Abstract
Single base substitutions can lead to missense mutations, silent mutations or intronic mutations, whose significance is uncertain. Aberrant splicing can occur due to mutations that disrupt or create canonical splice sites or splicing regulatory sequences. The assessment of their pathogenic role may be difficult, and is further complicated by the phenomenon of alternative splicing. We describe an HNPCC patient, with early-onset colorectal cancer and a strong family history of colorectal and breast tumors, who harbours a germ line MLH1 intronic variant (IVS9 c.790 +4A>T). The proband, together with 2 relatives affected by colorectal-cancer and 1 by breast cancer, have been investigated for microsatellite instability, immunohistochemical MMR protein staining, direct sequencing and Multiplex Ligation-dependent Probe Amplification. The effect of the intronic variant was analyzed both by splicing prediction software and by hybrid minigene splicing assay. In this family, we found a novel MLH1 germline intronic variant (IVS9 c.790 +4A>T) in intron 9, consisting of an A to T transversion, in position +4 of the splice donor site of MLH1. The mutation is associated with the lack of expression of the MLH1 protein and MSI in tumour tissues. Furthermore, our results suggest that this substitution leads to a complete skip of both exon 9 and 10 of the mutant allele. Our findings suggest that this intronic variant plays a pathogenic role.
Keywords: Intronic mutation, MLH1, Minigene assay, Colon cancer, Breast cancer
Introduction
Hereditary Non-Polyposis Colorectal Cancer (HNPCC), or Lynch syndrome, is an autosomal dominant inherited disorder that accounts for approximately 3–5% of all colorectal cancers [1]. Lynch syndrome is associated with germline mutations in MMR genes, in particular in MLH1 and MSH2, that have been found in up to 90% of HNPCC patients, and less frequently in MSH6, PMS2 and MSH3 [2–4]. MMR deficiency leads to accumulation of replication errors, typically at short repetitive DNA sequences in the tumour cells, giving rise to the molecular hallmark of HNPCC: the microsatellite instability (MSI).
Individuals carrying germ line mutations of MMR genes have a high lifetime risk of developing colorectal cancer as well as an increased risk of tumours in extracolonic sites such as endometrium, renal pelvis, ureter, stomach, small intestine, bile ducts, ovary and possibly breast [5, 6]. Immunoistochemistry of MLH1 and MSH2 in tumour tissue can be used as a complementary screening method, providing information about the presence or absence of the protein tested and, thus the specific gene defect [7].
As mutations in MLH1 and MSH2 are spread throughout the genes, without “hot spots”, the detection of a predisposing mutation requires entire genes to be screened. To date approximately 450 different mutations are listed in the database of the International Society for Gastrointestinal Hereditary Tumours (InSiGHT/LOVD database) (http://www.insight-group.org/mutations/) [3].
Many of these mutations (~50%) lead to truncated proteins (nonsense, frameshift, splice site mutations or large genomic deletions), and are considered pathogenic. However in Lynch syndrome families a substantial proportion of MLH1 and MSH2 mutations are single nucleotide substitutions, either within the coding sequence (missense, or silent mutations) or in intronic regions.
Whether these mutations affect the normal function of mismatch DNA repair proteins, and thus have a pathogenic role, is an essential piece of information in genetic counselling.
Traditionally, a missense mutation is considered more likely to be pathogenic when a highly conserved amino acid is involved, when its polarity is changed or when it is not present in a large number of controls. More recent studies have shown that some previously unclassified variants may interfere with normal splicing process. Aberrant splicing can occur due to mutations that disrupt the splice sites themselves or ESE or ESS motifs [8, 9, 12]. DNA mutations can affect RNA processing by destroying or creating target sequences for splicing regulatory proteins, thereby changing the complex of factors bound to the pre-mRNA. This can initially be assessed by bioinformatics methods that predict the creation or the destruction of protein binding sites.
Here we describe the case of an HNPCC patient with a strong family history, with a germ line MLH1 intronic variant: intron 9 (IVS9 c.790 +4A>T).
Patients and methods
Case report
The proband was a 24 year-old male (IV-1), diagnosed with mucinous adenocarcinoma of the right colon when he was 24, and underwent right hemicolectomy (pT3N1M0, G3).
