Abstract
Estrogen (E) plays a pivotal role in regulating the female reproductive system, particularly the ovary. However, the number and type of ovarian genes influenced by estrogen remain to be fully elucidated. In this study, we have utilized wild-type (WT) and aromatase knockout (ArKO; estrogen free) mouse ovaries as an in vivo model to profile estrogen dependent genes. RNA from each individual ovary (n = 3) was analyzed by a microarray-based screen using Illumina Sentrix Mouse WG-6 BeadChip (45,281 transcripts). Comparative analysis (GeneSpring) showed differential expression profiles of 450 genes influenced by E, with 291 genes up-regulated and 159 down-regulated by 2-fold or greater in the ArKO ovary compared to WT. Genes previously reported to be E regulated in ArKO ovaries were confirmed, in addition to novel genes not previously reported to be expressed or regulated by E in the ovary. Of genes involved in 5 diverse functional processes (hormonal processes, reproduction, sex differentiation and determination, apoptosis and cellular processes) 78 had estrogen-responsive elements (ERE). These analyses define the transcriptome regulated by E in the mouse ovary. Further analysis and investigation will increase our knowledge pertaining to how E influences follicular development and other ovarian functions.
Introduction
The importance of estrogen (E) in female reproductive endocrinology and in ovarian function has been well documented [1]. Estrogen signalling is primarily transduced by estrogen receptors (ER) α and β [2]. ER are members of a conserved superfamily of ligand activated transcription factors. The effects of E on ER are exerted through a complex array of convergent and divergent signaling pathways that mediate genomic events involved in regulation of mitogenesis, differentiation and apoptosis [3], [4]. The interaction of E and ER with specific DNA sequences called estrogen responsive elements (EREs), constitutes a primary genomic signaling pathway [3], [4]. The ERE-bound ER recruits an ensemble of co-factors responsible for the alteration of local chromatin structure and interaction with the basal transcription machinery.
Much effort has been invested in developing techniques to identify genes of interest, such as, Northern blot, semi-quantitative RT-PCR and serial analysis of gene expression (SAGE). Several candidate genes, such as cyclin D2 [5], growth and differentiation factor 9 (GDF-9) [5], forkhead box transcription factor (Foxo1) [6], inhibinα and inhibinβB [7] were shown to be regulated by E using these approaches. However, many downstream gene targets of E were unlikely to have been discovered using this approach. The aromatase knockout (ArKO) female mouse is deficient in aromatase activity postnatally and therefore is an excellent model to allow us to define E-dependent ovarian genes in adulthood. Using RT-PCR we identified genes usually associated with male reproduction such as Sox9, DAX1, liver homolog-1 (Lrh-1) and Mullerian-inhibiting substance [8] as being significantly increased [8] in the ArKO ovary. Steroidogenic enzymes such as 17α-OHase, 17ß-Hsd1, and 17β-Hsd3 mRNA's were also increased and the patterns of expression of the steroidogenic enzymes responsible for androgen biosynthesis in the ArKO ovaries correlated with increased serum testosterone levels in ArKO females [8].
The development of microarray technology now enables the simultaneous measurement of thousands of gene transcripts in a biological sample [9]. Therefore, in order to identify E-dependent genes in the ovary, this study utilized a microarray approach profiling wildtype (WT) and ArKO ovaries. The study aimed to 1) confirm the preliminary observations on estrogen dependent genes, 2) provide novel information about genes that can be used to unravel the mechanism of E in maintaining the female gonad, and 3) provide new insights into the regulation of ovarian follicular development.
Materials and Methods
Animals
Wild-type (WT) and ArKO mice on a J129/C57B6 background were maintained under specific pathogen-free (SPF) conditions, on a 12L∶12D regimen and fed ad libitum a soy free mouse chow (Glen Forrest Stockfeeders, Western Australia). All animal procedures were approved by a Monash University Animal Ethics Committee (Project number: MMCB2002/37) and were carried out in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Experimental design
The experimental approach is summarized in Figure 1. WT and ArKO mice, 16 weeks of age (n = 3/grp) were killed by CO2 asphyxiation. The abdomen was opened and both ovaries from each animal were collected and snap frozen in liquid nitrogen. RNA was extracted from individual ovaries using a phenol-chloroform-based method (Ultraspec; Fisher Biotech, Subiaco, Western Australia, Australia). Quantification of RNA concentration and purity was measured using the NanoDrop spectrophotometer (Thermo Scientific). The quality of the mouse RNA was ascertained using the Agilent Bioanalyser 2100 using the NanoChip protocol. A total of 500ng RNA was amplified and labeled using the Illumina TotalPrep RNA Amplification kit (Ambion) as per the manufacturer's instructions. A total of 1.5ug of labelled cRNA was then prepared for hybridisation to the Sentrix Mouse-6 Expression Beadchip (v1.1) by preparing a probe cocktail (cRNA @ 0.05ug/ul) that includes GEX-HYB Hybridisation Buffer (supplied with the beadchip). A total hybridisation volume of 30ul is prepared for each sample and 30ul loaded into a single array on the Sentrix Mouse-6 Expression Beadchip (v1.1). The Sentrix Mouse-6 Expression Beadchip (v1.1) allows for six samples and targets 48,500 unique well-documented RefSeq transcripts. A total of 6 different labelled samples can be loaded into 6 individual arrays per beadchip. The chip is hybridised at 58°C for 16 hours in an oven with a rocking platform. After hybridisation, the chip is washed using protocols outlined in the Illumina manual. Upon completion of the washing, the chips are then coupled with Cy3 and scanned in the Illumina BeadArray Reader. The scanner operating software, BeadStudio, converts the signal on the array into a text file for analysis. All data is MIAME compliant and the raw data has been deposited in a MIAME compliant database ArrayExpress (ArrayExpress accession: E-MEXP-2824). The Illumina microarray was performed by the Australian Genome Research Facility (AGRF) Melbourne, Australia.
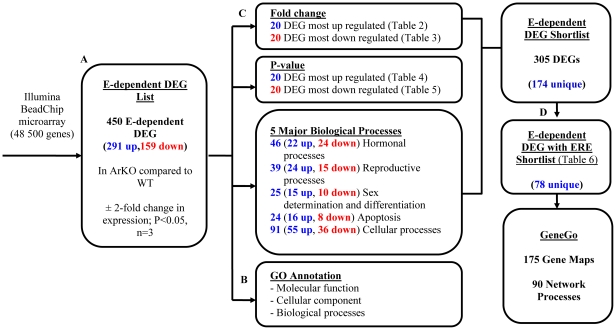
Figure 1. Flowchart of microarray data ranking and analysis.
A) 450 E-dependent DEG list were identified by microarray in ovaries of ArKO vs WT as having ±2-fold change expression with p-value<0.05. B) The E-dependent DEG list was annotated using Gene Ontology and analysed for molecular function, cellular component and biological processeses. C) E-dependent DEG from the list were ranked based upon fold change of up/down regulation (Fold change); significance of the change (P-value); 5 major GeneGo biological processes genes: Hormonal processes, Reproductive processes, Sex determination and differentiation, Apoptosis and, Cellular processes. D) Genes that possess EREs. Genes identified from the list using these methods were compiled to form an E-dependent DEG with ERE Shortlist which was used for downstream analysis of gene networks and pathways affected by E using GeneGo pathway analysis. The E-dependent DEG list can be found in Table S1.
