Abstract
Tandem repeats (microsatellites or SSRs) are molecular markers with great potential for plant genetic studies. Modern strategies include the transfer of these markers among widely studied and orphan species. In silico analyses allow for studying distribution patterns of microsatellites and predicting which motifs would be more amenable to interspecies transfer. Transcribed sequences (Unigene) from ten species of three plant families were surveyed for the occurrence of micro and minisatellites. Transcripts from different species displayed different rates of tandem repeat occurrence, ranging from 1.47% to 11.28%. Both similar and different patterns were found within and among plant families. The results also indicate a lack of association between genome size and tandem repeat fractions in expressed regions. The conservation of motifs among species and its implication on genome evolution and dynamics are discussed.
Keywords: SSR, EST, comparative genomics, molecular markers
Introduction
Microsatellites or SSRs (Simple sequence repeats) are DNA sequences formed by the tandem arrangement of nucleotides through the combination of one to six base pairs, being widely distributed in prokaryote and eukaryote genomes (Morgante and Olivieri, 1993; Tóth et al., 2000). Microsatellite regions tend to form loops or hairpin structures, leading to the slippage of DNA polymerase during replication, thereby provoking the insertion or deletion of nucleotides (Iyer et al., 2000). The expansion and/or contraction of microsatellites may lead to a gain or loss of gene function (Li et al., 2002, 2004a). Initially, it was suggested that the occurrence and distribution of microsatellites could be the result of random processes. However, new evidence indicates that the genomic distribution of these repeats had its origin in non-random processes (Bell, 1996; Li et al., 2004b). Microsatellites have been reported to correspond to 0.85% of Arabidopsis (Arabidopsis thaliana), 0.37% of maize (Zea mays subsp. mays), 3.21% of fugu fish (Fugu rubripes), 0.21% of the nematode Caenorhabditis elegans and 0.30% of yeast (Saccharomyces cerevisae) genomes (Morgante et al., 2002). Moreover, they constitute 3.00% of the human genome (Subramanian et al., 2003).
For microsatellites located in genic regions, 5'UTRs are hotspots for the presence of this type of repeats. It is known that the contraction and/or expansion of repeats found in 5'UTR regions alter the transcription and/or translation of these genes (Li et al., 2004b; Zhang et al., 2006a). Mutations in microsatellite loci found in 3'UTR regions are associated with gene silencing, transcript-cytosol exporting and splicing mechanism changes as well as the expression levels of flanking genes (Davis et al., 1997; Thornton et al., 1997; Philips et al., 1998; Conne et al., 2000). For coding sequences (CDS), the impact of mutations has been described as functional changes, loss of function and protein truncation (Li et al., 2004b). Although much has been reported on microsatellites frequencies in transcribed regions in plants (Temnykh et al., 2001; McCouch et al., 2002; Morgante et al., 2002; Thiel et al., 2003, Nicot et al., 2004; Kashi and King, 2006; Lawon and Zhang, 2006; Varshney et al., 2006; Zhang et al., 2006b), additional comparative or descriptive analysis can offer novel perspectives on their use as molecular markers. The genomic abundance of microsatellites, and their ability to associate with many phenotypes, make this class of molecular markers a powerful tool for diverse application in plant genetics. The identification of microsatellite markers derived from EST and/or cDNAs, and described as functional markers, represents an even more useful possibility for these markers when compared to those based on assessing anonymous regions (Varshney et al., 2005, 2006).
In order to provide information regarding the patterns of microsatellite occurrence and distribution on transcribed genome regions, non-redundant full-length cDNAs (fl-cDNAs) and/or ESTs belonging to ten plant species from three different families (Brassicaceae, Solanaceae and Poaceae) were used.
Material and Methods
Obtaining the expressed sequence
Files containing expressed sequences were obtained for the following families/species: Brassicaceae (Arabidopsis thaliana and Brassica napus), Solanaceae (Solanum lycopersicum and Solanum tuberosum) and Poaceae (Oryza sativa, Sorghum bicolor, Triticum aestivum, Zea mays, Saccharum officinarum and Hordeum vulgare), all deposited in the NCBI-Unigene data-base. Non-redundant yet representative sequences for all known genes in each species were selected. The sequences used in the present study were downloaded from the Unigene database in June, 2008.
Distribution of sequences in different transcribed regions
By using computer scripts developed in Perl language and based on the existing annotation for each of the cDNAs and/or ESTs sequences, the sequences were categorized as CDS, upstream and downstream regions, partitioned into fasta files and denominated CDS, 5' UTR and 3' UTR for each species. Since the annotation of introns was not part of the database, the repeats present in intronic regions were not considered in this study.
Location of tandem repeats
SSRLocator software was used (Maia et al., 2008) for the location of tandem repeats. Software options were adjusted to locate monomers, dimers, trimers, pentamers and hexamers containing a minimum of 10, 7, 5, 4 and 4 repeats, respectively. For mini-satellites, heptamer, octamer, nonamer and decamers containing a minimum of 3, 3, 3 and 2 repeats, respectively, were selected.
Results and Discussion
Distribution of sequences in UTRs and CDSs
The sequences, separated into coding (CDS) and untranslated (5'UTR and 3'UTR) regions, and distributed by number of sequences, amount (Mb) and average size (bp) for all the ten species, are shown in Table 1. On an average and in all of these, there were sequence fragments between 560 and 893 bp long, except for the A. thaliana and O. sativa databases, where they were longer, reaching averages of 1,447 and 1,490 bp, respectively. The number of sequences deposited in Unigene was the largest for both of the Poaceae species Z. mays and O. sativa, with 57,447 and 40,259, respectively. It is worthy of note that not all sequences deposited in this database contain 5'UTR and 3'UTR regions, for in some both types are found, whereas in others only one is (i.e., 5' or 3'UTR). The overall average sizes were found to be 130 bp for 5'UTR, 873 bp for CDS and 270 bp for 3'UTR regions. The total nucleotides allocated to each were, on an average, 0.9% for 5'UTR, 97.5% for CDS and 1.6% for 3'UTR. The only species with contrasting values was Arabidopsis, where 6.8%, 82.6% and 10.7% of total nucleotides were allocated to 5'UTR, CDS and 3'UTR regions, respectively.
Table 1.
