Abstract
Dynactin is a multisubunit protein complex required for the activity of cytoplasmic dynein. In Caenorhabditis elegans, although 10 of the 11 dynactin subunits were identified based on the sequence similarities to their orthologs, the p24/p22 subunit has not been detected in the genome. Here, we demonstrate that DNC-3 (W10G11.20) is the functional counterpart of the p24/p22 subunit in C. elegans. RNAi phenotypes and subcellular localization of DNC-3 in early C. elegans embryos were nearly identical to those of the known dynactin components. All other dynactin subunits were co-immunoprecipitated with DNC-3, indicating that DNC-3 is a core component of dynactin. Furthermore, the overall secondary structure of DNC-3 resembles to those of the mammalian and yeast p24/p22. We found that DNC-3 is required for the localization of the DNC-1/p150Glued and DNC-2/dynamitin, the two components of the projection arm of dynactin, to the nuclear envelope of meiotic nuclei in the adult gonad. Moreover, DNC-3 physically interacted with DNC-1 and DNC-2 and significantly enhanced the binding ability between DNC-1 and DNC-2 in vitro. These results suggest that DNC-3 is essential for the formation of the projection arm subcomplex of dynactin.
Introduction
The dynactin complex is an activator of cytoplasmic dynein, a major microtubule minus-directed motor protein. It is necessary to target dynein to specific cellular locations (Kardon & Vale 2009), to connect dynein to cargo (Holleran et al. 2001) and to increase processivity of the dynein motor activity (Karki & Holzbaur 1995; Vaughan & Vallee 1995; King & Schroer 2000). The dynein–dynactin complex is involved in a variety of cellular events, including membrane vesicle transport, mitotic spindle formation, centrosome separation and nuclear positioning (Kardon & Vale 2009).
Dynactin is a multiprotein complex of 1.2 MDa containing 11 different subunits and has two distinct structural domains; the projection arm that binds microtubules and motor proteins and the Arp1 rod that binds to cargo (Schroer 2004). The projection arm is composed of two copies of p150Glued (DCTN1) and p24/p22 (DCTN3) and four copies of dynamitin (p50/DCTN2). The Arp1 rod contains an octameric polymer of the actin-related protein Arp1, the actin-capping protein CapZ α/β, another actin-related protein Arp11, p62 (DCTN4), p25 (DCTN5) and p27 (DCTN6) (Schroer 2004).
Among the three components of the projection arm, p150Glued is the largest subunit and essential for the direct binding to microtubules and the dynein intermediate chain (Karki & Holzbaur 1995; Vaughan & Vallee 1995; Waterman-Storer et al. 1995). These binding activities are important for the effect of dynactin for the dynein processivity and microtubule anchoring at centrosomes (Quintyne et al. 1999; Quintyne & Schroer 2002). The second subunit, dynamitin, connects two structural domains of dynactin (Eckley et al. 1999). The smallest subunit of the dynactin complex, p24/p22, is the least conserved among the subunits (Karki et al. 1998; Pfister et al. 1998; Amaro et al. 2008). Human p24/p22 was shown to bind directly to p150Glued and dynamitin in vitro (Karki et al. 1998; Maier et al. 2008). In budding yeast, the p24/p22 homolog was genetically shown to be essential for maintaining the association of p150Glued with the dynactin complex (Amaro et al. 2008).
In the nematode Caenorhabditis elegans, some of the dynactin subunits have been identified based on the sequence similarity, and their functions were genetically analyzed. Loss-of-function phenotypes of DNC-1 (p150Glued), DNC-2 (Dynamitin), CAP-2 (CapZβ) and ARP-1 demonstrated that these proteins are required for microtubule-dependent events in early embryos, such as pronuclear migration, centrosome separation and mitotic spindle positioning (Skop & White 1998; Gonczy et al. 1999; Le Bot et al. 2003; Zhang et al. 2008). These phenotypes are similar to those by dynein knockouts (Gonczy et al. 1999). DNC-1, DNC-2 and ARP-1 localize to centrosomes during prometaphase, nuclear envelope, mitotic spindles, midbody remnants and P2-EMS cell borders (Skop & White 1998; Zhang et al. 2008; Ai et al. 2009). Other putative orthologs of dynactin components, except p24/p22, was detected in the C. elegans genome based on the amino acid sequence similarities, but their functions in vivo have not been well studied.
In this study, we genetically and biochemically demonstrate that W10G11.20/DNC-3 is the C. elegans ortholog of p24/p22, although this protein shows only a marginal sequence similarity to p24/p22 of other organisms. We also show that W10G11.20/DNC-3 is essential for the formation of dynactin complex by binding to both DNC-1 and DNC-2. Through this study, all the C. elegans dynactin subunits orthologs were biochemically confirmed to form a complex in vivo. Now that the complete subunits of dynactin were defined, C. elegans will provide an excellent model system to analyze the in vivo function and its regulatory mechanisms of the dynein–dynactin complex.
Results
W10G11.20 is required for microtubule-mediated events in early C. elegans embryos
In a large-scale protein–protein interaction analyses in C. elegans (Li et al. 2004), W10G11.20 was reported to interact with protein phosphatase 4 (PPH-4.1) that is required for centrosome maturation and establishment of chiasmata during meiotic prophase I (Sumiyoshi et al. 2002). The W10G11.20 gene encodes a protein with 171 amino acids containing coiled-coil structures, but a search by the Blast software did not detect any proteins with significant homologies.
To investigate the in vivo function of W10G11.20, the phenotypes by RNAi knockdown were analyzed. When maternal expression of W10G11.20 was inhibited by the soaking RNAi method (Maeda et al. 2001), the majority of embryos did not hatch (85%, 448/525), indicating that this gene is essential for embryonic development. To further analyze the function of W10G11.20, we investigated the phenotypes of live W10G11.20(RNAi) embryos using Nomarski microscopy.