Statistical analysis for microarray data
Prior to differential gene expression analysis, quality control diagnostic measurements were run on the raw data generated from the Illumina chips. 6 Sentrix Mouse-6 Expression Beadchip (v1.1) samples were normalised using lumi package (part of bioconductor) in R statistical software and were subsequently imported into GeneSpring GX software for further analysis. The significant E-dependent differentially expressed genes (DEG) were obtained from comparing KO samples to WT samples and were defined by a fold difference of 2.0 and a P-value cut-off of 0.05 (derived from ANOVA). Genes that met these parameters were included in the E-dependent DEG list and colour coded with blue representing down-regulation and red representing up-regulation of transcript when comparing KO samples to WT samples. The data normalisation and statistical analysis to generate E-dependent DEG list was performed by the AGRF Melbourne, Australia.
Bioinformatic analysis and downstream annotation
E-dependent DEG were ranked according to the methods described in the results section. Specific methods for Gene Ontology, GeneGo pathways analysis and Dragon estrogen response element (ERE) Finder version 6 are detailed below.
1. Gene Ontology (GO) [6] Analysis
E-dependent DEG lists were annotated according to the GO database (http://www.geneontology.org) using ontology categories for Molecular Function, Cellular Component and Biological Processes. The representation of E-dependent DEG within each ontology category was measured and statistical significance of the overlap was done using a hypergeometric p-value without multiple testing corrections, to determine the likelihood of coincidental overlap with ontology groups. A p-value lower than 0.05 indicates over representation of genes from the E-dependent DEG list within that particular category, suggestive of a functional effect. Annotation of the E-dependent DEG lists using GO was conducted by the AGRF Melbourne, Australia.
2. Dragon ERE Locator version 6
Nucleotide search from National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/nucleotide/) was used to retrieve the sequence information of the differentially expressed genes within the selected Biological Processes categories in the FASTA format. These sequences were then examined for the presence of ERE binding sites using the latest Dragon ERE Finder version 6.0 (http://apps.sanbi.ac.za/ere/index.php) [10] based upon information present in the transcriptional regulation, from patterns to profiles (TRANSFAC) database [11]. The detection algorithm was tested on several large datasets and achieved a sensitivity of 83% [10]. An ERE is defined as a site which contains the 17 basepairs consensus sequence with its flanking 20 nucleotides on either side against the promoter sequences of Eukaryotic Promoter Database (EPD) [10], [12], [13].
3. GeneGo pathway analysis
The E-dependent DEG list containing the 450 unique E-dependent DEG, complete with Illumina transcript identifiers, were uploaded from a Microsoft Excel spreadsheet onto Metacore 5.0 software (GeneGo pathways analysis) (http://www.genego.com). GeneGo recognizes the Illumina identifiers and maps the E-dependent DEG to the MetaCore™ data analysis suite, generating maps to describe common pathways or molecular connections between E-dependent DEG on the list. Graphical representations of the molecular relationships between genes were generated using the GeneGo pathway analysis, based upon processes showing significant (P<0.05) association.
Results
E-dependent Differentially expressed genes (DEG) in ArKO compared to WTs
Microarray analysis of whole genome expression in ArKO ovary compared to WT ovary identified 450 differentially expressed transcripts with a ±2-fold expression difference that was significant (p<0.05 according to t-tests, n = 3) (Table 1). This list is referred to as the E-dependent DEG list and can be found in full in the supplementary data. The 450 E-dependent DEGs represent ∼0.94% of the total number of transcripts present on the Illumina BeadChip. Of these 450 E-dependent DEG, 291 were up regulated (within a range of 2 to 47-fold change), and 159 DEG were down regulated (range between −2 to −15.7-fold change) in the absence of E.
Table 1. E-dependent DEG in ArKO ovary compared to WT ovary.
| Fold Change | Differential Expressed Genes (DEG) | ||
| Up | Down | total | |
| 47∼2.0 | 291 | - | 291 |
| −2.0∼−15.7 | - | 159 | 159 |
| Total | 450 | ||
Up; up regulated within designated fold range. Down; down regulated within designated fold range. All E-dependent DEG were significantly differentially expressed (p<0.05) (n = 3).
GO annotation of the E-dependent DEG list
We hypothesised that some of the E-dependent DEG would be involved in downstream molecular pathways important for maintaining the female gonad. In order to identify which types of global cellular processes or specific molecular functions were responsive to E, the E-dependent DEG list was annotated using the GO [6] database. Each of the 450 genes was assigned to Molecular Function, Cellular Component and Biological Process categories as designated by the GO database. GO categories significantly over-represented in the E-dependent DEG list are those that match a greater number of the E-dependent DEG than would be expected by chance (based upon comparison to the total number of genes in the genome assigned to the category).
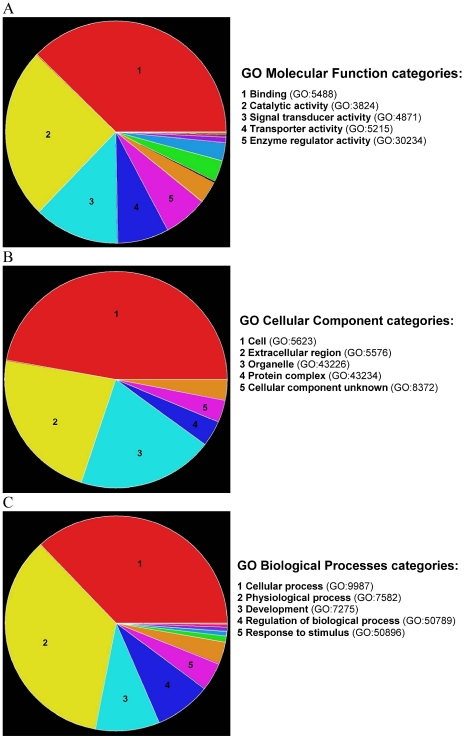
Figure 2A summarises the annotation of the E-dependent DEG list with a Molecular Function using GO. 388 of the 450 genes were matched to categories and the distribution of these 388 E-dependent DEG into GO Molecular Function categories. The five GO categories with the greatest proportion of E-dependent DEG were Binding, Catalytic activity, Signal Transducer activity, Transporter activity and Enzyme regulator activity.
Figure 2. Gene Ontology (GO) annotation E-dependent DEG list with Molecular Function, Cellular Component and Biological Processes categories.
A) Pie chart shows the distribution of the 388 E-dependent DEG that were matched to a Molecular Function using GO [6]. B) Pie chart shows the distribution of the 319 E-dependent DEG that were matched to a Cellular Component using GO [6]. C) Pie chart shows the distribution of the 302 E-dependent DEG that were matched to a Biological Processes using GO [6].
Figure 2B summarises the annotation of the E-dependent DEG list with a Cellular Component using GO. 319 of the 450 genes were matched to categories and the distribution of these 319 E-dependent DEG into GO Cellular Component categories. The five GO categories with the greatest proportion of E-dependent DEG were Cell, Extracellular region, Organelle, Protein complex and Cellular component unknown.
Figure 2C summarises the annotation of the E-dependent DEG list to GO Biological Processes. 302 of the 450 genes were matched to categories and the distribution of these 302 E-dependent DEG into GO Biological Process categories. The GO categories with the greatest proportion of E-dependent DEGs were Cellular process, Physiological process, Development, Regulation of Biological Process and Response to Stimulus.