Overall distribution (amounts and percentage) of expressed sequences in translated and non-translated regions.
| Expressed sequences
|
5' UTR
|
CDS
|
3' UTR
|
||||||||||||
| Total seq.1 | Total mb1 | Mean pb1 | Total seq.2 | % mb2 | Mean pb2 | Total seq.3 | % mb3 | Mean pb3 | Total seq.4 | % mb4 | Mean pb4 | ||||
| A. thaliana | 29,918 | 43.3 | 1,447 | 16,625 | 6.8 | 176 | 29,918 | 82.6 | 1,195 | 17,591 | 10.7 | 262 | |||
| B. napus | 26,285 | 20.3 | 773 | 216 | 0.1 | 74 | 26,285 | 99.7 | 770 | 242 | 0.2 | 204 | |||
| S. lycopersicum | 16,945 | 14.0 | 823 | 614 | 0.5 | 103 | 16,945 | 98.3 | 809 | 710 | 1.2 | 245 | |||
| S. tuberosum | 19,539 | 15.6 | 796 | 554 | 0.3 | 93 | 19,539 | 98.6 | 785 | 635 | 1.0 | 252 | |||
| O. sativa | 40,259 | 60.0 | 1,490 | 1,088 | 0.5 | 270 | 40,259 | 98.7 | 1,470 | 1,158 | 0.8 | 438 | |||
| S. bicolor | 13,547 | 9.5 | 699 | 68 | 0.1 | 115 | 13,547 | 99.7 | 697 | 82 | 0.2 | 244 | |||
| T. aestivum | 34,505 | 26.2 | 758 | 498 | 0.2 | 92 | 34,505 | 99.2 | 753 | 611 | 0.6 | 246 | |||
| Z. mays | 57,447 | 32.2 | 560 | 704 | 0.3 | 120 | 57,447 | 99.1 | 555 | 803 | 0.7 | 275 | |||
| S. officinarum | 15,586 | 12.7 | 815 | 48 | 0.1 | 160 | 15,586 | 99.8 | 813 | 54 | 0.1 | 273 | |||
| H. vulgare | 21,418 | 19.1 | 893 | 359 | 0.2 | 102 | 21,418 | 99.2 | 886 | 458 | 0.6 | 259 | |||
| Averages | 27,545 | 25.3 | 905 | 2,077 | 0.9 | 130 | 27,545 | 97.5 | 873 | 2,234 | 1.6 | 269.8 | |||
Expressed sequences: Total Seq.1 (Total number of cDNA sequences), Total mb1 (sum of base pairs of fl-cDNA sequences), Mean pb1 (average size of sequences – sum of base pairs divided by number of sequences (Total mb1 / Total Seq.1)). 5'UTR: Total Seq.2 (Total sequences containing 5'UTR regions), % mb2 (percentage of Total mb1 contained in 5'UTR regions), Mean pb2 (average size of 5'UTR sequences- sum of base pairs divided by the number of sequences (Total pb(% mb2) / Total Seq.2)). CDS: Total Seq.3 (Total sequences containing CDS regions), % mb3 (percentage of Total mb1 contained in CDS regions), Mean pb3 [average size of CDS sequences – sum of base pairs divided by number of sequences (Total pb(% mb3) / Total Seq.3)]. 3'UTR: Total Seq.4 (Total of sequences containing 3'UTR regions), % mb4 (percentage of Total mb1 contained in 3'UTR regions), Mean pb4 (average size of 3'UTR sequences – sum of base pairs divided by the number of sequences (Total pb(% mb4) / Total Seq.4)).
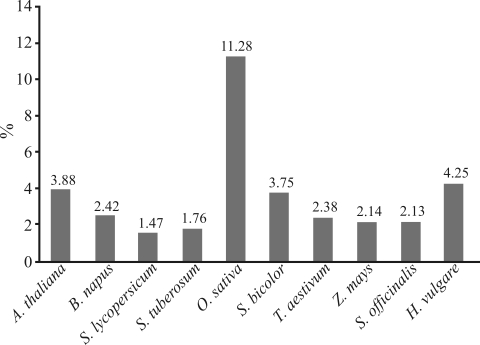
Percentage of expressed sequences with tandem repeats
On an average, 3.55% of analyzed sequences contain one or more loci with tandem repeats. The respective percentages for each species are shown in Figure 1. The highest were for rice (11.28%), and the lowest for the Solanaceae species S. lycopersicum and S. tuberosum, i.e., 1,47% and 1,76%, respectively. The percentage found for Arabidopsis (3.88%) is in agreement with other reports of between 3% and 5% (Cardle et al., 2000; Kumpatla and Mukhopadhyay, 2005). For B. napus, S. lycopersicon and S. tuberosum 2.42%, 1.47% and 1.76% of these sequences were found, respectively. However, different values (6.9%, 4.7% and 2.65%, respectively) have been reported (Kumpatla and Mukhopadhyay, 2005). For the Poaceae, a comparison of present results with former reports for H. vulgare (4.25% vs. 8.11%), Z. mays (2.14% vs. 1.5%), O. sativa (11.28% vs. 4.7%), S. officinarum (2.13% vs. 2.9%) and T. aestivum (2.38% vs. 7.5%) show a different range of values (Cordeiro et al., 2001; Kantety et al., 2002; Thiel et al., 2003; Nicot et al., 2004; Asp et al.., 2007). Nevertheless, all differences are within the 2-3 fold range.
Figure 1.
Percentage of expressed sequences containing tandem repeat loci.
The variations encountered in different reports are related to the strategy employed by the authors (software, repeat number and type defined for the search). However, by common agreement, microsatellite stretches with minimum sizes of 20 bp are present in approximately 2%-5% of cereal EST sequences (Varshney et al., 2005).
Frequency of tandem repeats in UTR and CDS regions
Results for total occurrence (total loci), percentage per region (the amount of loci per region divided by their total number) and frequencies (amount of loci per megabase) are shown separately for each species and by genic region (5'UTR, CDS and 3'UTR) in Table 2. In the 5'UTR and 3'UTR regions, 4.92% (529 loci) and 2.21% (237 loci), respectively, of all repeats were found in all the surveyed species (10,731 loci), with an average frequency of 1.3 and 0.7 loci/Mb, respectively. In coding regions (CDS), a higher occurrence of micro and minisatellites was detected, this reaching 92.86% of the total loci found (9,965 occurrences) with an average frequency of 35.1 loci/Mb. The higher percentage of repeats occurred in CDS regions as a consequence of the trimers present in this region. However, for Arabidopsis, high percentages of dimer (17.9%), trimer (19.3%) and total (44.5%) microsatellites were found in UTR regions, thus contrasting with the other species (Table 3). For the Rosaceae, between 44.3% and 53.2% of the microsatellites were found in UTR regions (Jung et al., 2005). For Arabidopsis, 81% and 26%, respectively, of dimers and trimers were found in UTR regions (Yu et al., 2004).