In the wild-type zygote, the male pronucleus locates to the posterior end, whereas the female pronucleus to the opposite side and migrates toward the male pronucleus (Fig. 1A,B). The pronuclei meet at approximately 70% of the embryo length (Fig. 1C), move to the center of the embryo and rotate 90° (Fig. 1D). Then, nuclear envelope breaks down and a bipolar spindle is assembled. In W10G11.20(RNAi) embryos, after the male and female pronuclei became visible (Fig. 1F), the migration of the male pronucleus did not occur (Fig. 1G,H). In some W10G11.20(RNAi) embryos, the female pronucleus partially migrated toward the male pronucleus, but never met with the male pronucleus. The nuclear envelope break down (NEBD) of the male pronucleus took place 1–2 min before that of the female pronucleus (Fig. 1I,J). After NEBD, bipolar spindle formation was not apparent by the DIC observation (Fig. 1J). In addition, W10G11.20(RNAi) embryos often had multiple female pronuclei (Table 1, Fig. 1K), indicating the defect of meiotic divisions. These phenotypes imply that W10G11.20 is required for microtubule-mediated events including pronuclear migration, bipolar spindle formation and oocyte meiosis. Therefore, we next examined the microtubule behaviors in W10G11.20(RNAi) embryos.
Figure 1.
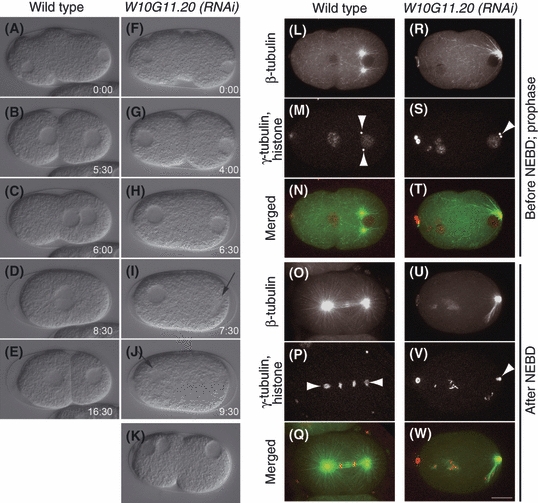
Early embryonic phenotypes in W10G11.20(RNAi) embryos. (A–K) DIC microscopy images of one-cell embryos. Time-series images of a wild-type embryo (A–E) and a W10G11.20(RNAi) embryo showing the defect of pronuclei meeting (F–J). (K) a W10G11.20(RNAi) embryo showing multiple female nuclei. Arrows in (I, J) indicate nuclear envelope break down (NEBD). (L–W) Confocal live images of embryos expressing GFP::β-tubulin, mCherry::γ-tubulin and mCherry::histone. (L–Q) Wild-type embryos. (R–W) W10G11.20(RNAi) embryos. (L–N, R–T) before NEBD. (O–Q, U–W) after NEBD. Arrowheads in (M, P, S, V) indicate centrosomes. Scale bar: 10 μm.
Table 1.
Early embryonic RNAi phenotypes of W10G11.20 and dnc-2
| Number of embryos analyzed | Embryos with single female pronucleus | Embryos with multiple female pronuclei | Average number of multiple female pronuclei† | Defect in male pronuclear migration‡ | No apparent defect in one-cell stage | |
|---|---|---|---|---|---|---|
| N2 | 13 | 13 | – | – | – | 13 |
| dnc-2(RNAi) | 21 | 16 | 5 | 2 | 20 | 1 |
| W10G11.20(RNAi) | 24 | 21 | 3 | 2 | 22 | 2 |
Average number of multiple female pronuclei was calculated among the embryos with multiple female pronuclei.
Male pronuclei fail to detach from cell cortex.
To concurrently visualize microtubules, centrosomes and chromosomes, we have constructed a strain that expresses GFP::β-tubulin, mCherry::γ-tubulin and mCherry::histone H2B (Toya et al. 2010). In the wild-type embryo, centrosomes separate to the opposite sides of the male pronucleus (Fig. 1M). In contrast, centrosomes in W10G11.20(RNAi) embryos did not separate and stay at the posterior to the male pronucleus (Fig. 1S). In the wild type, a bipolar mitotic spindle was observed after NEBD (Fig. 1P), but in W10G11.20(RNAi) embryos, it was never formed and two centrosomes stayed at the posterior side of the male pronucleus (Fig. 1V). These phenotypes are highly similar to those by the depletion of subunits of cytoplasmic dynein (DHC-1, dynein heavy chain) and dynactin (DNC-1/p150Glued, DNC-2/Dynamitin/p50, ARP-1/Arp1 and CAP-2/CapZβ) (Skop & White 1998; Gonczy et al. 1999; Le Bot et al. 2003) (Table 1). Based on these results, we speculated that W10G11.20 is likely to function with cytoplasmic dynein and/or dynactin.
Localizations of W10G11.20 in early embryos are similar to those of dynein and dynactin subunits
To observe the subcellular localization of W10G11.20 in adult gonads and early embryos, we generated transgenic lines that express GFP::W10G11.20::FLAG under the control of the pie-1 promoter that drives transcription in the germ-line. The FLAG-tag was included in the construct so that this transgenic line can also be used for affinity purification of W10G11.20-interacting proteins.
Live imaging analysis of early embryos using a spinning-disk confocal time-lapse microscopy revealed that GFP::W10G11.20::FLAG was present in the cytoplasm throughout cell cycle (Fig. 2A–F). As the cell cycle proceeds, the signal at the nuclear periphery became brighter, and it was also detected around centrosomes (Fig. 2A–C). In metaphase, when the nuclear membrane becomes partially permeable, GFP::W10G11.20::FLAG was enriched on both sides of condensed chromosomes (metaphase plates) (Fig. 2C). After cell division, GFP::W10G11.20::FLAG was temporally enriched at the cell boundaries (Fig. 2D). In addition, GFP::W10G11.20::FLAG localized to the female meiotic spindle (Fig. 2E,F). These localization patterns are similar to those of the cytoplasmic dynein subunit DHC-1 and the dynactin subunits, DNC-1, DNC-2, ARP-1 (Skop & White 1998; Gonczy et al. 1999; Zhang et al. 2008).
Figure 2.