Detailed four generation family history was collected. Overall, eight family members developed one or more colorectal cancers and, interestingly, two had breast cancer.
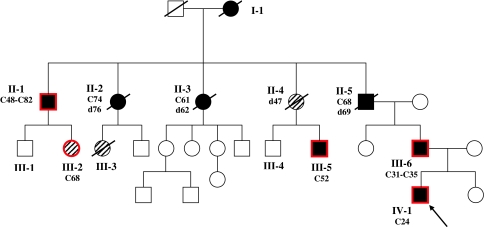
The proband’s family showed a typical HNPCC (Lynch syndrome) pedigree Fig. 1.
Fig. 1.
I-1: died at age 42 of colon cancer. II-1: 2 metachronous colon cancers at age 48 and 82. II-2: died at age 76 of colon cancer (onset age 74). II-3: died at age 62 of colon cancer (onset age 61). II-4: died at age 47 of breast cancer. II-5: died at age 69 of colon cancer (onset age 68). III-2: breast cancer (onset age 48). III-3: leukaemia. III-5: colon cancer (onset age 52). III-6: 2 metachronous colon cancers at age 31 and age 35. IV-1: colon cancer (onset age 24). Arrow indicates the proband. Red borders indicate carrier status
Tumour specimens of the proband and three relatives (II-1, III-2, III-6) were available at the “Dipartimento di Anatomia Patologica and were all examined by the same pathologist (I.B.). The purpose of the study was explained, and informed consent was obtained from participating subjects. Ethics committee approval was obtained.
Microsatellite instability analysis
MSI was performed for the proband (IV-1) and relatives II-1, III-2, III-6.
Genomic DNA was isolated from tumour samples and corresponding normal tissues as previously described [14]. Tumours were examined for MSI using the 5-marker panel recommended by the National Cancer Institute Workshop on MSI for Cancer Detection and Familial Predisposition [16]. Oligonucleotide primers were fluorescently labelled (5′-VIC, 5′-NED, 5′-6-FAM, 5′-6-FAM, 5′-PET), MLH1 IVS9 c.790 +4A>T and PCR products were evaluated with an ABI Prism 310 Genetic Analyzer based on automated capillary electrophoresis and automated sizing of the alleles by GeneScan 3.7 (Applied Biosystems, Foster City, CA). CAT25 microsatellite was also studied in all patients [17].
Expression of MLH1, MSH2, MSH6 and PMS2 proteins in tumour tissues
Tumour tissue was available for proband and patients II-1, III-2, III-6.
Expression of the MLH1 and MSH2 proteins was studied by immunohistochemistry (IHC) on 2-μm sections of Formalin-Fixed, Paraffin-Embedded (FFPE) tissues, following antigen retrieval as previously described [18]. Primary antibodies used for immunohistochemistry were: anti-MLH1: clone G168-728, PharMingen, San Diego, CA, 1:50 dilution; anti-MSH2: clone FE11, Oncogene Research Products, Cambridge, MA, 1:100 dilution; anti-MSH6: clone H-141: sc-10798; Santa Cruz Biotechnology, Inc, Santa Cruz, CA 1:250 dilution.
Mutation analysis of the MLH1, MSH2 and MSH6 genes
Mutation analysis was performed in patients IV-1 (proband), II-1, III-2, III-5 and III-6 and in two healthy relatives: III-1 and III-4.
Mutation analysis was performed on genomic DNA, isolated from peripheral blood lymphocytes, according to standard procedures. The 19 MLH1 exons, 16 MSH2 exons and 10 MSH6 exons, including flanking intronic regions, were individually amplified and directly sequenced as previously described [15]. Primer sequences are available from the corresponding Author upon request [14].
Multiplex ligation-dependent probe amplification analysis
The patient was also studied by Multiplex Ligation-dependent Probe Amplification (MLPA) analysis.
MLPA was performed with 200 ng of normal and tumour DNAs using the MRC-Holland (Amsterdam, Holland) MLH1/MSH2 and MSH6 SALSA MLPA Kits, according to the supplier’s protocol and as previously described [15].