Table 2.
Overall distribution of tandem repeat occurrences in translated and non-translated transcripts.
| 5' UTR
|
CDS
|
3' UTR
|
Total
|
|||||||||||
| Occurrence | % | ssr/Mb | Occurrence | % | ssr/Mb | Occurrence | % | ssr/Mb | Occurrence | ssr/Mb | ||||
| A. thaliana | 395 | 34.0 | 9.1 | 610 | 52.5 | 14.1 | 157 | 13.5 | 3.6 | 1,162 | 27 | |||
| B. napus | 1 | 0.2 | 0.0 | 632 | 99.5 | 31.1 | 2 | 0.3 | 0.1 | 635 | 31 | |||
| S. lycopersicum | 6 | 2.4 | 0.4 | 234 | 94.0 | 16.8 | 9 | 3.6 | 0.6 | 249 | 18 | |||
| S. tuberosum | 4 | 1.2 | 0.3 | 336 | 97.7 | 21.6 | 4 | 1.2 | 0.3 | 344 | 22 | |||
| O. sativa | 78 | 1.7 | 1.3 | 4,433 | 97.6 | 73.9 | 29 | 0.6 | 0.5 | 4,540 | 76 | |||
| S. bicolor | 3 | 0.6 | 0.3 | 505 | 99.4 | 53.3 | 0 | 0.0 | 0.0 | 508 | 54 | |||
| T. aestivum | 11 | 1.3 | 0.4 | 795 | 97.0 | 30.4 | 14 | 1.7 | 0.5 | 820 | 31 | |||
| Z. mays | 12 | 1.0 | 0.4 | 1,205 | 98.0 | 37.4 | 13 | 1.1 | 0.4 | 1,230 | 38 | |||
| S. officinarum | 0 | 0.0 | 0.0 | 332 | 100.0 | 26.1 | 0 | 0.0 | 0.0 | 332 | 26 | |||
| H. vulgare | 19 | 2.1 | 1.0 | 883 | 96.9 | 46.2 | 9 | 1.0 | 0.5 | 911 | 48 | |||
| Average | 529 | 4.9 | 1.3 | 9,965 | 92.9 | 35.1 | 237 | 2.2 | 0.7 | 10,731 | 37 | |||
Table 3.
Overall occurrence, in percentage, of microsatellite and minisatellite motifs in different sections of genic regions of ten plant species.
| Dimer
|
Trimer
|
Tetramer
|
Pentamer
|
Hexamer
|
||||||||||||||||||||
| Microssatélites | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | ||||
| A. thaliana | 13.6 | 4.0 | 4.3 | 21.9 | 14.6 | 38.6 | 4.7 | 58.0 | 1.0 | 0.9 | 1.0 | 2.9 | 2.1 | 0.8 | 1.9 | 4.7 | 0.9 | 5.8 | 0.4 | 7.1 | ||||
| B. napus | 0.2 | 40.8 | 0.2 | 41.2 | - | 35.9 | 0.2 | 36.1 | - | 4.4 | - | 4.4 | - | 4.3 | - | 4.3 | - | 9.1 | - | 9.1 | ||||
| S. lycopersicum | 0.4 | 17.7 | 2.0 | 20.1 | 0.4 | 40.2 | 0.8 | 41.4 | - | 4.4 | - | 4.4 | - | 6.0 | 0.8 | 6.8 | 0.8 | 17.3 | - | 18.1 | ||||
| S. tuberosum | 0.3 | 22.4 | 0.6 | 23.3 | 0.3 | 34.0 | - | 34.3 | - | 4.4 | - | 4.4 | - | 6.1 | 0.3 | 6.4 | - | 20.1 | - | 20.1 | ||||
| O. sativa | 0.5 | 14.9 | 0.3 | 15.7 | 0.7 | 53.9 | 0.1 | 54.7 | 0.0 | 6.0 | 0.1 | 6.1 | 0.3 | 9.3 | 0.1 | 9.7 | 0.1 | 10.3 | 0.0 | 10.4 | ||||
| S. bicolor | 0.2 | 18.5 | - | 18.7 | 0.2 | 35.2 | - | 35.4 | - | 10.2 | - | 10.2 | - | 14.6 | - | 14.6 | 0.2 | 18.1 | - | 18.3 | ||||
| T. aestevum | 0.5 | 26.5 | 0.4 | 27.3 | 0.5 | 34.0 | 0.5 | 35.0 | 0.2 | 13.3 | 0.1 | 13.7 | 0.1 | 11.3 | 0.6 | 12.1 | - | 7.6 | 0.1 | 7.7 | ||||
| Z. mays | 0.5 | 16.0 | 0.5 | 17.0 | 0.2 | 34.5 | - | 34.6 | 0.1 | 10.7 | 0.4 | 11.2 | 0.1 | 16.2 | 0.2 | 16.4 | 0.1 | 17.4 | - | 17.5 | ||||
| S. officinarum | - | 18.7 | - | 18.7 | - | 36.4 | - | 36.4 | - | 8.4 | - | 8.4 | - | 14.5 | - | 14.5 | - | 16.9 | - | 16.9 | ||||
| H. vulgare | 0.5 | 14.6 | 0.2 | 15.4 | 0.7 | 35.1 | 0.3 | 36.1 | 0.4 | 15.7 | 0.3 | 16.5 | 0.2 | 13.8 | 0.1 | 14.2 | 0.1 | 12.8 | - | 13.0 | ||||
| Average | 1.7 | 19.4 | 0.8 | 21.9 | 1.7 | 37.8 | 0.7 | 40.2 | 0.2 | 7.8 | 0.2 | 8.2 | 0.3 | 9.7 | 0.4 | 10.4 | 0.2 | 13.5 | 0.1 | 13.8 | ||||
| Heptamer
|
Octamer
|
Nonamer
|
Decamer
|
General
|
||||||||||||||||||||
| Minissatélites | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | 5'UTR | CDS | 3'UTR | Total | ||||
| A. thaliana | 1.0 | 0.9 | 0.8 | 2.8 | 0.6 | 0.3 | 0.3 | 1.2 | 0.1 | 1.0 | - | 1.1 | 0.1 | 0.2 | 0.1 | 0.3 | 34.0 | 52.5 | 13.5 | 100.0 | ||||
| B. napus | - | 3.8 | - | 3.8 | - | 0.5 | - | 0.5 | - | 0.6 | - | 0.6 | - | 0.2 | - | 0.2 | 0.2 | 99.5 | 0.3 | 100.0 | ||||
| S. lycopersicum | 0.8 | 8.4 | - | 9.2 | - | - | - | - | - | - | - | - | - | - | - | - | 2.4 | 94.0 | 3.6 | 100.0 | ||||
| S. tuberosum | 0.6 | 9.0 | 0.3 | 9.9 | - | 0.6 | - | 0.6 | - | 1.2 | - | 1.2 | - | - | - | - | 1.2 | 97.7 | 1.2 | 100.0 | ||||
| O. sativa | 0.1 | 2.0 | 0.0 | 2.1 | 0.0 | 0.2 | - | 0.2 | - | 0.7 | - | 0.7 | - | 0.3 | - | 0.3 | 1.7 | 97.6 | 0.6 | 100.0 | ||||
| S. bicolor | - | 2.4 | - | 2.4 | - | - | - | - | - | 0.4 | - | 0.4 | - | - | - | - | 0.6 | 99.4 | - | 100.0 | ||||
| T. aestevum | - | 3.5 | - | 3.5 | - | 0.2 | - | 0.2 | - | 0.4 | - | 0.4 | - | 0.1 | - | 0.1 | 1.3 | 97.0 | 1.7 | 100.0 | ||||
| Z. mays | 0.1 | 3.0 | - | 3.1 | - | - | - | - | - | 0.2 | - | 0.2 | - | - | - | - | 1.0 | 98.0 | 1.1 | 100.0 | ||||