Ranking of genes on the E-dependent DEG list to create an E-dependent DEG with ERE Shortlist
As described in Table 1, 450 E-dependent DEG were identified in ArKO ovaries that showed expression ±2-fold compared to WT, with a significance of p<0.05 (E-dependent DEG list, Table S1). In order to narrow the E-dependent DEG list to those most likely to represent direct biological E target genes, four approaches were used to create a ranked shortlist of E-dependent DEG (Figure 1). The approaches used were, 1) Fold Change: the E-dependent DEG list was sorted according to fold change in expression, to identify the genes with the greatest up regulation and down regulation; 2) P-value: the E-dependent DEG list was sorted according to the statistical significance of the expression difference; 3) GeneGo analysis: 5 major GeneGo biological processes genes: Hormonal processes, Reproductive processes, Sex determination and differentiation, Apoptosis and, Cellular processes and 4) Genes containing EREs: Dragon ERE Locator version 6 was used to identify genes that contain ERE promoters from the shortlisted genes from step 1, 2 and 3. Using these criteria the list was reduced to generate a 78 E-dependent DEG with ERE Shortlist (Table S2). The results of each sorting method (1–4) used in the generation of the E-dependent DEG with ERE Shortlist are detailed below:
1. Genes from the E-dependent DEG list showing greatest fold change in expression
The twenty genes found to be most up regulated and the twenty found to be most down regulated in the DEG List based upon fold change values are summarised in Table 2 and Table 3, respectively. The fold change values range from 47 to 6.3 for the most up regulated genes and from −15.7 to −4.8 for the most down regulated genes.
Table 2. Twenty most up-regulated genes in the E-dependent DEG list.
| Genbank | Gene symbol | Name | Fold change | P-value | |
| 1 | NM_178396 | Car12* | carbonic anyhydrase 12 | 47.04 | 0.00076 |
| 2 | NM_008491 | Lcn2* | lipocalin 2 | 20.61 | 0.00358 |
| 3 | XM_130383 | 8030411F24Rik* | RIKEN cDNA 8030411F24 | 13.46 | 0.00144 |
| 4 | XM_485085 | Defb36* | defensin beta 36 | 13.39 | 0.00125 |
| 5 | NM_011413 | C4* † | sex-limited protein | 12.93 | 0.00198 |
| 6 | XM_355911 | Pace4 | paired basic amino acid cleaving system 4 isoform 1 | 11.18 | 0.00008 |
| 7 | NM_008770 | Cldn11* | claudin 11 | 10.89 | 0.00001 |
| 8 | XM_357518 | LOC384244* | similar to Forkhead box protein L1 (Forkhead-related protein FKHL11) (Forkhead-related transcription factor 7) (FREAC-7) | 10.87 | 0.00255 |
| 9 | XM_130716 | Gm122* | hypothetical protein LOC209351 isoform 1 | 9.23 | 0.00438 |
| 10 | NM_009778 | C3 | complement component 3 | 8.91 | 0.00205 |
| 11 | NM_009979 | Cst9* | cystatin 9 | 8.73 | 0.00007 |
| 12 | NM_024285 | Bves* | blood vessel epicardial substance | 8.10 | 0.00720 |
| 13 | NM_009252 | Serpina3n* | serine (or cysteine) proteinase inhibitor, clade A, member 3N | 7.90 | 0.01640 |
| 14 | NM_144936 | BC018222* | transmembrane protein 45b | 7.89 | 0.00092 |
| 15 | NM_026323 | Wfdc2 | WAP four-disulfide core domain 2 | 6.84 | 0.00251 |
| 16 | NM_009301 | Svs5* | seminal vesicle secretion 5 | 6.67 | 0.00285 |
| 17 | NM_029325 | Spinlw1* | eppin | 6.65 | 0.00051 |
| 18 | NM_007878 | Drd4 | dopamine receptor 4 | 6.34 | 0.00205 |
| 19 | XM_133614 | Kctd14 | potassium channel tetramerisation domain containing 14 | 6.27 | 0.0141 |
| 20 | NM_007739 | Col8a1* | procollagen, type VIII, alpha 1 | 6.27 | 0.00854 |
Cut-off is 2 fold change in expression in ArKO compared to WT (n = 3), p-value≤0.05.
*denote genes have not been shown to express in the ovary.
Table 3. Twenty most down-regulated genes in the E-dependent DEG list.
| Genbank | Gene symbol | Name | Fold change | P-value | |
| 1 | NM_134066 | Akr1c18* | aldo-keto reductase family 1, member C18 | −15.67 | 0.00634 |
| 2 | NM_144890 | BC018465* | hypothetical protein LOC228802 | −11.96 | 0.00195 |
| 3 | NM_007409 | Adh1* | alcohol dehydrogenase 1 (class I) | −11.66 | 0.00031 |
| 4 | NM_010476 | Hsd17b7 | hydroxysteroid (17-beta) dehydrogenase 7 | −10.83 | 0.01950 |
| 5 | NM_176930 | C130076O07Rik* | neuronal cell adhesion molecule | −10.16 | 0.00779 |
| 6 | NM_013582 | Lhcgr | luteinizing hormone/choriogonadotropin receptor | −9.62 | 0.00012 |
| 7 | NM_009285 | Stc1 | stanniocalcin 1 | −8.93 | 0.00929 |
| 8 | NM_175293 | D630023F18Rik* | hypothetical protein LOC98303 | −6.99 | 0.00062 |
| 9 | NM_175556 | A930027K05Rik* | phospholipase C-like 4 | −6.94 | 0.01060 |
| 10 | NM_199303 | Bpil3* | bactericidal/permeability-increasing protein-like 3 precursor | −6.71 | 0.02000 |
| 11 | NM_009610 | Actg2 | actin, gamma 2, smooth muscle, enteric | −6.29 | 0.00215 |
| 12 | XM_129987 | Ccdc3* | coiled-coil domain containing 3 | −6.25 | 0.00030 |
| 13 | NM_023635 | Rab27a* | RAB27A protein | −6.21 | 0.04000 |
| 14 | NM_144551 | Trib2* | tribbles homolog 2 | −5.99 | 0.00013 |
| 15 | NM_130450 | Elovl6* | ELOVL family member 6, elongation of long chain fatty acids | −5.38 | 0.00170 |
| 16 | NM_019517 | Bace2* | beta-site APP-cleaving enzyme 2 | −5.13 | 0.04830 |
| 17 | NM_022315 | Smoc2* | secreted modular calcium-binding protein 2 | −5.10 | 0.00359 |
| 18 | NM_027865 | Tmem25* | transmembrane protein 25 | −5.08 | 0.00694 |
| 19 | NM_001004365 | Arp3b* | ARP3 actin-related protein 3 homolog B | −5.08 | 0.00020 |
| 20 | NM_011797 | Car14* | carbonic anhydrase 14 | −4.83 | 0.00004 |
Cut-off is 2 fold change in expression in ArKO compared to WT (n = 3), p-value≤0.05.
*denote genes have not been shown to express in the ovary.
2. Genes from the E-dependent DEG list showing the most significant changes in gene expression
The twenty genes found to be most significantly up regulated compared to control and the twenty found to be most significantly down regulated compared to control, based upon the lowest p-values, are summarised in Table 4 and Table 5, respectively. The p-values range from 0.000001 to 0.000214 for the most up regulated genes and from 0.000004 to 0.000203 for the most down regulated genes.