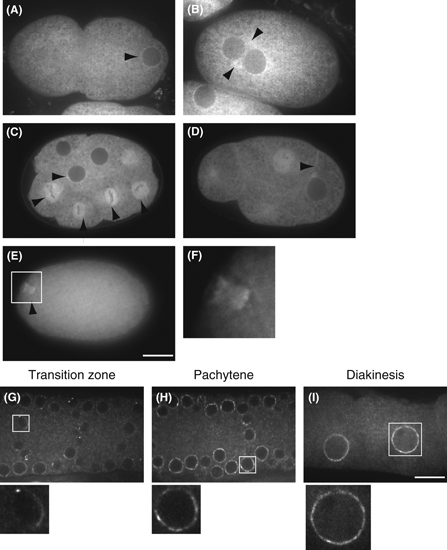
Subcellular localization of GFP::W10G11.20. (A–F) Early embryos. Arrowheads indicate the nuclear envelope (A), centrosomes (B, C), mitotic spindle (C), cell border (D) and meiotic spindle (E). Panel F shows magnified views of the indicated regions in panel E. (G–I) Adult gonads: transition zone (G), pachytene (H) and diakinesis (I) stages. Lower panels show magnified views of the indicated regions in upper panels. Scale bar: 10 μm.
W10G11.20 localizes to the nuclear envelope of the adult germ cells
As some subunits of dynein and dynactin was shown to be present at the nuclear periphery in the C. elegans germ-line (Malone et al. 2003; Sato et al. 2009; Zhou et al. 2009), localization of W10G11.20 in the adult hermaphrodite gonads was observed using the transgenic line expressing GFP::W10G11.20::FLAG described earlier. While GFP::W10G11.20::FLAG was undetectable in the distal gonad where germ cells are mitotic, it was detected as bright dots at the periphery of nuclei (a few dots per nucleus) in the transition zone (Fig. 2G). After the pachytene stage, it localized to the entire periphery of the nuclei (Fig. 2H,I).
The localization of W10G11.20 at the nuclear periphery was also confirmed by antibody staining. We raised polyclonal antibodies against W10G11.20 and stained the gonads of wild-type adult hermaphrodites. The W10G11.20 staining coincided with that of KT23 monoclonal antibody that recognizes nuclear membrane (Takeda et al. 2008) (Fig. S1A,B in Supporting Information). This signal by anti-W10G11.20 antibody was not detected in the gonad of W10G11.20(RNAi) worms, confirming that this signal is specific for the W10G11.20 protein (Fig. S1C,D in Supporting Information). This localization at the nuclear periphery in adult gonads is similar to the reported localization of dynein heavy chain (DHC-1) and p150Glued (DNC-1) (Malone et al. 2003; Sato et al. 2009; Zhou et al. 2009).
Taken together, the loss-of-function phenotypes and subcellular localization in the gonad and early embryos of W10G11.20 are highly similar to those of the subunits of the dynein–dynactin, strongly indicating that W10G11.20 functions closely with this complex.
W10G11.20 is a component of the dynactin complex
Above-mentioned analyses raised the possibility that W10G11.20 may be a component of the dynein–dyn-actin complex, although no sequence similarity to any of the known subunits was detected. In C. elegans, 10 of 11 dynactin subunits were detected in the genome. Among them, DNC-1/p150Glued, DNC-2/Dynamitin, ARP-1/Arp1 and CAP-2/CapZβ were genetically characterized (Skop & White 1998; Gonczy et al. 1999; Le Bot et al. 2003). Six other subunits (Actin, CAP-1/CapZα, DNC-4/p62, Y71F9AL.14/p25, Y54E10A.5/p27 and C49H3.8/Arp11) were reported based on their sequence similarity to the corresponding subunits (Waddle et al. 1993; Eckley et al. 1999; Eckley & Schroer 2003) (Table 2). The only subunit whose ortholog unfound was p24/p22. Because the molecular mass of W10G11.20 (19.4k) is similar to p24/p22, we suspected that W10G11.20 might be the functional counterpart of p24/p22.
Table 2.
Complete components of the Caenorhabditis elegans dynactin complex identified by mass spectrometry
| # of fragment | C. elegans protein name† | Coverage (%)‡ | Mammalian homologs | BlastpE value§ | References |
|---|---|---|---|---|---|
| 1 | DNC-1 | 53 | p150Glued (DCTN1) | 2e-104 | Skop & White (1998); Le Bot et al. (2003) |
| 4 | DNC-2 | 50 | Dynamitin (p50/DCTN2) | 6e-15 | Skop & White (1998) |
| 2 | W10G11.20 (DNC-3) | 63 | p24 (DCTN3) | Not significant | This study |
| 3 | DNC-4 | 26 | p62 (DNTN4) | 2e-43 | Eckley et al. (1999) |
| 3 | C49H3.8 (ARP-11) | 49 | Arp11 (ACTR10) | 6e-31 | Eckley & Schroer (2003) |
| 3,4,5 | Actin | 33 | actin | ||
| 4 | ARP-1 | 59 | Arp1 (ACTR1A) | 3e-162 | Le Bot et al. (2003); Zhang et al. (2008) |
| 5 | CAP-1 | 50 | CapZα | 3e-81 | Waddle et al. (1993) |
| 5 | CAP-2 | 35 | CapZβ | 1e-112 | Waddle et al. (1993); Le Bot et al. (2003) |
| 6 | Y54E10A.5 (DNC-6) | 46 | p27 (DCTN6) | 7e-29 | Eckley et al. (1999) |
| 6 | Y71F9AL.14 (DNC-5) | 25 | p25 (DCTN5) | 6e-30 | Eckley et al. (1999) |
Genes named in this study are indicated with parentheses.
Percent coverage in the mass spectrometric analysis of the immunoprecipitation shown in Fig. 3.
E values of Blastp against human orthologs.