| S. officinarum | - | 4.2 | - | 4.2 | - | - | - | - | - | 0.6 | - | 0.6 | - | 0.3 | - | 0.3 | - | 100.0 | - | 100.0 | ||||
| H. vulgare | 0.1 | 3.6 | - | 3.7 | - | 0.2 | - | 0.2 | - | 1.0 | - | 1.0 | - | - | - | - | 2.1 | 96.9 | 1.0 | 100.0 | ||||
| Average | 0.3 | 4.1 | 0.1 | 4.5 | 0.1 | 0.2 | 0.0 | 0.3 | 0.0 | 0.6 | - | 0.6 | 0.0 | 0.1 | 0.0 | 0.1 | 4.4 | 93.3 | 2.3 | 100.0 | ||||
In the present study, a very high percentage of microsatellites in 5'UTRs were detected in Arabidopsis, with a frequency of 9.1 loci/Mb. These repeats represented 34% of all the 1,162 found in the 29,918 sequences analyzed in this species. The second and third highest frequencies of repeats in these regions were encountered in the species O. sativa and H. vulgare, with an average 1.3 and 1.0 loci/Mb, respectively (Table 2).
Many studies indicate the UTR regions as being more abundant in microsatellites than CDS regions (Morgante et al., 2002). In the present work, 92.86% of microsatellite loci in CDS regions are due to a deficiency in annotation when separating translated from non-translated fractions in the Unigene transcript database.
As observed for 5'UTRs, contrasting values were also found in 3'UTR regions. Much higher values were encountered in Arabidopsis (an average of 3.6 loci/Mb) when compared to those below 0.6 loci/Mb in the remaining species (Table 2).
On considering the overall occurrence of 5'UTRs, 3'UTRs and CDSs in all species, the average frequency observed is 37 loci/Mb. Values normally range from 18 loci/Mb in tomato to 76 in rice. Average frequency values per family are 29.0 loci/Mb in the Brassicaceae, 19.9 in the Solanaceae and 45.4 in the Poaceae (Table 2).
Several reports have indicated values higher than those found in this study, i.e., 112-133 loci/Mb in barley, 133 loci/Mb in maize, 94-161 loci/Mb in wheat, 158-169 loci/Mb in sorghum, 161 loci/Mb in rye, 256-277 loci/Mb in rice and 133 loci/Mb in Arabidopsis (Varshney et al., 2002; Thiel et al., 2003; Parida et al., 2006). In Citrus species, values as high as 507 loci/Mb have been described in EST sequences (Palmieri et al., 2007). Values as high as 125 loci/Mb were also noted in Brassica rapa (Hong et al., 2007). Frequency values closer to our study have been reported for the CDS regions in Rosa chinensis (Rose), Prunus dulcis (Almond), Prunus persica (Peach) and Arabidopsis, with values ranging from 39 to 78 loci/Mb (Jung et al., 2005).
Percentage occurrence of different microsatellite types in the UTR and CDS regions
The detailed percentage values for each repeat type in the diverse sections of a genic region are listed for each species in Table 3. The average occurrence of dimer microsatellites in all the species was 21.9%, the majority of these loci being present in the CDS regions. The average percentage of dimer occurrence for each family was 31.5% in Brassicaceae, 21.7% in Solanaceae and 18.8% in Poaceae species. The percentage values for dimer microsatellites in CDS regions ranged from 4.0% in Arabidopsis to 40.8% in B. napus. An interesting feature which seems to be specific for the Arabidopsis genome is the high occurrence of dimer microsatellites in the 5' and 3' UTR regions (13,6% and 4,3%, respectively). In the Poaceae, dimer microsatellites ranged from 15.4% in barley to 27.3% in wheat (Table 3). Other studies indicated that the highest dimer occurrence rates are generally associated with 5'UTR regions (Morgante et al., 2002; Lawson and Zhang, 2006; Hong et al., 2007), but one should bear in mind that this prevalence in CDS regions may be a consequence of deficient database annotation. Trimer microsatellites were found in 40.2% of the sequences, with a high predominance in CDS regions. The species with higher trimer values were Arabidopsis, rice and tomato, with 58.0%, 54.7% and 41.4% of occurrence, respectively. The average percentage of trimers within each family was 47.0% in the Brassicaceae, 37.8% in the Solanaceae and 38.7% in the Poaceae. Among Poaceae species, the highest percentage of trimer occurrence was found in rice (54.7%) and the lowest in maize (34.6%). In Brassicaceae, trimers were found more frequently in Arabidopsis (58.0%) and less so in B. napus (36.1%) (Table 3).
