molecular parameters and molecular mechanics. Quant. Struct.-Act. Relat. 1985;4:166–172. [Google Scholar]
- 25.Waller CL, McKinney JD. Comparative molecular field analysis of polyhalogenated dibenzo-p-dioxins, dibenzofuranes, and biphenyls. J. Med. Chem. 1992;35:3660–3666. doi: 10.1021/jm00098a010. [DOI] [PubMed] [Google Scholar]
- 26.Waller CL, McKinney JD. Three-dimensional quantitative-activity relationships of dioxins and dioxin-like compounds: model validation and Ah receptor characterization. Chem. Res. Toxicol. 1995;8:847–858. doi: 10.1021/tx00048a005. [DOI] [PubMed] [Google Scholar]
- 27.Poso A, Tuppurainen K, Ruuskanen J, Gynther J. Binding of some dioxins and dibenzofuranes to the Ah receptor. A QSAR model based on comparative molecular field analysis (CoMFA) J. Mol. Struct. (Theochem) 1993;282:259–264. [Google Scholar]
- 28.Kafafi SA, Afeefy HY, Said HK, Hakimi JM. A new structure-activity model for Ah receptor binding. Polychlorinated dibenzo-p-dioxins and dibenzofuranes. Chem. Res. Toxicol. 1992;5:856–862. doi: 10.1021/tx00030a020. [DOI] [PubMed] [Google Scholar]
- 29.Mekenyan OG, Veith GD, Call DJ, Ankley GT. A QSAR evaluation of Ah receptor binding of halogenated aromatic xenobiotics. Environ. Health Persp. 1996;104:1302–1310. doi: 10.1289/ehp.961041302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraschini E, Bonati L, Pitea D. Molecular polarizability as a tool for understanding the binding properties of polychlorinated dibenzo-p-dioxins: definition of a reliable computational procedure. J. Phys. Chem. 1996;100:10564–10569. [Google Scholar]
- 31.Pearson RG. Absolute electronegativity and hardness correlated with molecular orbital theory. Proc. Natl. Acad. Sci. USA. 1986;83:8440–8441. doi: 10.1073/pnas.83.22.8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parr RG, Szentpály LV, Liu S. Electrophilicity index. J. Am. Chem. Soc. 1999;121:1922–1924. [Google Scholar]
- 33.Chattaraj PK, Maiti B, Sarkar U. Philicity: a unified treatment of chemical reactivity and selectivity. J. Phys. Chem. A. 2003;107:4973–4975. [Google Scholar]
- 34.Sarkar U, Padmanabhan J, Parthasarathi R, Subramanian V, Chattaraj PK. Toxicity analysis of polychlorinated dibenzofuranes through global and local electrophilicities. J. Mol. Struct. (Theochem) 2006;758:119–125. [Google Scholar]
- 35.Arulmozhiraja S, Morita M. Structure–activity relationships for the toxicity of polychlorinated dibenzofuranes: approach through density functional theory-based descriptors. Chem. Res. Toxicol. 2004;17:348–356. doi: 10.1021/tx0300380. [DOI] [PubMed] [Google Scholar]
- 36.Behnisch PA, Hosoe K, Sakai S-I. Brominated dioxin-like compounds: in vitro assessment in comparisons to classical dioxin-like compounds and other polyaromatic compounds. Environ. Int. 2003;29:861–877. doi: 10.1016/s0160-4120(03)00105-3. [DOI] [PubMed] [Google Scholar]
- 37.Piskorska-Pliszcynska, Keys B, Safe S, Newman MS. The cytosolic receptor binding affinities and AHH induction potencies of 29 polynuclear aromatic hydrodrocarbons. Toxicol. Lett. 1986;34:67–74. doi: 10.1016/0378-4274(86)90146-3. [DOI] [PubMed] [Google Scholar]