Splice site predictions
We used NNSPLICE 0.9 [19], NetGene2 [20], SpliceView [21] and ASSP [22] tools for the splice sites predictions. Among the tools to predict the ESE, ISS, ESS and ISE motifs we use SpliceAid [23], ESEfinder [9], RESCUE-ESE [24] and SplicingRainbow [25] tools. Many of these tools form the Alamut 1.5 tool (www.interactive-biosoftware.com).
MLH1 transcript analysis
Total RNA was purified from peripheral-blood lymphocytes by the RNeasy Mini Kit and the QIAamp DNA Blood Mini Kit (both from Qiagen, Hilden, Germany) according to the instructions of the manufacturer. cDNA was synthesized from 2 to 3 μg by random hexamer-primed system for RT-PCR (Invitrogen), according to the manufacture’s protocol. The EX7-EX11 fragment was obtained according to standard PCR protocols. Sequences of PCR primers are available from the corresponding Author, primer forward (7F) 5′-GTATTCAGTACACAATGCAGG-3′ and primer reverse (11R) 5′-GCACATTCTGGGGACTGATTTC-3′ [26]. The PCR cycling conditions included the following cycles:1 cycle at 95°C for 5 min; 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; 1 extension cycle at 72°C for 7 min. Aliquots of the RT-PCR products were separated on a 3% agarose gel and visualized with ethidium bromide on an UV imaging system. Bands, corresponding to shorter amplified fragments, were excised from the gels and extracted by QIAquick Gel Extraction Kit (Qiagen, hilden, Germany). The purified cDNA fragments were sequenced directly, using Applied Biosystem ABI PRISM 310 automated sequencer and the Big Dye terminator cycle sequencing Ready Reaction-Kit (Applied Biosystems), as described. The analysis was also conducted in the cDNA of normal controls.
Loss of heterozygosity analysis of tumor tissue
LOH analysis was performed in the colorectal cancer tissue samples from the proband RNA (cDNA).
Total tumor RNA was extracted from FFPE splices with, in which 70% or more of tumor tissue, using Trizol (Invitrogen). After homogenization, proteinase K was added and samples incubated at 55°C for 30 min. Total RNA was extracted according to the manufacturer’s instructions. Samples were DNAse treated using Turbo DNAfree (Ambion) for 30 min at 37°C. Reverse transcription was performed from 2.6 μg of total RNA.
Tumor cDNA was amplified using the same primers used for the MLH1 transcript analysis. The forward primer was labelled with the NED Fluorochrome at the 5′. The PCR conditions are the same as in: “MLH1 transcript analysis”. The PCR products were subjected to fragment analysis on ABI Prism 310 Genetic Analyzer based on automated capillary electrophoresis and automated sizing of the alleles by GeneScan 3.7 (Applied Biosystems). The analysis was also carried out on a cDNA normal control. The electropherograms were compared.
Hybrid minigene splicing assay
To generate the pCalfa Minigene construct, the alfa globin gene from exon 1 to exon 3 was cloned in pCDNA3.1(+) vector (Invitrogen) in two steps in order to retain the vector single restriction sites EcoR1, BamH1 and Xho1 for the following cloning. Human genomic DNA was amplified to generate a fragment containing MLH1 exon 10 with inronic flanking sequences and cloned in pCalfa between BamH1 and Xho1 restriction sites. Patient genomic DNA was amplified to generate fragments that contain MLH1 exon 9 wild-type (wt) and mutant with intronic flanking sequences and cloned in pCalfa between the EcoR1 and BamH1 restriction sites. The hybrid minigenes pCalfaMwt and pCalfaM+4t were then analyzed for splicing. One microgram of each minigene was transfected into 3 × 105 HeLa cells with 5 μl Lipofectamine transfection Reagent (Invitrogen). Following RNA extraction (RNeasy plus kit, Qiagen) and RT-PCR (Reverse Transcription System, Promega) the PCR analysis was performed using specific primers for the pCalfa minigene. The size of the PCR amplified cDNA bands obtained when analysed by agarose gel electrophoresis will be the sum of each exon included in the transcript which confirms alternative splicing events.