On an average, tetramers represented 8.2% of the microsatellites, with average frequencies of 3.4%, 4.4% and 11.0% in Brassicaceae, Solanaceae and Poaceae, respectively. Among the Brassicaceae, a less than one-fold difference in frequencies was observed between Arabidopsis (2.9%) and B. napus (4.4%). In Poaceae, a 2.7-fold difference was found between rice (6.1%) and barley (16.5%).
On an average, pentamers represented 10.36% of the microsatellites, with average frequencies of 4.5%, 6.6% and 13.6% in the Brassicaceae, Solanaceae and Poaceae, respectively (Table 3). Less than one-fold differences were found between Brassicaceae and Solanaceae species. Nevertheless, in the Poaceae a 1.7-fold difference was found between rice (9.7%) and maize (16.5%).
On an average, hexamers represented 13.8% of the microsatellites, with average frequencies of 8.1%, 19.1% and 13% in the Brassicaceae, Solanaceae and Poaceae, respectively. In the Poaceae, a 2.4-fold difference was found between wheat (7.7%) and sorghum (18.3%).
Mini-satellite frequencies were also assessed from the available data (Table 3). On an average, heptamers represented 4.5% of the total occurrence (mini-satellite plus microsatellite). These types of repeats were more common in the Solanaceae family (9.6%). In both the Brassicaceae and Poaceae, the average frequencies of heptamers were 3.3% and 3.2%, respectively. Octamers were more frequent in the Brassicaceae (0.8%), when compared to the Solanaceae (0.3%) and Poaceae (0.1%). Nonamers were also more frequent in the Brassicaceae (0.9%), when compared to the Solanaceae (0.6%) and Poaceae (0.5%). Decamers were comparatively less frequent than other mini-satellites, reaching frequencies of 0.2%, 0.1% and zero in the Brassicaceae, Poaceae and Solanaceae, respectively (Table 3).
There are several studies proclaiming EST sequences containing microsatellites. For the Poaceae (rice, maize, sorghum, barley and wheat), frequencies ranging from 16.6 to 40% for dimers, 41 to 78% for trimers, 2.6 to 14% for tetramers, 0.4 to 18.9% for pentamers and below 1% for hexamers (Varshney et al., 2002; Thiel et al., 2003; La Rota et al., 2005; Parida et al., 2006) have been reported. In the case of Arabidopsis, frequencies of dimers (36.5%), trimers (62.1%), tetramers (1.1%), pentamers (0.15%) and hexamers (0.13%) have been noted (Parida et al., 2006).
Most frequent motifs
Dimers and trimers
Motif frequencies per species and average frequency per family are listed in Tables 4 and 5. For dimers, differences were observed within and between families. As regards the Brassicaceae, AG/CT and GA/TC dimer motifs were the most frequent, reaching 9.69% and 8.89% of observations within the family. A 6.9-fold difference was the case for AG/CT between Arabidopsis (2.46%) and B. napus (16.93%). Moreover, as to the GA/TC motif, an almost 10-fold difference was found between Arabidopsis (1.64%) and B. napus (16.14%). Other reports have shown that AG/GA motifs were the most frequent in Arabidopsis (Cardle et al., 2000; Morgante et al., 2002; Lawson and Zhang, 2006; Parida et al., 2006) and AT/TA in B. rapa (Hong et al., 2007). Among the Solanaceae, AT/AT and TA/TA motifs were the most frequent, with frequencies of 8.29% and 5.69%, respectively. In Solanaceae ESTs, frequencies between 20%-25% and 15%-20% were found for AG and AT dimers, respectively (Kumptla and Mukhopadhyay, 2005). In the Poaceae, the most frequent motifs were AG/CT and GA/TC, with average percentages of 6.72% and 5.61%, respectively. In still other studies, frequencies ranging from 38%-50% were the rule for the AG motif in maize, barley, rice, sorghum and wheat (Kantety et al., 2002; Morgante et al., 2002; Varshney et al., 2002; Thiel et al., 2003; Yu et al., 2004; La Rota et al., 2005) and frequencies of 50% for the AC motif in barley (Varshney et al., 2002). GA has also been shown to be the most abundant motif in grasses (Temnykh et al., 2001; Kantety et al., 2002; Nicot et al., 2004; Parida et al., 2006). In all the species that were analyzed in the present study, the lowest frequencies were found for those motifs formed by guanine and cytosine (CG/GC), which were even absent in Brassicaceae and Solanaceae species.
Table 4.
Distribution of di-, tri- and tetramer motifs, percentage occurrence per species and average occurrence per family.