Table 4. Twenty most significant differentially up-regulated genes in the E-dependent DEG list.
| Genbank | Gene symbol | Name | Fold change | P-value | |
| 1 | NM_145219 | Lgi3* | leucine-rich repeat LGI family, member 3 | 5.70 | 0.000001 |
| 2 | NM_008770 | Cldn11* | claudin 11 | 10.89 | 0.000006 |
| 3 | NM_009893 | Chrd | chordin | 2.28 | 0.000016 |
| 4 | AK033210 | 8030463A06Rik* | hypothetical protein LOC414083 | 2.25 | 0.000020 |
| 5 | XM_148974 | BC034090* | hypothetical protein XP_148974 isoform 1 | 2.11 | 0.000024 |
| 6 | NM_021564 | Fetub* | fetuin beta | 2.91 | 0.000032 |
| 7 | NM_177547 | Cisk; 2510015P22Rik; A330005P07Rik* | serum/glucocorticoid regulated kinase 3 | 2.02 | 0.000046 |
| 8 | NM_009979 | Cst9* | cystatin 9 | 8.73 | 0.000067 |
| 9 | NM_028295 | Pdir* | protein disulfide isomerase associated 5 | 5.43 | 0.000075 |
| 10 | XM_355911 | Pace4 | paired basic amino acid cleaving system 4 isoform 1 | 11.18 | 0.000078 |
| 11 | XM_129211 | Psat1* | phosphoserine aminotransferase 1 | 2.49 | 0.000081 |
| 12 | NM_018784 | Siat10* | alpha2,3-sialyltransferase VI | 2.13 | 0.000087 |
| 13 | NM_031380 | Fstl3 | follistatin-like 3 | 3.00 | 0.000090 |
| 14 | XM_110968 | D11Ertd686e* | dynein, axonemal, heavy chain 9 isoform 1 | 3.13 | 0.000110 |
| 15 | NM_013589 | Ltbp2 | latent transforming growth factor beta binding protein 2 | 2.93 | 0.000118 |
| 16 | NM_029173 | 4930519N16Rik* | nucleoredoxin | 2.54 | 0.000138 |
| 17 | NM_008968 | Ptgis | prostaglandin I2 (prostacyclin) synthase | 3.75 | 0.000153 |
| 18 | NM_177547 | Cisk; 2510015P22Rik; A330005P07Rik* | serum/glucocorticoid regulated kinase 3 | 2.08 | 0.000163 |
| 19 | NM_007752 | Cp* | ceruloplasmin | 4.66 | 0.000214 |
| 20 | NM_011594 | Timp2 | tissue inhibitor of metalloproteinase 2 | 3.00 | 0.000214 |
Genes up regulated in the E-dependent DEG list were ranked from most significant to least significant according to fold change. Cut-off is 2 fold change in expression in ArKO compared to WT (n = 3), p-value ≤0.05.
*denote genes have not been shown to express in the ovary.
Table 5. Twenty most significant differentially down-regulated genes in the E-dependent DEG list.
| Genbank | Gene symbol | Name | Fold change | P-value | |
| 1 | NM_176950 | 9230107O10Rik* | hypothetical protein LOC319579 | −2.58 | 0.000004 |
| 2 | NM_010417 | Heph* | hephaestin isoform 1 | −2.54 | 0.000020 |
| 3 | NM_019919 | Ltbp1* | latent transforming growth factor beta binding protein 1 isoform a | −3.23 | 0.000036 |
| 4 | NM_011797 | Car14* | carbonic anhydrase 14 | −4.83 | 0.000036 |
| 5 | NM_010544 | Ihh | Indian hedgehog | −2.98 | 0.000113 |
| 6 | NM_013582 | Lhcgr | luteinizing hormone/choriogonadotropin receptor | −9.62 | 0.000117 |
| 7 | NM_153546 | Oact1* | O-acyltransferase (membrane bound) domain containing 1 | −3.17 | 0.000118 |
| 8 | AK080168 | ENSMUSG00000055015* | −3.27 | 0.000118 | |
| 9 | XM_358333 | E030049G20Rik* | Nck-associated protein 5 | −2.16 | 0.000119 |
| 10 | NM_011146 | Pparg | peroxisome proliferator activated receptor gamma | −2.83 | 0.000119 |
| 11 | NM_144551 | Trib2* | tribbles homolog 2 | −5.99 | 0.000131 |
| 12 | NM_007904 | Ednrb | endothelin receptor type B | −3.33 | 0.000148 |
| 13 | NM_025363 | 1110001J03Rik* | hypothetical protein LOC66117 | −2.61 | 0.000151 |
| 14 | NM_011535 | Tbx3* | T-box 3 protein isoform 1 | −3.42 | 0.000163 |
| 15 | NM_010497 | Idh1 | isocitrate dehydrogenase 1 (NADP+), soluble | −2.74 | 0.000164 |
| 16 | NM_181990 | Cklfsf2a* | chemokine-like factor super family 1 | −2.78 | 0.000175 |
| 17 | NM_019739 | Foxo1 | forkhead box O1a | −2.87 | 0.000176 |
| 18 | NM_008289 | Hsd11b2 | hydroxysteroid 11-beta dehydrogenase 2 | −2.11 | 0.000177 |
| 19 | NM_001004365 | Arp3b* | ARP3 actin-related protein 3 homolog B | −5.08 | 0.000202 |
| 20 | NM_010570 | Irs1 | insulin receptor substrate 1 | −2.16 | 0.000203 |
Genes down regulated in the E-dependent DEG list were ranked from most significant to least significant according to fold change. Cut-off is 2 fold change in expression in ArKO compared to WT (n = 3), p-value ≤0.05.
*denote genes have not been shown to express in the ovary.
3. GeneGo
Having narrowed the E target genes identified using the Illumina microarray down to a workable shortlist of 78 E-dependent DEG with ERE (Table S2); the molecular interactions between shortlisted genes were determined. Of the total set of 78 genes, ∼37 (∼47%) were not previously documented as ovarian genes, as determined by an extensive literature review and cross-checking with online databases (PubMed and Ovarian Kaleidoscope Database). Therefore, the identification of known and unknown genes could reveal how E affects regulation in folliculogenesis. We used GeneGo Pathways Analysis software to analyse the E-dependent DEG Shortlist and create ‘networks’ best connecting the 78 unique genes.