Results
Microsatellite instability analysis
The proband (IV-I) showed high microsatellite instability (MSI-H): three out of the five loci studied (Bethesda panel) resulted altered. Similar results were observed in three affected relatives: patient II-1 and patient III-2 exhibited microsatellite instability in two out of the five markers (MSI-H), patient III-6 showed microsatellite instability in four out of the five markers (MSI-H). The CAT25 microsatellite analysis showed instability in the proband and in relatives II-1, III-2, and III-6 (Table 1).
Table 1.
Results’summary (MSI: Microsatellite instability; IHC: Immunohistochemistry; wt: wild type)
| Subjects | Tumor type | MSI: altered loci (Bethesda panel) | Locus CAT25 | IHC | MLH1 mutation | ||
|---|---|---|---|---|---|---|---|
| MLH1 | MSH2 | MSH6 | |||||
| II-1 | Colon | BAT25, BAT26 | Unstable | − | + | + | Intron 9-IVS9 c.790 +4A>T |
| III-1 | Healthy subject | Not done (Healthy subject) | Not done (Healthy subject) | Not done (Healthy subject) | wt | ||
| III-2 | Breast | BAT25, BAT26 | Unstable | − | + | + | Intron 9-IVS9 c.790 +4A>T |
| III-4 | Healthy subject | Not done (Healthy subject) | Not done (Healthy subject) | Not done (Healthy subject) | wt | ||
| III-5 | Colon | Tissue not available | Tissue not available | Tissue not available | Intron 9-IVS9 c.790 +4A>T | ||
| III-6 | Colon | BAT25, BAT26, D2S123, D5S346 | Unstable | − | + | + | Intron 9-IVS9 c.790 +4A>T |
| IV-1 | Colon | BAT25, BAT26, D2S123 | Uns | − | + | + | Intron 9-IVS9 c.790 +4A>T |
The subjects have been described in the pedigree Fig. 1
Expression of MLH1, MSH2 and MSH6 proteins in tumour tissues
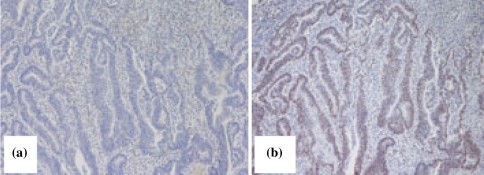
Study of protein expression was conducted in tumour specimens of patients II-1, III-2, III-6 and IV-1 (proband). In all cases, we observed lack of expression of MLH1 and normal staining of MSH2 and MSH6 (Table 1; Fig. 2).
Fig. 2.
Immunohistochemical staining for MLH1 (a), and MSH2 (b), proteins showing loss of MLH1 expression in the proband’s colon cancer
Mutation analysis of the MLH1, MSH2 and MSH6 genes
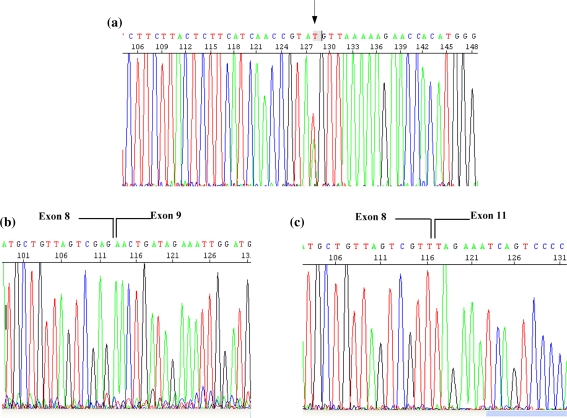
The sequence analysis of the entire coding regions of MSH2 and MSH6 did not show any mutation. An intronic variant was found in MLH1, IVS9 c.790 +4A>T, an A to T transversion, affecting position +4 of the splice donor site of MLH1/exon 9 (Fig. 3a). This variant was present in the proband and his relatives II-1, III-2, III-5, III-6 (Fig. 1).
Fig. 3.
Sequence analysis of the MLH1 IVS9 c.790 +4A>T. The figure a shows the electropherogram of genomic DNA mutation. The arrow indicate the intronic variation. The figures b and c shows the electropherogram of wt cDNA (b), and the electropherogram of cDNA fragment, lacking both exon 9 and 10 (c)
This mutation is not reported in InSiGHT/LOVD mutation database and was not found in either 100 healthy individuals, nor in 175 colorectal cancer patients.