| Brassicaceae
|
Solanaceae
|
Poaceae
|
|||||||||||||||
| Ara | Bra | Average | Lyc | Sol | Average | Ory | Sor | Tri | Zea | Sac | Hor | Average | |||||
| Dimers | |||||||||||||||||
| AG/CT | 2.46 | 16.93 | 9.69 | AT/AT | 8.55 | 8.04 | 8.29 | AG/CT | 6.38 | 5.15 | 9.06 | 6.56 | 6.63 | 6.57 | 6.72 | ||
| GA/TC | 1.64 | 16.14 | 8.89 | TA/TA | 5.13 | 6.25 | 5.69 | GA/TC | 5.46 | 5.35 | 10.19 | 5.15 | 3.92 | 3.62 | 5.61 | ||
| AT/AT | 1.80 | 4.11 | 2.96 | GA/TC | 1.71 | 4.76 | 3.24 | AT/AT | 1.31 | 1.39 | 1.01 | 1.83 | 2.71 | 1.25 | 1.58 | ||
| TA/TA | 0.98 | 2.22 | 1.60 | AG/CT | 3.42 | 2.98 | 3.20 | CA/TG | 0.56 | 2.38 | 2.89 | 0.75 | 1.51 | 1.36 | 1.57 | ||
| GT/AC | 0.49 | 0.79 | 0.64 | GT/AC | 0.00 | 0.60 | 0.30 | GT/AC | 0.59 | 2.38 | 2.77 | 0.50 | 1.20 | 1.36 | 1.47 | ||
| CA/TG | 0.16 | 0.79 | 0.48 | CA/TG | 0.00 | 0.30 | 0.15 | TA/TA | 0.92 | 1.98 | 1.26 | 1.58 | 2.41 | 0.57 | 1.45 | ||
| GC/GC | 0.00 | 0.00 | 0.00 | GC/GC | 0.00 | 0.00 | 0.00 | GC/GC | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.23 | 0.09 | ||
| CG/CG | 0.00 | 0.00 | 0.00 | CG/CG | 0.00 | 0.00 | 0.00 | CG/CG | 0.07 | 0.00 | 0.13 | 0.00 | 0.00 | 0.11 | 0.05 | ||
| Trimers | |||||||||||||||||
| GAA/TTC | 12.13 | 4.59 | 8.36 | GAA/TTC | 3.85 | 5.65 | 4.75 | CCG/CGG | 11.41 | 5.15 | 2.52 | 5.81 | 4.22 | 6.23 | 5.89 | ||
| AAG/CTT | 9.51 | 3.96 | 6.73 | AGA/TCT | 3.85 | 5.36 | 4.60 | CGC/GCG | 10.47 | 4.75 | 3.02 | 5.98 | 6.02 | 4.87 | 5.85 | ||
| AGA/TCT | 8.85 | 4.59 | 6.72 | ATA/TAT | 5.13 | 3.57 | 4.35 | GCC/GGC | 6.11 | 4.95 | 3.27 | 5.81 | 6.93 | 3.28 | 5.06 | ||
| ATC/GAT | 7.54 | 2.22 | 4.88 | AAT/ATT | 4.27 | 2.98 | 3.62 | CAG/CTG | 1.87 | 2.77 | 2.64 | 2.41 | 3.31 | 2.60 | 2.60 | ||
| TCA/TGA | 4.59 | 2.37 | 3.48 | AAG/CTT | 3.42 | 3.57 | 3.50 | GCA/TGC | 1.47 | 2.77 | 2.01 | 2.16 | 1.81 | 2.83 | 2.17 | ||
| CAA/TTG | 4.75 | 1.90 | 3.33 | TAA/TTA | 2.99 | 1.19 | 2.09 | CTC/GAG | 3.77 | 1.19 | 1.89 | 1.49 | 2.41 | 2.15 | 2.15 | ||
| ATG/CAT | 4.43 | 1.74 | 3.08 | CAA/TTG | 2.14 | 1.19 | 1.66 | AGC/GCT | 1.47 | 2.18 | 1.26 | 1.16 | 2.41 | 2.27 | 1.79 | ||
| AAC/GTT | 4.10 | 1.27 | 2.68 | CTC/GAG | 2.14 | 0.60 | 1.37 | AGG/CCT | 2.50 | 1.19 | 1.89 | 1.24 | 0.30 | 1.59 | 1.45 | ||
| ACA/TGT | 3.93 | 1.11 | 2.52 | CAG/CTG | 2.14 | 0.60 | 1.37 | GGA/TCC | 2.57 | 0.99 | 1.13 | 1.74 | 1.20 | 0.79 | 1.41 | ||
| GGA/TCC | 3.44 | 0.79 | 2.12 | TCA/TGA | 0.85 | 1.79 | 1.32 | AAG/CTT | 1.51 | 0.59 | 1.64 | 0.41 | 0.30 | 1.59 | 1.01 | ||
| AGG/CCT | 1.31 | 2.06 | 1.68 | ACA/TGT | 1.71 | 0.89 | 1.30 | CAA/TTG | 0.29 | 0.40 | 3.02 | 0.41 | 1.20 | 0.68 | 1.00 | ||
| CTC/GAG | 1.15 | 2.22 | 1.68 | CAC/GTG | 2.14 | 0.30 | 1.22 | CCA/TGG | 1.38 | 1.39 | 0.38 | 0.75 | 0.60 | 1.13 | 0.94 | ||
| ACC/GGT | 2.13 | 0.63 | 1.38 | ATC/GAT | 1.71 | 0.60 | 1.15 | CGA/TCG | 1.58 | 0.99 | 0.38 | 1.58 | 0.30 | 0.34 | 0.86 | ||
| CCA/TGG | 1.48 | 1.11 | 1.29 | CCA/TGG | 0.85 | 1.19 | 1.02 | CAC/GTG | 0.99 | 0.79 | 0.75 | 0.58 | 0.90 | 1.13 | 0.86 | ||
| CAC/GTG | 1.31 | 0.32 | 0.81 | CCG/CGG | 1.71 | 0.30 | 1.00 | GAC/GTC | 0.99 | 0.40 | 0.50 | 1.00 | 1.20 | 0.68 | 0.80 | ||
| GCA/TGC | 0.16 | 0.95 | 0.56 | GGA/TCC | 0.85 | 0.89 | 0.87 | AGA/TCT | 1.35 | 0.20 | 1.01 | 0.33 | 0.60 | 0.79 | 0.71 | ||
| TAA/TTA | 0.00 | 0.95 | 0.47 | ACC/GGT | 0.43 | 1.19 | 0.81 | GAA/TTC | 1.40 | 0.40 | 1.64 | 0.17 | 0.00 | 0.68 | 0.71 | ||