To test this possibility, we examined whether W10G11.20 directly interacts with other dynactin components. The extract from the transgenic worm that expresses GFP::W10G11.20::FLAG was immunoprecipitated using anti-FLAG antibody, and proteins co-precipitated with GFP::W10G11.20::FLAG were determined by mass spectrometry (Fig. 3). GFP::W10G11.20::FLAG and ten dynactin components described earlier(DNC-1, DNC-2, DNC-4, Y71F9AL.14/p25, Y54E10A.5/p27, ARP-1, C49H3.8/Arp11, CAP-1, CAP-2 and actin) were detected with a significant sequence coverage (Table 2). Furthermore, in reciprocal immunoprecipitation experiments using worm extracts (wild type, GFP::DNC-2 or GFP::ARP-1 transgenic lines) and antibodies against DNC-1 or GFP, co-precipitation of W10G11.20 with DNC-1, GFP::DNC-2 and GFP::ARP-1 was confirmed (Fig. S2 in Supporting Information). These results strongly suggest that W10G11.20 is a component of dynactin complex and that it is the ortholog of the p24/p22 subunit.
Figure 3.
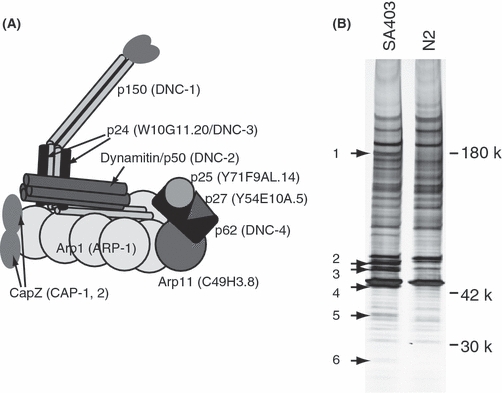
W10G11.20 is a component of the dynactin complex. (A) Diagram of the mammalian dynactin complex. Caenorhabditis elegans homologs are shown in parentheses. (B) Co-immunoprecipitation of the dynacitn subunits with FLAG-tagged W10G11.20. Silver stained 5%–20% gradient polyacrylamide gel is shown. Arrows indicate the proteins specifically precipitated from the extract of the GFP::W10G11.20::FLAG transgenic line, which were analyzed by mass spectrometry.
To further confirm that W10G11.20 corresponds to p24/p22, we compared the peptide sequences of W10G11.20 with human p24/p22 and the budding yeast p24/p22 ortholog, Ldb18 (Fig. 4). Low similarities that were missed by the Blast search were detected (18% and 16% identity, 39% and 43% similarity with W10G11.20, respectively), and all three proteins contain 5–6 coiled-coil domains (predicted using the COILS algorithm) in the similar locations (Fig. 4). Taken together, we concluded that W10G11.20 is the C. elegans ortholog of the dynactin component p24/p22 and named it DNC-3. [In addition, p25 ortholog Y71F9AL.14, p27 ortholog Y54E10A.5 and Arp11 ortholog C49H3.8 were named DNC-5, DNC-6 and ARP-11, respectively (Table 2)].
Figure 4.
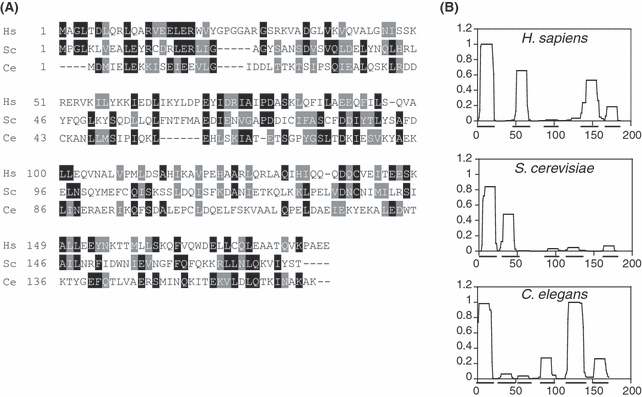
Sequence and structural features of W10G11.20 are similar to those of human p24/p22 and yeast Ldb18. (A) Multiple alignment of human p24/p22 (Hs), Ldb18 (Sc) and W10G11.20 (Ce). The alignment was generated by clustalW program. Identical residues are indicated by black boxes, and similar residues are shaded in gray. (B) p24/p22, Ldb18 and W10G11.20 have 5–6 coiled-coil domains. Coiled-coil structures were predicted using the COILS algorithm with a 14-residue window.
DNC-3 is required for the localization of other dynactin components at the nuclear envelope
It was previously reported that dynein localizes to the periphery of germ cell nuclei and required for the arrangement of nuclei in the gonad (Zhou et al. 2009). We observed whether dynactin is also necessary for the gonad architecture. In the wild-type adult hermaphrodites, germ-line nuclei are arranged at the periphery of the gonad. In contrast, germ-line nuclei were disarrayed from the surface of the gonad in the dnc-1(RNAi), dnc-2(RNAi) and dnc-3(RNAi) worms (Fig. 5). These results demonstrated that the dynactin complex is required for the formation of regular arrays of germ-line nuclei to the surface of the gonads, which is consistent with the requirement of dynein in this process.
Figure 5.
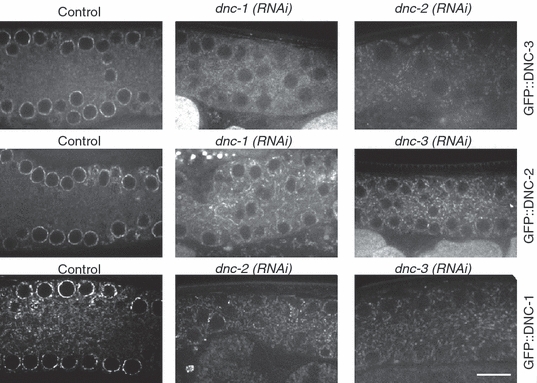
DNC-3/W10G11.20 is required for localization of DNC-1 and DNC-2 at the nuclear envelope in gonads. Confocal images of the pachytene stage region in adult gonads are shown. From top to bottom: images of a gonad expressing GFP::DNC-3, GFP::DNC-2 and GFP::DNC-1. The left panels show the gonads without RNAi treatment. The middle and right panels show images of gonads with dnc-1 (RNAi), dnc-2 (RNAi) or dnc-3 (RNAi), as indicated. Scale bar: 10 μm.