The substitution has been searched for in two healthy relatives: (III-1 and III-4) and was not detected (Table 1).
MLPA analysis
The MLPA analysis for MLH1 MSH2 and MSH6, did not show any large genomic rearrangements.
Splice site prediction
The mutation strongly weakens the 5′ splice site according to NNSPLICE, NetGene2 and SpliceView tools whereas ASSP does not detect either real or mutated site. Further predictions show that there are not strong cryptic 5′ splice sites in a window of 200 bp straddling the mutation so it seems more probable the occurrence of an exon skipping rather than an alternative 5′ splice site event. SpliceAid database shows that the mutation creates a binding sequence for ETR-3 splicing factor over the 5′ splice site [26, 27]. This prediction advises the competition between ETR-3 and snRNP U1 factors for recognition of the 5′ splice site. ESEfinder, RESCUE-ESE and Splicing Rainbow tools did not detect any alterations.
MLH1 transcript analysis
In the proband and in his relatives (with the mutation) we RT-PCR amplified from peripheral lymphocyte cDNA, a 352 bp fragment of the MLH1 open reading frame (ORF), including exons 7–11. In addition to the expected band corresponding to the amplified product, we found two lower molecular weight fragments: the 239-bp fragment, lacking exon 9, and the 145 bp fragment, lacking boths exon 9 and 10 (Fig. 3b, c). The band corresponding to fragment lacking both exons 9 and 10 showed the same intensity of the wt band, while the band corresponding to fragment lacking exon 9, exhibited a weaker intensity.
We conducted the same analysis on cDNA from healthy volunteers. In addition to the product amplified from full-length cDNA, there were two other products, resulting from the skipping of exon 9 and both exons 9 and 10, as in patients’ cDNA.
However we found a significant difference, in the proportion of the three splicing isoforms. The patient’s cDNA, showed two bands, wt and the skipping exons 9 +10, with the same intensity and a third band, corresponding to the skipping of exon 9, with lower intensity. The control cDNA showed three bands of different intensity: very high for wt and significantly lower for the fragments skipping the exons 9 +10 and exon 9 alone.
Loss of heterozygosity analysis of tumor tissue
To clarify whether the patient’s tumor samples displayed allelic loss, cDNA was obtained from tumor and amplified along with cDNA from the patient’s normal tissue and from a normal control. The tumor sample showed only the mutated product (145 bp) with complete loss of the wt product. Amplification and Genescan analysis of cDNA from patient’s normal tissue showed two alleles of equal heights (352 and 145 bp).
The cDNA of the healthy control showed a fragment corresponding to wt transcript (352 bp) and two fragments of less intensity: a fragment showing the same molecular weight of mutated fragment of the tumor DNA (145 bp) and a fragment showing a molecular weight corresponding to deletion of MLH1 exon 9 (239 bp).
Hybrid minigene splicing assay
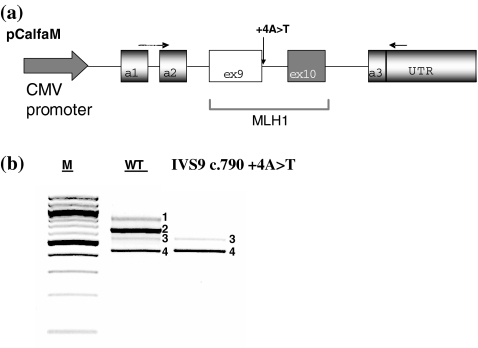
Transfection of the construct carrying the wild-type MLH1 intron 9 minigene (Fig. 4a), pCalfaM, in HeLa cells, followed by RT-PCR amplification generated a predominant transcript of 646 bp containing exon 9 and exon 10 (Fig. 4b). The 439 and 552 bp bands consisted of exon 9 and 10 or exon 9 alone skipping analogous to that already observed in tissues from normal individuals [11]. An additional transcript of 799 bp was observed, representing a splicing event where a cryptic 5′ss, 101 nucleotides upstream of the MLH1 exon 9 acceptor site is used instead of the alfa globin exon 2 donor site. The activation of the cryptic 5′ss is not represented in nature.
Fig. 4.