- 38.Puzyn T, Falandysz J, Jones PD, Giesy JP. Quantitative structure activity relationships for the prediction of relative in vitro potencies (REPs) for chloronaphthalenes. J. Environ. Sci. Health. 2007;42:573–590. doi: 10.1080/10934520701244326. [DOI] [PubMed] [Google Scholar]
- 39.Safe S, Bandiera S, Sawyer T, Zmudzka B, Mason G, Romkes M, Denomme MA, Sparling J, Okey AB, Fujita T. Effects of structure on bidning to the 2,3,7,8-TCDD: receptor and AHH induction. Halogenated biphenuyls. Environ. Health Perspect. 1985;61:21–33. doi: 10.1289/ehp.856121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denison MS. Unpublished results. [Google Scholar]
- 41.Mekenyan O, Dimitrov D, Nikolova N, Karabunarliev S. Conformational Coverage by a Genetic Algorithm. Chem. Inf. Comput. Sci. 1999;39:997. [Google Scholar]
- 42.Pavlov T, Todorov M, Serafimova R, Aladjov H, Mekenyan O. Conformational coverage by a genetic algorithm: saturation of conformational space. J. Chem. Inf. Model. 2007;47:851–863. doi: 10.1021/ci700014h. [DOI] [PubMed] [Google Scholar]
- 43.Steward J. MOPAC: a semiempirical molecular orbital program. J. Comput.-Aided Mol. Des. 1990;4:1–105. doi: 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- 44.Steward J. Fujitsu Limited, 9-3, Nakase 1-Chome, Mihama-ku.Chiba-City, Chiba 261, Japan and Stewart Computational Chemistry. Colorado Springs, CO; 15210 Paddington Circle: 1993. MOPAC 93. [Google Scholar]
- 45.Mekenyan O, Ivanov JM, Karabunarliev S, Bradbury S, Ankley G, Karcher W. A computationally-based hazard identification algorithm that incorporates ligand flexibility. 1. Identification of potential androgen receptor ligands. Environ. Sci. Technol. 1997;31:3702–3711. [Google Scholar]
- 46.Mekenyan O, Nikolova N, Karabunarliev S, Bardbury S, Ankley G, Hansen B. New developments in a hazard identification algorithm for hormone receptor ligands. Quant. Struct.-Act. Relat. 1999;18:139–153. [Google Scholar]
- 47.Mekenyan OG, Nikolova N, Schmieder P, Veith GD. COREPA-M: a Multi-Dimensional Formulation of COREPA. QSAR Comb. Sc. 2004;23:5–18. [Google Scholar]
- 48.Dimitrov S, Dimitrova G, Pavlov T, Dimitrova N, Patlewicz G, Niemela J, Mekenyan O. A Stepwise Approach for Defining the Applicability Domain of SAR and QSAR Models. J. Chem. Inf. Model. 2005;45:839–849. doi: 10.1021/ci0500381. [DOI] [PubMed] [Google Scholar]
- 49.Procopio M, Lahm A, Tramontano A, Bonati L. A model for recognition of polychlorinated dibenzo-p-dioxins by the aryl hydrocarbon receptor. Eur. J. Biochem. 2002;269:13–18. doi: 10.1046/j.0014-2956.2002.02619.x. [DOI] [PubMed] [Google Scholar]
- 50.Gasiewicz T, Kende AS, Rucci G, Whitney B, Willey JJ. Analysis of structural requirements for Ah Receptor Antagonist Activity: Ellipticines, Flavones, and Related Compounds. Biochem. Pharmacol. 1996;52:1787–1803. doi: 10.1016/s0006-2952(96)00600-4. [DOI] [PubMed] [Google Scholar]
- 51.Henry EC, Kende AS, Rucci G, Totleben MJ. Flavone Antagonists Bind Competitively with 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (TCDD) to the Aryl Hydrocarbon Receptor But Inhibit Nuclear Uptake and Transformation. Mol. Pharmacol. 1999;55:716–725. [PubMed] [Google Scholar]