We next tested whether DNC-3 is necessary for the localization of other subunits of dynactin complex at the nuclear envelope. In untreated lines, GFP::DNC-1 and GFP::DNC-2 localized to the nuclear envelope of pachytene or diakinesis stage nuclei in the adult gonads. In contrast, their localization was largely impaired in dnc-3(RNAi) worms (Fig. 5). In the reciprocal experiments, GFP::DNC-3::FLAG did not localize to the nuclear envelope in dnc-1(RNAi) and dnc-2(RNAi) gonads (Fig. 5) and embryos (data not shown). These results demonstrated that localization of dynactin components DNC-1, DNC-2 and DNC-3 at the nuclear envelope are mutually dependent, implying that the targeting of the dynactin complex to the specific subcellular loci is determined not by a single subunit, but by a complete functional complex.
DNC-3 physically interacts with DNC-2 and DNC-1 and connects them together
Mutually dependent subcellular localization among DNC-1, DNC-2 and DNC-3 and the co-immunoprecipitation results indicate that these proteins are likely to physically interact with each other. Direct interaction between DNC-1, DNC-2 and DNC-3 was examined in vitro using the recombinant proteins expressed in Escherichia coli. GST pull-down assays demonstrated that GST-DNC-3 directly interacts with 6xHis-DNC-1 and 6xHis-DNC-2 (Fig. 6B). Using the truncated versions of DNC-3 tagged with GST, we found that the C-terminal half of the DNC-3 was sufficient to interact with 6xHis-DNC-2, whereas both N-terminal and C-terminal regions of DNC-3 were required to interact with 6xHis-DNC-1 (Fig. 6B).
Figure 6.
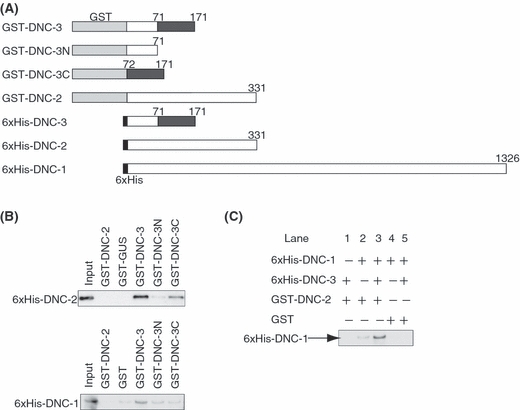
DNC-3/W10G11.20 physically interacts with DNC-1 and DNC-2. (A) Diagram of the constructed DNC recombinant proteins. Light gray and black bars indicate the GST and 6xHis tag, respectively. Dark gray bars indicate the C-terminal region of DNC-3. Numbers indicate amino acid. (B) Upper panel: Pull-down assay for 6xHis-tagged DNC-2 by GST-tagged proteins as indicated. Lower panel: Pull-down assay for 6xHis-tagged DNC-1 by GST-tagged proteins as indicated. (C) Pull-down assay for 6xHis-DNC-1 by either GST-DNC-2 or GST with or without 6xHis-DNC-3.
Our in vitro assay 6xHis-DNC-1 did not bind to GST-DNC-2 efficiently (Fig. 6B). Because DNC-3 bound to both DNC-1 and DNC-2, we speculated that DNC-3 connects both proteins to form a tertiary complex. To test this model, we examined the binding of DNC-1 and DNC-2 in the presence and absence of DNC-3. Whereas direct interaction of 6xHis-DNC-1 and GST-DNC-2 was very weak, the presence of 6xHis-DNC-3 dramatically enhanced the interaction (Fig. 6C lane 3). Taken together, we concluded that DNC-3 connects DNC-1 with DNC-2 efficiently to form the dynactin complex.
Discussion
In C. elegans, while 10 of 11 dynactin subunits are detectable in the genome based on the sequence similarities (Eckley et al. 1999; Eckley & Schroer 2003), the ortholog of p24/p22 has not been clearly defined. Previously, W02A2.2 was reported as the candidate C. elegans ortholog of p24/p22 based on a Blast search result (Eckley & Schroer 2003), but its sequence similarity to p24/p22 was very low and no further supporting evidence have been presented. In this study, based on four lines of evidence, we concluded that DNC-3/W10G11.20 is the C. elegans ortholog of p24/p22. First, the knockout phenotypes of dnc-3 were virtually equivalent to those of the known dynactin components. Second, the subcellular localization of DNC-3 coincided with other dynactin subunits. Third, DNC-3 co-immunoprecipitated with all other ten dynactin subunits and directly bound to DNC-1/p150Glued and DNC-2/dynamitin. Finally, the overall size and secondary structure of DNC-3 is similar to those of human and yeast p24/p22. We analyzed all the major coprecipitated bands with DNC-3 by mass spectrometry, and no other uncharacterized proteins were identified. Collectively, although the similarity of primary amino acid sequence is marginal, DNC-3 is most likely to be the p24/p22 ortholog in C. elegans.
Our in vitro assay demonstrated that DNC-3 function as an adaptor to connect DNC-1 with DNC-2. Consistently, in budding yeast, disruption of the p24/p22 homolog LDB18 results in reduction of Jnm1 (homolog of DNC-2/dynamitin)-Nip100 (homolog of DNC-1/p150Glued) interaction by approximately 95% (Amaro et al. 2008). It was also reported that mammalian p150Glued (DNC-1 ortholog), p50 (DNC-2 ortholog) and p24/p22 (DNC-3 ortholog) form a stable complex, whereas direct interaction between p150Glued and dynamitin was very weak or undetectable (Karki et al. 1998; Moore et al. 2008). Thus, the essential role p24/p22/DNC-3 in the assembly of a dynactin to link p150Glued/DNC-1 and dynamitin/DNC-2 is evolutionarily conserved.