Minigene Functional Splicing Assay. a The hybrid minigene (pCalfaM) used in transient transfection splicing assays in HeLa cells is shown. Exons are indicated as boxes. Introns are indicated as lines. MLH1 Exon 9 and 10 (white and gray box) were tested for splicing efficiency by using specific primers (arrows). b RT-PCR products from transfection experiments. RNA splicing variants corresponding to wt (2) exon 9 exclusion (3) and both exon 9 and 10 exclusion (4) are shown. (1) indicate a splice variant generated as an artefact of the minigene. M is the 100 bp marker
Transfection of the mutant minigene pCalfaM_IVS9 +4A>T in HeLa cells showed aberrant splicing with a minority of the transcript of 552 bp lacking exon 9 and the majority lacking both exon 9 and exon 10 (Fig. 4b).
Discussion
Numerous mutations leading to truncated proteins have been found in the mismatch repair genes MLH1 and MSH2 and are involved in Lynch syndrome.
Some patients carry DNA changes considered genetic variants of unknown clinical significance (Unclassified Variants (UVs) with uncertainty about their cancer risks. This is often the case for synonymous, missense and intronic variants or when the nucleotide change affects or creates a (putative) splice-site.
It is not easy to clarify whether an UV contributes to the disease phenotype or merely represents a rare polymorphism. Several criteria are used to help in classifying these variants as pathogenic, if they: (1) segregate with disease; (2) are absent in controls; (3) result in a change of aminoacid polarity or size; (4) cause the substitution of an amino acid in a domain that is evolutionary highly conserved and/or is shared between proteins belonging to the same protein family; (5) exhibit altered function in an in vitro assay.
The correct classification of an UV is of obvious importance when assessing the cancer risk for the proband and his family.
We report a MLH1 intronic mutation, in intron 9 donor site (IVS9 c.790 +4A>T) affecting the inclusion of both exon 9 and exon 10.
The family studied fulfilled Amsterdam I criteria; all affected family members, in which we could perform the genetic test, carried the same MLH1 intronic mutation (IVS9 c.790 +4A>T). Moreover they all showed MSI-H and the lack of expression of MLH1 protein in tumour specimens. The mutation was not found in two unaffected relatives of the proband and in 100 healthy individuals.
In this family there were two cases of breast cancer (II-4, III-2). We could not examine a tumor specimen of the II-4 patient (she died approximately 30 years ago), but she was an obligate mutation carrier. Both cases of breast cancer were diagnosed at an age compatible with what is reported in literature for HNPCC associated breast cancers [28–31]. The inclusion of breast cancer in HNPCC is controversial but there are recent studies supporting this hypothesis. Although breast cancer is a common tumor and there may be phenocopies, approximately half of the cases in HNPCC families [30, 31] show lack of staining for the protein encoded by the mutated gene and the tumor samples show microsatellite instability (similar to our patient III-2). It is of note that HNPCC associated breast cancers seem to be more common in MLH1 mutated families [28].
The bioinformatic predictions show weakening of the 5′ splice site. Generally, the weakening of a strong 5′ splice site makes the presence of additional alternative splicing forms lacking an exon or with alternative 5′ splice site more probable. The lack of cryptic 5′ splice sites near the mutation supports the former event that is exon skipping. Besides the weakening of the 5′ss, the SpliceAid tool prediction, showed the competition between ETR-3 and snRNP U1 factors for recognition of the 5′ss, enhances the hypothesis that the mutation gives rise to exon 9 skipping event in all alternative transcripts. Since the exon 9 length is not a multiple of 3, its skipping event would give rise to a frameshift resulting in premature stop codons. The intronic mutation can therefore dramatically alter MLH1 exon 9 inclusion levels.
On the basis of the in silico predictions, we experimentally investigated the presence of aberrant splicing due to the IVS9c.790 +4A>T mutation. The RNA analysis, from blood lymphocytes of all patients considered, showed, in addition to the wt band, one shorter fragment lacking exons 9 + 10 of and an additional band lacking exon 9.
Although several Authors have previously described deletion of exon 9 +10 and deletion of exon 9, as normal alternative MLH1 transcripts [10, 13], their relative intensity appeared significantly lower than that of normal-sized fragments (wt) [11].