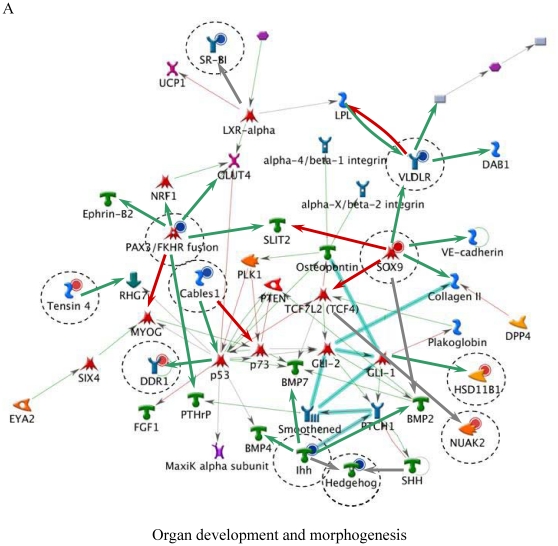
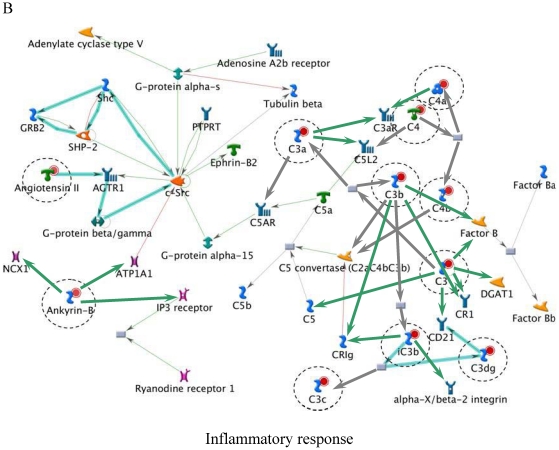
Table 6 lists the top five most relevant networks identified by the GeneGo software. All the molecules listed within each network are connected based upon their known relationships or functions within the GeneGo Pathways Knowledge Base. The gene content of the E-dependent DEG Shortlist is used as the input list for generation of biological networks using the Analyze Networks (AN) algorithm. This is a variant of the shortest paths algorithm with main parameters of 1) relative enrichment with the E-dependent DEG Shortlist, and 2) relative saturation of networks with canonical pathways (g-Score). These networks are built upon the E-dependent DEG Shortlist and unique for the E-dependent DEG Shortlist (Detailed network object legend, Figure S1). For instance, Network A (Figure 3) with the highest g-Score (43.64) (Table 6) contained 11 genes. The top processes associated with this network included ‘organ development’, ‘anatomical structure morphogenesis’ and ‘organ morphogenesis’. Both Network B and C (Figure 4 and 5) were noted as having genes that are related to ‘response to stimulus’ and ‘inflammatory responses’. Similarly, Network D (Figure 6) further emphasised processes associated with organ development, system development and anatomical structure development. Network E (Figure 7) was associated with cellular processes, such as, regulation of multicellular organismal process, regulation of cell migration and regulation of cellular component movement. All Networks: A to E (Figure 3, 4, 5, 6, 7) complemented processes that were identified in the GO analysis.
Table 6. Top five gene networks identified by GeneGo Pathways Analysis using the E-dependent DEG Shortlist of 78 unique genes.
| Processes | p-value | z-Score | g-Score | |
| A | Organ morphogenesis (58.7%), organ development (73.9%), anatomical structure morphogenesis (63.0%) | 1.56e-21 | 39.89 | 43.64 |
| B | Complement activation (23.3%), activation of plasma proteins involved in acute inflammatory response (23.3%), response to stimulus (86.7%) | 1.59e-19 | 37.44 | 41.19 |
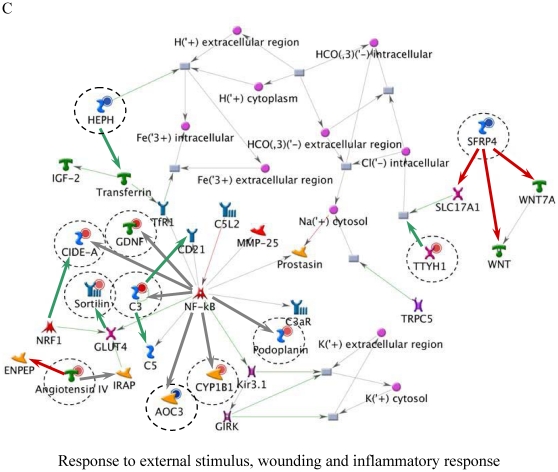
| C | Response to external stimulus (50.0%), response to wounding (40.0%), inflammatory response (33.3%) | 1.20e-21 | 40.32 | 40.32 |
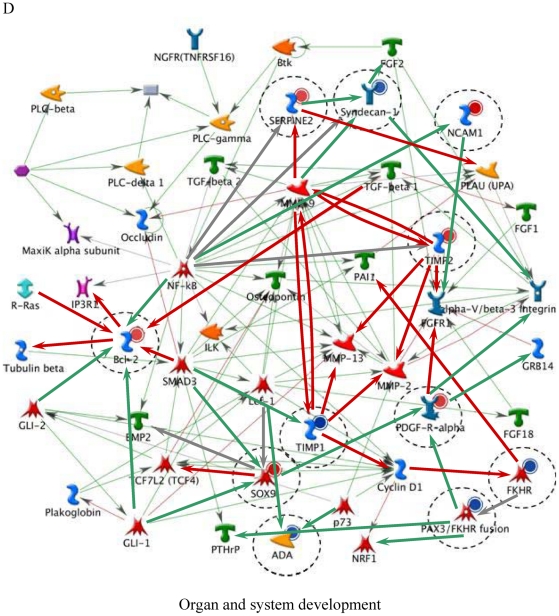
| D | Organ development (79.2%), system development (85.4%), anatomical structure development (85.4%) | 2.01e-21 | 39.47 | 39.47 |
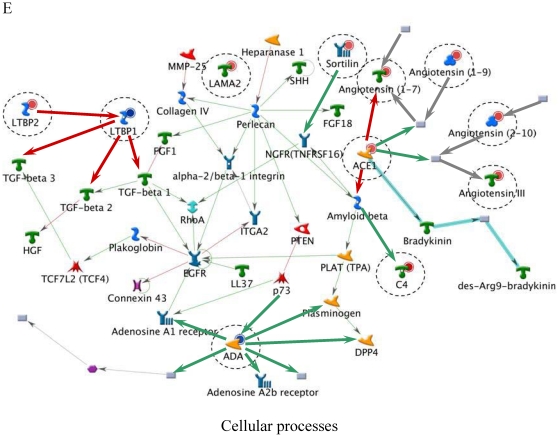
| E | Regulation of multicellular organismal process (64.7%), regulation of cell migration (38.2%), regulation of cellular component movement (38.2%) | 2.51e-17 | 34.06 | 35.31 |
The molecules that make up the top 5 Networks (A–E) identified using the E-dependent DEG with ERE Shortlist and ranked by GeneGo according to the g-Score are shown. The g-Score modifies the z-Score based on the number of Canonical Pathways used to build the network. If a network has a high g-Score, it is saturated with expressed genes (from z-Score) and it contains many Canonical Pathways. Sorting the table by this value essentially enables you to sort the table by two factors at once. The number of genes involved within the processes is shown in percentage.
Figure 3. Molecular network of identifying E-dependent DEG with ERE Shortlist genes in ArKO ovary.
Solid connecting lines represent a direct relationship between two molecules: activation marked as green solid line, inhibition marked as red solid line and unspecified marked as gray solid line. Thick cyan lines indicate the fragments of canonical pathways. Up-regulated genes are marked with red circles; down-regulated with blue circles. Genes with ERE are encircled. (Please refer to Figure S1 for detailed network object legend).
Figure 4. Molecular network of identifying E-dependent DEG with ERE Shortlist genes in ArKO ovary.
Solid connecting lines represent a direct relationship between two molecules: activation marked as green solid line, inhibition marked as red solid line and unspecified marked as gray solid line. Thick cyan lines indicate the fragments of canonical pathways. Up-regulated genes are marked with red circles; down-regulated with blue circles. Genes with ERE are encircled. (Please refer to Figure S1 for detailed network object legend).