In interphase in early embryos and meiotic prophase I in the adult gonads, dynactin localizes to the nuclear envelope. While bovine p150Glued has the ability to bind to the nuclear pore complex by itself (Payne et al. 2003), our data showed that DNC-1, DNC-2 and DNC-3 are mutually dependent for their localization to the nuclear envelope. Thus, in C. elegans, none of these subunits are sufficient for localizing to the nuclear envelope by itself, and other components might be responsible. Notably, dynein/dynactin does not accumulate on the nuclear envelope in budding yeast and fission yeast, both of which apparently lack p62, p25 p27 subunits that form “the pointed-end complex” subdomain of dynactin (McMillan & Tatchell 1994; Kahana et al. 1998; Eckley & Schroer 2003). Therefore, one possibility might be that the pointed-end complex or any of its components may be involved in the nuclear envelope targeting in C. elegans.
In metazoans, the dynein–dynactin complex is involved in a variety of cellular events including centrosome separation, mitotic spindle formation and nuclear migration/positioning (Gonczy et al. 1999; Morris 2003). In addition, dynein–dynactin is required for efficient homologous chromosome paring during meiotic prophase in some organisms including fission yeast (Yamashita et al. 1997; Niccoli et al. 2004) and C. elegans (Sato et al. 2009). In the C. elegans germ cells undergoing homologous chromosome paring, dynein and nuclear envelope proteins ZYG-12 and SUN-1 form patch-like structures (Sato et al. 2009), which is similar to the ones observed for GFP::DNC-3::FLAG. Additionally, PPH-4.1 (protein phosphatase 4), a DNC-3 interactor identified by yeast 2-hybrid analysis (Li et al. 2004), is also reported to be required for meiotic recombination (Sumiyoshi et al. 2002). Currently, it is not well understood whether dynactin activity is regulated by protein phosphorylation, but it is an intriguing possibility that the phosphorylation state of DNC-3 is regulated by PPH-4.1. It will be of future interest to examine the interaction between these proteins in the process of homolog pairing and meiotic recombination.
While dynein/dynactin activity is essential for mitosis in metazoans including C. elegans and vertebrates, it is dispensable for mitosis in budding and fission yeast (McMillan & Tatchell 1994; Yamashita et al. 1997). In budding yeast, dynein/dynactin is required for nuclear positioning and short-range nuclear movement, but not for chromosome segregation (McMillan & Tatchell 1994). In fission yeast, the expression of dynein/dynactin is limited in meiosis (Yamashita et al. 1997). In addition to the difference in the requirement for mitosis, yeasts lack p62, p25 and p27 subunits of the dynactin complex. Although yeasts have been extensively used for genetic studies of dynactin, these differences in requirements and components imply that dynactin might have acquired new physiological roles and regulatory mechanisms in the course of evolution of metazoans. In this study, we revealed that all the dynactin components are conserved between C. elegans and mammals. Now that all the C. elegans dynactin subunits were defined, it will be more feasible to dissect the role of each dynactin component by combining biochemistry and genetics, for example, in the complex assembly and in the modulation of its activity. We believe that C. elegans will provide an excellent metazoan model system to study the in vivo functions and regulatory mechanisms of the dynein–dynactin complex.
Experimental procedures
Worm strains
The following strains were used: N2, wild type; SA403, unc-119(ed3);tjIs191[pie-1 promoter::gfp-TEV::dnc-3::FLAG unc-119(+)]; SA155, unc-119(ed3);tjIs11[pie-1 promoter::gfp::dnc-2 unc-119(+)]; WH257, unc-119(ed3);ojIs66[pie-1 promoter::gfp::dnc-1 unc-119(+)]; WH258, unc-119(ed3);ojIs57[pie-1 promoter::gfp::dnc-2 unc-119(+)]; WH259, unc-119(ed3);ojIs47[pie-1 promoter::gfp::arp-1 unc-119(+)]; SA250, unc-119(ed3);tjIs54[pie-1 promoter::gfp::tbb-2 pie-1 promoter::2xmCherry::tbg-1 unc-119(+);tjIs57[pie-1 promoter::mCherry::H2B unc-119(+)] (Toya et al. 2010).
RNAi
RNAi was carried out by the soaking method (Maeda et al. 2001). dsRNA was prepared from cDNA clones, as described (Sumiyoshi et al. 2002). The cDNA clones (gifts from Yuji Kohara) used were as follows: yk1299g08 (dnc-1), yk705c10 (dnc-2). The following primers were used to amplify W10G11.20/dnc-3 cDNA (T7 promoter sequences in bold). Forward, TAATACGACTCACTATAGGGATGGACATGATCGAGCTTGA, reverse, TAATACGACTCACTATAGGGTTTAGCTTTTGCGTTGATTTTC. Worms were soaked in 2 mg/mL dsRNA solutions and incubated at 20 or 24 °C for 24 h. The worms were then cultured at 20 or 24 °C and observed 24 h after recovery from the dsRNA soaking.
Construction of fluorescent strains
Construction of the strain SA250 expressing GFP::β-tubulin, mCherry::γ-tubulin and mCherry::Histone are described in (Toya et al. 2010). SA403 that expresses GFP-TEV::DNC-3::FLAG was constructed by high-pressure particle bombardment (Praitis et al. 2001) of the DP38 worms with the plasmid pMTN1G_W10G11.20. The plasmid was constructed as follows. The full length of W10G11.20 cDNA was cloned into pENTR/TEV/D-TOPO entry vector (Invitrogen, Carlsbad, CA, USA) using the following primers: forward, CACCATGGACATGATCGAGCTTGA and reverse, TTATTTATCGTCATCGTCTTTGTAGTCTTTAGCTTTTGCGTTGATTTTC. The forward primer has four additional bases right before the start codon for TOPO cloning (in bold). The reverse primer contains 24 bases coding FLAG-tag right before the STOP codon (in bold). The insert was recombined into the pMTN1G plasmid (Toya et al. 2010) by LR reaction.
Live imaging microscopy
For observations of pronuclear movement and bipolar spindle formation, adult hermaphrodites in Egg buffer were dissected on coverslips, which were inverted onto 2% agarose pads and the embryos were analyzed by time-lapse DIC microscopy. Images were taken on Axioplan 2 (Zeiss, Carl Zeiss, Jena, Germany) microscope equipped with a CoolSNAP HQ CCD camera (Photometrics, Tucson, AZ, USA) using a 63×, 1.2 NA C-Apochromat objective lens.