The band corresponding to the deletion of exon 9 +10, from healthy subjects (100 controls) was of much lower intensity than the band in our patient. Whilst in our proband and his affected relatives the intensity of the bands relative to cDNA products was equal between the wt gene and the mutated gene A study of the functional role of the deletion of exons 9 +10, predicted that it does not contribute to MMR function in vivo since it failed to restore MMR in 293T [32].
Pagenstecher et al. published an intronic variant similar to ours: c.790 +4A>G, which showed high correlation with phenotype, but they did not complete functional studies and concluded that its pathogenicity was uncertain [33].
In our proband the loss of heterozygosity analysis showed complete loss of wt allele in cDNA from tumor tissue and two alleles of equal molecular weight, corresponding to the wt allele and to the mutant allele.
We also studied the effect of the IVS9c.790 +4A>T mutation by a functional splicing assay. Transient transfection of the minigene carrying the patient mutation in HeLa cells showed aberrant splicing with approximately 10% exon 9 skipping and 90% of both exons 9 and 10 skipping. Although the results resemble the patients aberrant splicing in tumor affected tissue, transfection of the minigene in other cell lines showed different proportions of the splicing variants (data not shown) suggesting that there is a strong influence of the splicing factors context on the splicing outcome. We also detected a band in the wt minigene corresponding to the partial retention of the minigene intron between alfa globin exon 2 and MLH1 exon 3. The intron retention resulting from the activation of a cryptic donor site, could be because of the altered genomic context in the minigene system, as the cryptic donor site is never used by the MLH1 endogenous gene. The fact that we obtained similar results even in different minigene contexts (data not shown) strongly suggests a predisposition to disease caused by deep intronic variations in MLH1 intron 8, since the creation of acceptor splice sites upstream of the cryptic donor site may result in pseudoexon inclusion as observed for the minigene.
In conclusion, we studied one patient and three relatives from a family with the hallmarks of HNPCC. All the patients studied (three with colon cancer and one with breast cancer) exhibited the same mutation and the laboratory features are, in our opinion, compatible with the pathogenicity of the c.790 +4A>T mutation.
The reclassification of this variant as deleterious has significant implications in the management of this family.
Acknowledgments
The Authors acknowledge Prof. Giovanni Principato., M.D. for helpful discussion and support.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- HNPCC
Hereditary Non-Polyposis Colorectal Cancer
- MMR
MisMatch Repair
- MSI
MicroSatellite Instability
- IHC
Immunohistochemistry
- InSiGHT
International Society for Gastrointestinal Hereditary Tumours
- ESE
Exonic splice enhancer
- ESS
Exonic splice silencer
- ISS
Intronic splicing silencer
- FFPE
Formalin-fixed, paraffin-embedded
- MLPA
Multiplex ligation-dependent probe amplification
- ISE
Intronic splice enhancer
- Wt
Wild type
References
- 1.Piñol V, Castells A, Andreu M, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association: accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 2.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 3.Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition–update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 5.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 6.Merg A, Lynch HT, Lynch JF, et al. Hereditary colorectal cancer-part II. Curr Probl Surg. 2005;42:267–333. doi: 10.1067/j.cpsurg.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Strate LL, Syngal S. Hereditary colorectal cancer syndromes. Cancer Causes Control. 2005;16:201–213. doi: 10.1007/s10552-004-3488-4. [DOI] [PubMed] [Google Scholar]
- 8.Arnold S, Buchanan DD, Barker M, et al. Classifying MLH1 and MSH2 Variants using Bioinformatic prediction, Splicing Assay, Segregation, and Tumor Characteristics. Hum Mutat. 2009;30:757–770. doi: 10.1002/humu.20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartegni L, Wang J, Zhu Z, et al. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charbonnier F, Martin C, Scotte M, et al. Alternative splicing of MLH1 messenger RNA in human normal cells. Cancer Res. 1995;55:1839–1841. [PubMed] [Google Scholar]