| ACT/AGT | 0.66 | 0.16 | 0.41 | GCA/TGC | 0.43 | 0.89 | 0.66 | ACC/GGT | 1.29 | 0.40 | 0.88 | 0.33 | 0.60 | 0.23 | 0.62 | ||
| AAT/ATT | 0.16 | 0.63 | 0.40 | ATG/CAT | 0.85 | 0.30 | 0.58 | ACG/CGT | 0.79 | 1.39 | 0.13 | 0.50 | 0.60 | 0.11 | 0.59 | ||
| CAG/CTG | 0.33 | 0.32 | 0.32 | AGC/GCT | 0.43 | 0.30 | 0.36 | ACA/TGT | 0.14 | 0.20 | 1.89 | 0.08 | 0.60 | 0.45 | 0.56 | ||
| AGC/GCT | 0.33 | 0.32 | 0.32 | GTA/TAC | 0.00 | 0.60 | 0.30 | ATC/GAT | 0.32 | 0.59 | 0.38 | 0.25 | 0.00 | 0.57 | 0.35 | ||
| GAC/GTC | 0.33 | 0.32 | 0.32 | GAC/GTC | 0.43 | 0.00 | 0.21 | TCA/TGA | 0.32 | 0.20 | 0.38 | 0.00 | 0.30 | 0.57 | 0.29 | ||
| CCG/CGG | 0.16 | 0.47 | 0.32 | ACT/AGT | 0.43 | 0.00 | 0.21 | AAC/GTT | 0.14 | 0.00 | 0.88 | 0.17 | 0.30 | 0.00 | 0.25 | ||
| GCC/GGC | 0.00 | 0.47 | 0.24 | CGC/GCG | 0.00 | 0.30 | 0.15 | ATG/CAT | 0.25 | 0.20 | 0.25 | 0.08 | 0.00 | 0.57 | 0.22 | ||
| ATA/TAT | 0.00 | 0.47 | 0.24 | GCC/GGC | 0.00 | 0.30 | 0.15 | ATA/TAT | 0.14 | 0.40 | 0.50 | 0.17 | 0.00 | 0.00 | 0.20 | ||
| GTA/TAC | 0.33 | 0.00 | 0.16 | AAC/GTT | 0.00 | 0.30 | 0.15 | AAT/ATT | 0.25 | 0.00 | 0.13 | 0.17 | 0.30 | 0.11 | 0.16 | ||
| CTA/TAG | 0.33 | 0.00 | 0.16 | AGG/CCT | 0.00 | 0.00 | 0.00 | ACT/AGT | 0.11 | 0.59 | 0.13 | 0.00 | 0.00 | 0.00 | 0.14 | ||
| CGA/TCG | 0.16 | 0.16 | 0.16 | CGA/TCG | 0.00 | 0.00 | 0.00 | TAA/TTA | 0.18 | 0.00 | 0.13 | 0.41 | 0.00 | 0.00 | 0.12 | ||
| CGC/GCG | 0.00 | 0.00 | 0.00 | ACG/CGT | 0.00 | 0.00 | 0.00 | GTA/TAC | 0.07 | 0.20 | 0.38 | 0.00 | 0.00 | 0.00 | 0.11 | ||
| ACG/CGT | 0.00 | 0.00 | 0.00 | CTA/TAG | 0.00 | 0.00 | 0.00 | CTA/TAG | 0.09 | 0.20 | 0.13 | 0.00 | 0.00 | 0.00 | 0.07 | ||
| Tetramers | |||||||||||||||||
| AAGA/TCTT | 0.33 | 0.47 | 0.40 | TAAA/TTTA | 0.85 | 0.89 | 0.87 | CCTC/GAGG | 0.09 | 0.40 | 0.50 | 0.17 | 0.00 | 0.79 | 0.32 | ||
| AAAC/GTTT | 0.33 | 0.32 | 0.32 | TTAA/TTAA | 0.85 | 0.30 | 0.58 | AGGA/TCCT | 0.14 | 0.00 | 0.13 | 0.17 | 0.60 | 0.57 | 0.27 | ||
| GAAA/TTTC | 0.33 | 0.32 | 0.32 | AAGA/TCTT | 0.43 | 0.60 | 0.51 | CATC/GATG | 0.27 | 0.00 | 0.50 | 0.25 | 0.00 | 0.57 | 0.26 | ||
| AGGA/TCCT | 0.16 | 0.16 | 0.16 | AAAG/CTTT | 0.00 | 0.60 | 0.30 | CACG/CGTG | 0.09 | 0.20 | 0.13 | 0.08 | 0.60 | 0.45 | 0.26 | ||
| CAAA/TTTG | 0.16 | 0.16 | 0.16 | AGAT/ATCT | 0.00 | 0.60 | 0.30 | AAAG/CTTT | 0.14 | 0.20 | 0.00 | 0.08 | 0.90 | 0.23 | 0.26 | ||
| CATA/TATG | 0.16 | 0.16 | 0.16 | AAAT/ATTT | 0.43 | 0.00 | 0.21 | ATGC/GCAT | 0.00 | 0.00 | 0.38 | 0.33 | 0.00 | 0.79 | 0.25 | ||
| AAAG/CTTT | 0.00 | 0.32 | 0.16 | AATT/AATT | 0.43 | 0.00 | 0.21 | CATA/TATG | 0.14 | 0.00 | 0.50 | 0.41 | 0.30 | 0.11 | 0.24 | ||
| AACA/TGTT | 0.00 | 0.32 | 0.16 | ATTA/TAAT | 0.43 | 0.00 | 0.21 | TCCA/TGGA | 0.11 | 0.00 | 0.50 | 0.50 | 0.00 | 0.34 | 0.24 | ||
| ACAA/TTGT | 0.00 | 0.32 | 0.16 | CCTC/GAGG | 0.43 | 0.00 | 0.21 | CTGC/GCAG | 0.02 | 0.59 | 0.38 | 0.33 | 0.00 | 0.11 | 0.24 | ||
| GAGC/GCTC | 0.16 | 0.00 | 0.08 | TCTG/CAGA | 0.43 | 0.00 | 0.21 | CTCC/GGAG | 0.07 | 0.20 | 0.13 | 0.17 | 0.60 | 0.23 | 0.23 | ||
Ara (Arabidopsis thaliana), Bra (Brassica napus), Lyc (Solanum lycopersicum), Sol (Solanum tuberosum), Ory (Oryza sativa), Sor (Sorghum bicolor), Tri (Triticum aestivum), Zea (Zea mays), Sac (Saccharum officinarum) and Hor (Hordeum vulgare).
Table 5.