SA250 strain was observed as previously described (Toya et al. 2010). For the observation of GFP-tagged DNC proteins in embryos, they were mounted on an agarose pads as described earlier. For observation of GFP-tagged DNC proteins in adult gonads, worms were paralyzed with 1 mg/mL of tetramisole in M9 buffer and mounted on an agarose pad. Embryos or gonads were observed with at 24 °C with an UPlanApo 100× 1.40 NA oil immersion lens by using a CSU-X1 spinning-disk confocal system (Yokogawa Electric Corp., Tokyo, Japan) mounted on an IX71 inverted microscope (Olympus, Tokyo, Japan). The specimens were illuminated with a 488 or 561 nm diode-pumped solid-state laser (25–35 mW; CVI Melles Griot). Images were acquired with an Orca-R2 12-bit/16-bit cooled CCD camera (Hamamatsu Photonics, Shizuoka, Japan), and the acquisition system was controlled by MetaMorph software (Molecular Devices, Inc., Downingtown, PA, USA). Fluorescence images were acquired by 2 × 2 binning in the camera and were processed with MetaMorph software and Adobe Photoshop (Adobe Systems Inc., San Jose, CA, USA).
Immunoprecipitation and mass spectrometry
Young adults worms expressing GFP-TEV::DNC-3::FLAG were grown synchronously on EPP plates and bleached to obtain embryos. Embryos were washed in lysis buffer (50 mm Hepes at pH7.4, 1 mm EGTA, 1 mm MgCl2, 100 mm KCl, 10% glycerol and 0.05% NP40) and frozen in liquid N2. Approximately 3 millions of embryos were suspended in 1 mL of lysis buffer containing a protease inhibitor cocktail (Roche, Basel, Switzerland) supplemented with 1 mm PMSF and lysed by sonication. After centrifugation at 20 000 g for 10 min, 20 μg of anti-FLAG antibody preincubated with 50 μL of Protein G-dynabeads was mixed with the supernatant for 3 h at 4 °C. The immunoprecipitates were collected by magnets and washed with 1 ml of lysis buffer 3 times and eluted by 200 μm of FLAG peptides (Sigma Chemical Co., St Louis, MO, USA) for 90 min. All of the eluates were precipitated with acetone and lysed in SDS loading buffer, then electrophoresed on 5%–20% SDS–PAGE and visualized with silver staining. Bands specifically detected in the extract from the FLAG-tagged strain were cut out of the gel and analyzed by mass spectrometry.
Protein structure analysis
Sequences of W10G11.20/DNC-3, yeast homolog of p24/p22, Ldb18 and human p24/p22 were aligned by clustalW (Thompson et al. 1994) (http://clustalw.ddbj.nig.ac.jp). The coiled-coil structures of those proteins were predicted by COILS algorithm (Lupas et al. 1991) (http://www.ch.embnet.org/software/COILS_form.html).
Preparation of anti-W10G11.20/DNC-3 antibody
Anti-W10G11.20/DNC-3 rabbit serum was made against GST-fused N-terminal truncated W10G11.20/DNC-3 (49–171). The W10G11.20/DNC-3 fragment was amplified from a cDNA library and cloned into the entry vector pENTR201 (Invitrogen). The entry clone was recombined with pDEST15 (Invitrogen) by LR recombination. The obtained plasmid was used for protein expression in E. coli, and GST-W10G11.20/DNC-3 was purified using GST-sepharose (GE Healthcare, Pollards Wood, UK).
Immunofluorescence of adult gonads
For observation of endogenous W10G11.20/DNC-3 in adult gonads using anti-W10G11.20/DNC-3 antibody, young adult worms of the wild type (N2) were paralyzed on MAS-coated glass slides (MATSUNAMI, Osaka, Japan), and the head regions were cut off at the level of the pharynx using a razor. Gonad arms that were extruded from the worm body and attached to the glass slide were used for immunofluorescence observation. Gonads were fixed and stained as described (Sumiyoshi et al. 2002) by using a 1 : 200 dilution of mouse anti-KT23 to stain nuclear envelope (Takeda et al. 2008) or a 1 : 300 dilution of rabbit anti-W10G11.20/DNC-3. The secondary antibodies used were a 1 : 500 dilution of Alexa- 488-conjugated goat anti-mouse IgG and a 1 : 500 dilution of Alexa-568-conjugated goat anti-rabbit IgG (Invitrogen). Slides were counterstained with 2 mg/mL DAPI. For the image acquisition, the same setting with the live imaging was used.
Construction of plasmids for expression of GST-fused dynactin
The DNC-1, 2 and 3 coding DNA fragments were amplified from a cDNA library and cloned into pENTR201 (Invitrogen). N- or C-terminal truncated DNC-3 was made as follows. The truncation point was determined by changes of secondary structure predicted by Chou–Fasman analysis (Chou & Fasman 1978). The PCR-amplified DNC-3N (1–71) or DNC-3C (72–171) cDNA fragments were cloned into the pDONR201 entry vector via BP recombination. The entry clones were recombined into pDEST15 for GST or pDEST17 for 6xHis (Invitrogen) via LR reaction.
GST pull-down assay and immunoblotting
The GST-tagged proteins were expressed in 3 mL of E. coli culture and lysed by sonication in 1 mL of binding buffer (50 mm Tris–HCl (pH7.4), 300 mm NaCl, 0.01% Tween20, 1 mm dithiothreitol) containing a protease inhibitor cocktail (Roche) supplemented with 1 mm PMSF. The lysates were centrifuged at 20 000 g for 10 min, and the supernatant were incubated for 2 h at 4 °C with MagneGST Glutathione Particles (Promega, Madison, WI, USA). After washing with binding buffer four times, approximately 100 μg of GST-tagged proteins that bound to MagneGST Glutathione Particles were used in each assay. 6xHis-tagged proteins were also expressed in 3 mL of E. coli culture, and the lysates were prepared as described for GST-tagged proteins. One hundred microliters of the lysate was added to the GST-tagged proteins that immobilized on MagneGST Glutathione Particles, then incubated for 2 h at 4 °C. After washing with binding buffer four times, GST-tagged proteins and interactors were eluted by 100 mm reduced glutathione. The eluted proteins were separated by SDS–PAGE and detected by immunoblotting. Immunoblotting were carried out by SNAPi.d. (Millipore, Billerica, MA, USA). All primary antibodies, anti-DNC-1 (gift from Dr. Skop) or anti-His-tag (Abgent, San Diego, CA, USA) for detection of DNC-2, were diluted 1 : 1000, the HRP-conjugated goat anti-rabbit secondary antibody (GE Healthcare) was diluted 1 : 5000 and the signal was detected by using ECL plus chemiluminescence (GE Healthcare).