- 11.Genuardi M, Viel A, Bonora D, et al. Characterization of MLH1 and MSH2 alternative splicing and its relevance to molecular testing of colorectal cancer susceptibility. Hum Genet. 1998;102:15–20. doi: 10.1007/s004390050648. [DOI] [PubMed] [Google Scholar]
- 12.Xia L, Shen W, Ritacca F, et al. A truncated hMSH2 transcript occurs as a common variant in the population: implications for genetic diagnosis. Cancer Res. 1996;56:2289–2292. [PubMed] [Google Scholar]
- 13.Kohonen-Corish M, Ross VL, Doe WF, et al. RNA-based mutation screening in hereditary nonpolyposis colorectal cancer. Am J Hum Genet. 1996;59:818–824. [PMC free article] [PubMed] [Google Scholar]
- 14.Scartozzi M, Bianchi F, Rosati S, et al. Mutations of hMLH1 and hMSH2 in patients with suspected hereditary nonpolyposis colorectal cancer: correlation with microsatellite instability and abnormalities of mismatch repair protein expression. J Clin Oncol. 2002;20:1203–1208. doi: 10.1200/JCO.20.5.1203. [DOI] [PubMed] [Google Scholar]
- 15.Bianchi F, Galizia E, Porfiri E, et al. A missense germline mutation in exon 7 of the MSH2 gene in a HNPCC family from Center-Italy. Fam Cancer. 2007;6:97–102. doi: 10.1007/s10689-006-9110-z. [DOI] [PubMed] [Google Scholar]
- 16.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 17.Bianchi F, Galizia E, Catalani R, et al. CAT25 is a mononucleotide marker to identify HNPCC patients. J Mol Diagn. 2009;11:248–252. doi: 10.2353/jmoldx.2009.080155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcus VA, Madlensky L, Gryfe R, et al. Immunohistochemistry for hMLH1 and hMSH2: a practical test for DNA mismatch repair-deficient tumors. Am J Surg Pathol. 1999;23:1248–1255. doi: 10.1097/00000478-199910000-00010. [DOI] [PubMed] [Google Scholar]
- 19.Reese MG, Eeckman FH, Kulp D, et al. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 20.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-O. [DOI] [PubMed] [Google Scholar]
- 21.Rogozin IB, Milanesi L. Analysis of donor splice sites in different eukaryotic organisms. J Mol Evol. 1997;45:50–59. doi: 10.1007/PL00006200. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Marín A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 23.Piva F, Giulietti M, Nocchi L, et al. SpliceAid: a database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics. 2009;25:1211–1213. doi: 10.1093/bioinformatics/btp124. [DOI] [PubMed] [Google Scholar]
- 24.Fairbrother WG, Yeh RF, Sharp PA, et al. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 25.Stamm S, Riethoven JJ, Le Texier V, et al. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charlet BN, Logan P, Singh G, et al. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9:649–658. doi: 10.1016/S1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 27.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrow E, Robinson L, Alduai JW, et al. Cumulative incidence of extracolonic cancers in Lynch syndrome a report of 121 families with proven mutations. Clin Genet. 2009;75:141–149. doi: 10.1111/j.1399-0004.2008.01125.x. [DOI] [PubMed] [Google Scholar]
- 29.Shanley S, Fung C, Milliken J, et al. Breast cancer immunohistochemistry can be useful in triage of some HNPCC families. Fam Cancer. 2009;8:251–255. doi: 10.1007/s10689-008-9226-4. [DOI] [PubMed] [Google Scholar]
- 30.Jensen UB, Sunde L, Timshel S, et al. Mismatch repair defective breast cancer in the hereditary nonpolyposis colorectal cancer syndrome. Breast Cancer Res Treat. 2010;120:777–782. doi: 10.1007/s10549-009-0449-3. [DOI] [PubMed] [Google Scholar]
- 31.Walsh MD, Buchanan DD, Cummings MC, et al. Lynch syndrome-associated breast cancers: clinicopathologic characteristics of a case series from the colon cancer family registry. Clin Cancer Res. 2010;16:2214–2224. doi: 10.1158/1078-0432.CCR-09-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trojan J, Zeuzem S, Randolph A, et al. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122:211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- 33.Pagenstecher C, Wehner M, Friedl W, et al. Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet. 2006;119:9–22. doi: 10.1007/s00439-005-0107-8. [DOI] [PubMed] [Google Scholar]