Figure 5. Molecular network of identifying E-dependent DEG with ERE Shortlist genes in ArKO ovary.
Solid connecting lines represent a direct relationship between two molecules: activation marked as green solid line, inhibition marked as red solid line and unspecified marked as gray solid line. Thick cyan lines indicate the fragments of canonical pathways. Up-regulated genes are marked with red circles; down-regulated with blue circles. Genes with ERE are encircled. (Please refer to Figure S1 for detailed network object legend).
Figure 6. Molecular network of identifying E-dependent DEG with ERE Shortlist genes in ArKO ovary.
Solid connecting lines represent a direct relationship between two molecules: activation marked as green solid line, inhibition marked as red solid line and unspecified marked as gray solid line. Thick cyan lines indicate the fragments of canonical pathways. Up-regulated genes are marked with red circles; down-regulated with blue circles. Genes with ERE are encircled. (Please refer to Figure S1 for detailed network object legend).
Figure 7. Molecular network of identifying E-dependent DEG with ERE Shortlist genes in ArKO ovary.
Solid connecting lines represent a direct relationship between two molecules: activation marked as green solid line, inhibition marked as red solid line and unspecified marked as gray solid line. Thick cyan lines indicate the fragments of canonical pathways. Up-regulated genes are marked with red circles; down-regulated with blue circles. Genes with ERE are encircled. (Please refer to Figure S1 for detailed network object legend).
Discussion
In this study, a genome wide transcriptional profiling microarray analysis was used for the first time to specify E-dependent genes or associated targets of E action in the mouse ovary. First, our studies led us to identify a set of 450 E-dependent ovarian genes whose biological roles may have significant relevance to functional and structural development of the ovary in mice. Second, we identified a set of 78 of these genes were most likely to have direct biological E targets and out of these, approximately 37 genes were not previously described in the ovary. Female ArKO mice are infertile due to failure to ovulate [14]. They have elevated levels of circulating gonadotrophins and testosterone, severely underdeveloped uteri [14], [15], [16] and increased adiposity [17], [18]. The ovaries possess hemorrhagic cysts and apparent sex-reversal of the ovarian somatic cells; that is granulosa cells become Sertoli-like cells [8], [20], [21]. The ArKO ovaries also displayed an infiltration of macrophages and contain substantial collagen deposition, a characteristic of tissues that are fibrotic [14]. It was logical to ask the question: were there changes in genes associated with these processes?
Masculinization
One of the major genes associated with masculinization, Sox9 [19] was confirmed to be expressed in the ArKO ovary from our microarray analysis results. Early in gonad development soon after the expression of SRY begins, Sox9 expression is strongly upregulated in Sertoli cells; Sox9 expression is maintained in the testis, whereas it is downregulated in the ovary [21]. The presence of the Sertoli cell marker Sox9 in the ArKO ovary confirmed the presence of testicular cell-like cells [19], [20]. 17α-Hydroxylase (17α-OHase) and 17β-Hydroxysteroid Dehydrogenase type-3 (17β-Hsd3, the Leydig cell- specific) are two important steroidogenic enzymes in the production of testosterone. ArKO mice also showed increased levels of 17α-OHase and 17β-Hsd3 at 10 weeks of age, when Sertoli- and Leydig-like cells are beginning to populate the ovary [19] which were correlated with high serum testosterone levels [8]. Based on the known mechanisms maintaining Sertoli cell development and function in the normal (Wt testis), we identified two very intriguing novel targets of E that were significantly increased in the ArKO ovary, Claudin-11 (Cldn11) and platelet-derived growth factor receptor α (Pdgfrα). Cldn11 is a protein component in tight junctions between Sertoli cells which are important for the maintenance of the blood–testis barrier [22,23] and Pdgfrα was identified previously as a key player downstream of Sry in testis organogenesis and Leydig cell differentiation [24]. In the ArKO ovary, granulosa cells become Sertoli-like cells [8], [19], [20] and interstitial cells were identified ultrastructurally as Leydig cells [19]. However, to date, the exact role of E in the somatic cell trans/de-differentiation is unclear, although the fact that granulosa and interstitial cells express ER [25] points to a direct action on the gonadal somatic cells to maintain a ovarian phenotype. It is possible that the oocyte, which also expresses ER [26], plays a part in maintaining the ovarian phenotype with many recorded cases of premature oocyte loss leading to transdifferentiation of granulosa cells into Sertoli-like cells [8], [19]. We therefore, hypothesized that E actively down regulates genes involved in masculinization to suppress the testicular phenotype, and that this action could be either direct on the somatic cells or via the oocyte or both.
Folliculogenesis
From the morphological and stereological assessments made in previous studies [14], [19], follicular growth beyond the preantral stage is unsuccessful in the absence of E, with most succumbing to atresia, developing into hemorrhagic cysts or taking on a testicular phenotype (as discussed above). We hypothesized that this phenomenon occurs due to the loss of oocytes in the ArKO ovary and this was supported by the array of genes that were identified in this study. For example, Paired basic amino acid cleaving system 4 isoform 1 (Pace4) or Proprotein convertase subtilisin/kexin type 6 (Pcsk6) is a proprotein convertase required for producing active forms of important TGFβ ligands in the ovary [27]. The levels of this enzyme were increased in the ArKO ovary. Steady state mRNA levels of Pcsk6 in granulosa cells are normally required to be suppressed by the oocyte during preantral to antral follicle transition [28]. Thus, Pcsk6 represents another granulosa cell transcript regulated by oocytes as well as E, but exhibiting a novel expression pattern during follicular development. Cytochrome P450 lanosterol 14 alpha-demethylase (Cyp51), which was decreased in the ArKO ovary, has long been known for reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. Recent studies on Cyp51 show that it promotes the initiation of meiosis in early folliculogenesis [29], [30]. The decrease in expression of Cyp51 in the ArKO ovary implies that E has a role in controlling early follicular development. This observation is also supported in primate studies by Zachos et al. [31], in which fetal baboons deprived of E showed a 50% reduction in the number of primordial follicles which was restored by E2 administration. Similar data for the ArKO mouse ovary has not been reported, although it has been shown that E influences the breakdown of oocyte nests prior to the formation of primordial follicles [32], [33].
Novel Network Processes
A significant over representation of genes involved in organ development and morphogenesis, inflammatory responses and cellular processes were identified in both the Gene Ontology annotation and GeneGo Pathway analyses of differentially expressed genes in ArKO ovary.
1. Hedgehog signalling
One of the affected pathways in the ArkO ovary is the Hedgehog-Patched (HH) signalling pathway, which is involved in ovarian follicle development. Expression of HH ligands, Sonic hedgehog (Shh), Indian hedgehog (Ihh) and Desert hedgehog (Dhh) are developmentally regulated in the adult ovary [36]. Other components of the HH pathway such as Hedgehog receptors (PTCH1 and PTCH2) and signal transducer Smoothened (SMO) are expressed by granulosa cells of the follicle [34]. Hedghog signalling in the mammalian ovary plays a role in the communication between granulosa cells and the developing theca cells [35]. A recent study by Ren and colleagues [36], showed that Amhr2cre/+SmoM2 mutant mice that constitutively expresses the dominant active form of SMO in the ovary, were anovulatory because of a dramatic reduction in the muscle cells of the theca layer surrounding the developing follicles. These muscle cells within the theca layer play a role in the release of the oocyte at the time of ovulation. One of the smooth muscle markers, actin, gamma 2, smooth muscle, enteric (Actg2) was found to be significantly decreased in the Amhr2cre/+SmoM2 mutant mice [36] and this was also the case in the ArKO ovary (Table 4), suggesting E might play a role in regulating the smooth muscle cells within the ovary during ovulation. In our study, Ihh mRNA expression was found to be decreased in the ArKO mouse ovary (Table 4). The reduced Ihh production in the ArKO ovary implies an inhibition of HH signalling and a hormonal regulation of the Hedgehog system (Figure 3).