Acknowledgments
We are grateful to K. Nagata and Y. Iida for helpful supports for plasmid constructions and dsRNA synthesis; K. Shinmyozu (RIKEN CDB, Mass Spectrometry Analysis Unit) for mass spectrometric analysis; Drs J. White and A. Skop for generously providing α-DNC-1 antibody. We also thank Drs. J. Nakayama and T. Uruno for technical advice. This work was supported by MEXT KAKENHI 17017038 and JSPS KAKENHI 19671003 to A.S., and JSPS KAKENHI 21570209 to M. Toya. M. Toya is a RIKEN Special Postdoctoral Researcher. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Supporting Information/Supplementary material
The following Supporting Information can be found in the online version of the article:
Figure S1 Localization of W10G11.20 in adult hermaphrodite gonads.
Figure S2 DNC-3/W10G11.20 was co-immunoprecipitated with DNC-1, GFP::DNC-2 and GFP::ARP-1.
Additional Supporting Information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ai E, Poole DS, Skop AR. RACK-1 directs dynactin-dependent RAB-11 endosomal recycling during mitosis in Caenorhabditis elegans. Mol. Biol. Cell. 2009;20:1629–1638. doi: 10.1091/mbc.E08-09-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro IA, Costanzo M, Boone C, Huffaker TC. The Saccharomyces cerevisiae homolog of p24 is essential for maintaining the association of p150Glued with the dynactin complex. Genetics. 2008;178:703–709. doi: 10.1534/genetics.107.079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J. Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Schroer TA. Interactions between the evolutionarily conserved, actin-related protein, Arp11, actin, and Arp1. Mol. Biol. Cell. 2003;14:2645–2654. doi: 10.1091/mbc.E03-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran EA, Ligon LA, Tokito M, Stankewich MC, Morrow JS, Holzbaur EL. beta III spectrin binds to the Arp1 subunit of dynactin. J. Biol. Chem. 2001;276:36598–36605. doi: 10.1074/jbc.M104838200. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, Silver PA. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J. Biol. Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- Karki S, LaMonte B, Holzbaur EL. Characterization of the p22 subunit of dynactin reveals the localization of cytoplasmic dynein and dynactin to the midbody of dividing cells. J. Cell Biol. 1998;142:1023–1034. doi: 10.1083/jcb.142.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- Le Bot N, Tsai MC, Andrews RK, Ahringer J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 2003;13:1499–1505. doi: 10.1016/s0960-9822(03)00577-3. [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Maier KC, Godfrey JE, Echeverri CJ, Cheong FK, Schroer TA. Dynamitin mutagenesis reveals protein–protein interactions important for dynactin structure. Traffic. 2008;9:481–491. doi: 10.1111/j.1600-0854.2008.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J. Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Nuclear positioning: the means is at the ends. Curr. Opin. Cell Biol. 2003;15:54–59. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Niccoli T, Yamashita A, Nurse P, Yamamoto M. The p150-Glued Ssm4p regulates microtubular dynamics and nuclear movement in fission yeast. J. Cell Sci. 2004;117:5543–5556. doi: 10.1242/jcs.01475. [DOI] [PubMed] [Google Scholar]
- Payne C, Rawe V, Ramalho-Santos J, Simerly C, Schatten G. Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. J. Cell Sci. 2003;116:4727–4738. doi: 10.1242/jcs.00784. [DOI] [PubMed] [Google Scholar]
- Pfister KK, Benashski SE, Dillman JF, III, Patel-King RS, King SM. Identification and molecular characterization of the p24 dynactin light chain. Cell Motil. Cytoskeleton. 1998;41:154–167. doi: 10.1002/(SICI)1097-0169(1998)41:2<154::AID-CM6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Schroer TA. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 2002;159:245–254. doi: 10.1083/jcb.200203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi E, Sugimoto A, Yamamoto M. Protein phosphatase 4 is required for centrosome maturation in mitosis and sperm meiosis in C. elegans. J. Cell Sci. 2002;115:1403–1410. doi: 10.1242/jcs.115.7.1403. [DOI] [PubMed] [Google Scholar]
- Takeda K, Watanabe C, Qadota H, Hanazawa M, Sugimoto A. Efficient production of monoclonal antibodies recognizing specific structures in Caenorhabditis elegans embryos using an antigen subtraction method. Genes Cells. 2008;13:653–665. doi: 10.1111/j.1365-2443.2008.01195.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M, Iida Y, Sugimoto A. Imaging of mitotic spidnle dynamics in Caenorhabditis elegans embryos. Methods Cell Biol. 2010;97:359–372. doi: 10.1016/S0091-679X(10)97019-2. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle JA, Cooper JA, Waterston RH. The alpha and beta subunits of nematode actin capping protein function in yeast. Mol. Biol. Cell. 1993;4:907–917. doi: 10.1091/mbc.4.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc. Natl Acad. Sci. USA. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Watanabe Y, Yamamoto M. Microtubule-associated coiled-coil protein Ssm4 is involved in the meiotic development in fission yeast. Genes Cells. 1997;2:155–166. doi: 10.1046/j.1365-2443.1997.1100307.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Skop AR, White JG. Src and Wnt signaling regulate dynactin accumulation to the P2-EMS cell border in C. elegans embryos. J. Cell Sci. 2008;121:155–161. doi: 10.1242/jcs.015966. [DOI] [PubMed] [Google Scholar]
- Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 2009;186:229–241. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