2. Inflammatory response and fibrosis
It was not surprising to see the inflammatory responses network filling two of the top 5 network processes (Figure 4 and 5) in the ArKO ovary. Previous studies have shown an increased infiltration of macrophages and deposition of collagen in the ArKO ovary, suggesting that in the absence of E, the ArKO ovary begins to degenerate [19]. Mast cells, macrophages and granulocytes usually accumulate in rat ovaries during the preovulatory period [37]. Their presence in ArKO ovaries indicates that the chemokine signals for ovulation including an increase in the abundance of the immune mediators have been activated, but the follicles have been unable to respond. The increased deposition of collagen in the absence of E suggests that matrix metalloproteinases (MMPs), including the collagenases, have not been activated. This was reflected in the network processes (Figure 6), as MMP-2, MMP-9, tissue inhibitors of MMP 1 (TIMP1) and TIMP2 were either directly or indirectly affected in the absence of E. Detailed studies on the hormonal regulation, site of synthesis, and absolute requirement on individual MMPs and TIMPs, for successful ovulation remain to be elucidated. The improved ovarian morphology in ArKO mice treated with E [8], correlated with a decrease in macrophages, mast cells and collagen again implicating the direct or indirect effects of E on the extracellular matrices of the ovary.
3. WNT signalling
WNT signaling influences cellular differentiation and proliferation in the embryonic and adult ovaries. In this study, we identified a significant decrease in Secreted frizzled-related protein 4 (SFRP4) mRNA expression in the ArKO mouse ovary. SFRP4 is a member of the Frizzled like cystein rich domain family and a modulator of non canonical WNT signalling [38]. It has been shown that SFRP4 transcripts co-localize in the ovary to sites of Fz-1, Wnt4, and Fz-4 expression, suggesting that SFRP4 may regulate signaling through these factors in the ovary [38], [39], [40]. Both SFRP4 and Fz-1 are expressed in granulosa cells of large follicles suggesting a potential regulation by SFRP-4 of Fz-1 signals that may impact granulosa cell differentiation or expression of genes involved in follicle rupture and ovulation [38], [40]. Wnt4 is a member of the WNT family of intracellular growth and differentiation factors, which regulate several key developmental steps. Deficiency of Wnt4 leads to partial female to male sex reversal and a marked reduction in oocyte number [41]. In addition, Fz-4 deficient mice showed defective corpora lutea [42]. It is therefore, tempting to speculate that E is regulating SFRP4 to maintain the ovarian phenotype, The ArKO model will be a useful tool to further study the effect of loss of estrogen on WNT signaling.
Other genes of interest
This study identified genes with known or suspected roles in ovarian functions as stated above. Interestingly, there were 37 genes from the E-dependent DEG Shortlist that possessed EREs that were completely novel to ovarian function. The roles of these genes in ovarian function will require further research and verification. There were also genes (DAX1 [8], MIS [8], ERα, [8], ERβ [8], GDF9 [43], and Cyclin D2 [5]) that were expected to be regulated (up or down) in the absence of E as a result of previous studies, which were not detected in the present study. It is likely that some genes will be missed, because they may be affected by E at earlier time-points or their expression may be below the level of detection in this study.
Conclusion
Our success in identifying E-dependent ovarian genes validates the usefulness of whole-genome arrays. We identified more than double the number of E-dependent ovarian/oocyte factors previously reported. The E-dependent ovarian genes identified in this study provide attractive candidates for studying female infertility [44]. In addition, analyses can be easily expanded by inclusion of additional expression profiles, such as, a set of E2 replacement ArKO ovarian genes or refined by identifying which ER subtype is responsible for specific E-dependent ovarian genes. Such study will be required in order to define more precisely mechanisms of ovarian function, as well as provides new targets for the development of contraceptives and highlights genes previously uncharacterised in the ovary, that may be involved in the pathophysiology of female reproductive disorders. Furthermore, these E-dependent expression datasets will enable further gene discovery focusing on microRNAs and non-coding RNAs.
Supporting Information
E-dependent DEG list.
(0.91 MB DOC)
78 most significant differentially genes that are most likely to represent direct biological E target genes.
(0.13 MB DOC)
Network object legend.
(3.18 MB TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The financial support of the National Health and Medical Research Council of Australia (Regkeys 494802 and 441101) is acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, et al. Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci U S A. 1998;95:15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, et al. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 5.Robker RL, Richards JS. Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol Endocrinol. 1998;12:924–940. doi: 10.1210/mend.12.7.0138. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Rudd MD, Hernandez-Gonzalez I, Gonzalez-Robayna I, Fan HY, et al. FSH and FOXO1 regulate genes in the sterol/steroid and lipid biosynthetic pathways in granulosa cells. Mol Endocrinol. 2009;23:649–661. doi: 10.1210/me.2008-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charpentier AH, Bednarek AK, Daniel RL, Hawkins KA, Laflin KJ, et al. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 2000;60:5977–5983. [PubMed] [Google Scholar]
- 8.Britt KL, Stanton PG, Misso M, Simpson ER, Findlay JK. The effects of estrogen on the expression of genes underlying the differentiation of somatic cells in the murine gonad. Endocrinology. 2004;145:3950–3960. doi: 10.1210/en.2003-1628. [DOI] [PubMed] [Google Scholar]
- 9.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 10.Bajic VB, Tan SL, Chong A, Tang S, Strom A, et al. Dragon ERE Finder version 2: A tool for accurate detection and analysis of estrogen response elements in vertebrate genomes. Nucleic Acids Res. 2003;31:3605–3607. doi: 10.1093/nar/gkg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Praz V, Perier R, Bonnard C, Bucher P. The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression data. Nucleic Acids Res. 2002;30:322–324. doi: 10.1093/nar/30.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang S, Tan SL, Ramadoss SK, Kumar AP, Tang MH, et al. Computational method for discovery of estrogen responsive genes. Nucleic Acids Res. 2004;32:6212–6217. doi: 10.1093/nar/gkh943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britt KL, Drummond AE, Cox VA, Dyson M, Wreford NG, et al. An age-related ovarian phenotype in mice with targeted disruption of the Cyp 19 (aromatase) gene. Endocrinology. 2000;141:2614–2623. doi: 10.1210/endo.141.7.7578. [DOI] [PubMed] [Google Scholar]
- 15.Britt KL, Drummond AE, Dyson M, Wreford NG, Jones ME, et al. The ovarian phenotype of the aromatase knockout (ArKO) mouse. J Steroid Biochem Mol Biol. 2001;79:181–185. doi: 10.1016/s0960-0760(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 16.Fisher CR, Graves KH, Parlow AF, Simpson ER. Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. Proc Natl Acad Sci U S A. 1998;95:6965–6970. doi: 10.1073/pnas.95.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones ME, Thorburn AW, Britt KL, Hewitt KN, Misso ML, et al. Aromatase-deficient (ArKO) mice accumulate excess adipose tissue. J Steroid Biochem Mol Biol. 2001;79:3–9. doi: 10.1016/s0960-0760(01)00136-4. [DOI] [PubMed] [Google Scholar]
- 18.Jones ME, Thorburn AW, Britt KL, Hewitt KN, Wreford NG, et al. Aromatase-deficient (ArKO) mice have a phenotype of increased adiposity. Proc Natl Acad Sci U S A. 2000;97:12735–12740. doi: 10.1073/pnas.97.23.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Britt KL, Kerr J, O'Donnell L, Jones ME, Drummond AE, et al. Estrogen regulates development of the somatic cell phenotype in the eutherian ovary. FASEB J. 2002;16:1389–1397. doi: 10.1096/fj.01-0992com. [DOI] [PubMed] [Google Scholar]
- 20.Liew SH, Drummond AE, Jones ME, Findlay JK. The lack of estrogen and excess luteinizing hormone are responsible for the female ArKO mouse phenotype. Mol Cell Endocrinol. 2010 doi: 10.1016/j.mce.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 22.Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S. Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J Cell Biol. 1999;145:579–588. doi: 10.1083/jcb.145.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellani A, Ji J, Mauduit C, Deschildre C, Tabone E, et al. Developmental and hormonal regulation of the expression of oligodendrocyte-specific protein/claudin 11 in mouse testis. Endocrinology. 2000;141:3012–3019. doi: 10.1210/endo.141.8.7625. [DOI] [PubMed] [Google Scholar]
- 24.Brennan J, Tilmann C, Capel B. Pdgfr-alpha mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 2003;17:800–810. doi: 10.1101/gad.1052503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couse JF, Lindzey J, Grandien K, Gustafsson JA, Korach KS. Tissue distribution and quantitative analysis of estrogen receptor-alpha (ERalpha) and estrogen receptor-beta (ERbeta) messenger ribonucleic acid in the wild-type and ERalpha-knockout mouse. Endocrinology. 1997;138:4613–4621. doi: 10.1210/endo.138.11.5496. [DOI] [PubMed] [Google Scholar]
- 26.Wu TC, Wang L, Wan YJ. Expression of estrogen receptor gene in mouse oocyte and during embryogenesis. Mol Reprod Dev. 1992;33:407–412. doi: 10.1002/mrd.1080330406. [DOI] [PubMed] [Google Scholar]
- 27.Constam DB, Robertson EJ. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev. 2000;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz FJ, Sugiura K, Eppig JJ. Regulation of Pcsk6 expression during the preantral to antral follicle transition in mice: opposing roles of FSH and oocytes. Biol Reprod. 2008;78:176–183. doi: 10.1095/biolreprod.107.063537. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Xu B, Zhou B, Zhang C, Yang J, et al. Reducing CYP51 inhibits follicle-stimulating hormone induced resumption of mouse oocyte meiosis in vitro. J Lipid Res. 2009;50:2164–2172. doi: 10.1194/jlr.M800533-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Xu B, Xie H, Zhou B, Ouyang H, et al. Lanosterol metabolic product(s) is involved in primordial folliculogenesis and establishment of primordial follicle pool in mouse fetal ovary. Mol Reprod Dev. 2009;76:514–521. doi: 10.1002/mrd.20973. [DOI] [PubMed] [Google Scholar]
- 31.Zachos NC, Billiar RB, Albrecht ED, Pepe GJ. Developmental regulation of baboon fetal ovarian maturation by estrogen. Biol Reprod. 2002;67:1148–1156. doi: 10.1095/biolreprod67.4.1148. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Breen K, Pepling ME. Estrogen can signal through multiple pathways to regulate oocyte cyst breakdown and primordial follicle assembly in the neonatal mouse ovary. J Endocrinol. 2009;202:407–417. doi: 10.1677/JOE-09-0109. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Jefferson WN, Newbold RR, Padilla-Banks E, Pepling ME. Estradiol, progesterone, and genistein inhibit oocyte nest breakdown and primordial follicle assembly in the neonatal mouse ovary in vitro and in vivo. Endocrinology. 2007;148:3580–3590. doi: 10.1210/en.2007-0088. [DOI] [PubMed] [Google Scholar]
- 34.Russell MC, Cowan RG, Harman RM, Walker AL, Quirk SM. The hedgehog signaling pathway in the mouse ovary. Biol Reprod. 2007;77:226–236. doi: 10.1095/biolreprod.106.053629. [DOI] [PubMed] [Google Scholar]
- 35.Wijgerde M, Ooms M, Hoogerbrugge JW, Grootegoed JA. Hedgehog signaling in mouse ovary: Indian hedgehog and desert hedgehog from granulosa cells induce target gene expression in developing theca cells. Endocrinology. 2005;146:3558–3566. doi: 10.1210/en.2005-0311. [DOI] [PubMed] [Google Scholar]
- 36.Ren Y, Cowan RG, Harman RM, Quirk SM. Dominant activation of the hedgehog signaling pathway in the ovary alters theca development and prevents ovulation. Mol Endocrinol. 2009;23:711–723. doi: 10.1210/me.2008-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brannstrom M, Mayrhofer G, Robertson SA. Localization of leukocyte subsets in the rat ovary during the periovulatory period. Biol Reprod. 1993;48:277–286. doi: 10.1095/biolreprod48.2.277. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh M, Mulders SM, Friis RR, Dharmarajan A, Richards JS. Expression and localization of secreted frizzled-related protein-4 in the rodent ovary: evidence for selective up-regulation in luteinized granulosa cells. Endocrinology. 2003;144:4597–4606. doi: 10.1210/en.2003-0048. [DOI] [PubMed] [Google Scholar]
- 39.Hsieh M, Johnson MA, Greenberg NM, Richards JS. Regulated expression of Wnts and Frizzleds at specific stages of follicular development in the rodent ovary. Endocrinology. 2002;143:898–908. doi: 10.1210/endo.143.3.8684. [DOI] [PubMed] [Google Scholar]
- 40.Drake JM, Friis RR, Dharmarajan AM. The role of sFRP4, a secreted frizzled-related protein, in ovulation. Apoptosis. 2003;8:389–397. doi: 10.1023/a:1024181203729. [DOI] [PubMed] [Google Scholar]
- 41.Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 42.Hsieh M, Boerboom D, Shimada M, Lo Y, Parlow AF, et al. Mice null for Frizzled4 (Fzd4−/−) are infertile and exhibit impaired corpora lutea formation and function. Biol Reprod. 2005;73:1135–1146. doi: 10.1095/biolreprod.105.042739. [DOI] [PubMed] [Google Scholar]
- 43.Britt KL, Saunders PK, McPherson SJ, Misso ML, Simpson ER, et al. Estrogen actions on follicle formation and early follicle development. Biol Reprod. 2004;71:1712–1723. doi: 10.1095/biolreprod.104.028175. [DOI] [PubMed] [Google Scholar]
- 44.Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E-dependent DEG list.
(0.91 MB DOC)
78 most significant differentially genes that are most likely to represent direct biological E target genes.
(0.13 MB DOC)
Network object legend.
(3.18 MB TIF)