Distribution of penta- to decamers motifs, percentage occurrence per species and average occurrence per family.
| Brassicaceae
|
Solanaceae
|
Poaceae
|
|||||||||||||||
| Ara | Bra | Average | Lyc | Sol | Average | Ory | Sor | Tri | Zea | Sac | Hor | Average | |||||
| Pentamers | |||||||||||||||||
| GAAAA/TTTTC | 0.16 | 0.47 | 0.32 | AAAAT/ATTTT | 0.85 | 0.30 | 0.58 | CTCTC/GAGAG | 0.34 | 0.59 | 0.00 | 0.25 | 0.30 | 0.68 | 0.36 | ||
| AAAAT/ATTTT | 0.16 | 0.32 | 0.24 | AAAAG/CTTTT | 0.85 | 0.00 | 0.43 | GAGGA/TCCTC | 0.32 | 0.00 | 0.38 | 0.17 | 0.00 | 0.57 | 0.24 | ||
| AAAAC/GTTTT | 0.00 | 0.47 | 0.24 | AGAAG/CTTCT | 0.43 | 0.30 | 0.36 | CTTCC/GGAAG | 0.07 | 0.20 | 0.25 | 0.17 | 0.60 | 0.11 | 0.23 | ||
| CAAAA/TTTTG | 0.33 | 0.00 | 0.16 | ATAAA/TTTAT | 0.43 | 0.30 | 0.36 | GGAGA/TCTCC | 0.25 | 0.20 | 0.13 | 0.33 | 0.00 | 0.34 | 0.21 | ||
| GAATC/GATTC | 0.00 | 0.32 | 0.16 | GAAAA/TTTTC | 0.43 | 0.30 | 0.36 | AGGAG/CTCCT | 0.29 | 0.20 | 0.13 | 0.33 | 0.00 | 0.23 | 0.20 | ||
| AAATA/TATTT | 0.16 | 0.00 | 0.08 | CAAAC/GTTTG | 0.00 | 0.60 | 0.30 | AGAGG/CCTCT | 0.32 | 0.00 | 0.25 | 0.17 | 0.00 | 0.34 | 0.18 | ||
| ACAAA/TTTGT | 0.16 | 0.00 | 0.08 | AAATA/TATTT | 0.43 | 0.00 | 0.21 | CTCCC/GGGAG | 0.16 | 0.00 | 0.13 | 0.17 | 0.60 | 0.00 | 0.18 | ||
| ACAAC/GTTGT | 0.16 | 0.00 | 0.08 | AAATC/GATTT | 0.43 | 0.00 | 0.21 | CACCA/TGGTG | 0.00 | 0.00 | 0.38 | 0.33 | 0.30 | 0.00 | 0.17 | ||
| ACTAG/CTAGT | 0.16 | 0.00 | 0.08 | AACTG/CAGTT | 0.43 | 0.00 | 0.21 | AGAAG/CTTCT | 0.09 | 0.20 | 0.25 | 0.00 | 0.00 | 0.45 | 0.17 | ||
| TGTTC/GAACA | 0.16 | 0.00 | 0.08 | AATAA/TTATT | 0.43 | 0.00 | 0.21 | AGGGG/CCCCT | 0.18 | 0.00 | 0.25 | 0.08 | 0.00 | 0.45 | 0.16 | ||
| Hexamers | |||||||||||||||||
| GATGAA/TTCATC | 0.33 | 0.16 | 0.24 | GGTGGA/TCCACC | 0.00 | 2.38 | 1.19 | CGGCGA/TCGCCG | 0.38 | 0.20 | 0.13 | 0.25 | 0.30 | 0.11 | 0.23 | ||
| AAAACA/TGTTTT | 0.00 | 0.47 | 0.24 | GAAGTA/TACTTC | 0.85 | 0.60 | 0.72 | GCACCA/TGGTGC | 0.09 | 0.00 | 0.25 | 0.17 | 0.60 | 0.00 | 0.19 | ||
| AAGGAG/CTCCTT | 0.33 | 0.00 | 0.16 | AGCAGG/CCTGCT | 0.85 | 0.30 | 0.58 | AGGCGG/CCGCCT | 0.25 | 0.20 | 0.13 | 0.25 | 0.00 | 0.23 | 0.17 | ||
| AGCCTC/GAGGCT | 0.33 | 0.00 | 0.16 | CAGCAA/TTGCTG | 0.43 | 0.60 | 0.51 | CCGACG/CGTCGG | 0.09 | 0.00 | 0.00 | 0.17 | 0.60 | 0.11 | 0.16 | ||
| ATCACC/GGTGAT | 0.33 | 0.00 | 0.16 | CCAACA/TGTTGG | 0.85 | 0.00 | 0.43 | CCGTCG/CGACGG | 0.18 | 0.00 | 0.13 | 0.17 | 0.30 | 0.11 | 0.15 | ||
| ATGAAG/CTTCAT | 0.33 | 0.00 | 0.16 | CCTATC/GATAGG | 0.85 | 0.00 | 0.43 | GCCTCC/GGAGGC | 0.18 | 0.40 | 0.13 | 0.17 | 0.00 | 0.00 | 0.14 | ||
| CATCAC/GTGATG | 0.33 | 0.00 | 0.16 | GGATGA/TCATCC | 0.85 | 0.00 | 0.43 | GCCACC/GGTGGC | 0.02 | 0.40 | 0.00 | 0.00 | 0.30 | 0.11 | 0.14 | ||
| CCTCCA/TGGAGG | 0.33 | 0.00 | 0.16 | AGGAAG/CTTCCT | 0.43 | 0.30 | 0.36 | CGGCGC/GCGCCG | 0.05 | 0.59 | 0.00 | 0.17 | 0.00 | 0.00 | 0.13 | ||
| CCTGAG/CTCAGG | 0.33 | 0.00 | 0.16 | ATGAAG/CTTCAT | 0.43 | 0.30 | 0.36 | CGACGC/GCGTCG | 0.07 | 0.40 | 0.00 | 0.33 | 0.00 | 0.00 | 0.13 | ||
| GAATCC/GGATTC | 0.33 | 0.00 | 0.16 | CAACCT/AGGTTG | 0.43 | 0.30 | 0.36 | GGAGCC/GGCTCC | 0.00 | 0.20 | 0.13 | 0.17 | 0.30 | 0.00 | 0.13 | ||
| Heptamers | |||||||||||||||||
| ACACAAA/TTTGTGT | 0.33 | 0.00 | 0.16 | CTTCTCT/AGAGAAG | 0.85 | 0.00 | 0.43 | CCGCCGC/GCGGCGG | 0.18 | 0.20 | 0.00 | 0.00 | 0.00 | 0.11 | 0.08 | ||
| GAGAGAA/TTCTCTC | 0.16 | 0.16 | 0.16 | GATCTCC/GGAGATC | 0.85 | 0.00 | 0.43 | CGCCGCC/GGCGGCG | 0.02 | 0.20 | 0.25 | 0.00 | 0.00 | 0.00 | 0.08 | ||
| AGAGAGA/TCTCTCT | 0.00 | 0.32 | 0.16 | AAAAAAT/ATTTTTT | 0.43 | 0.30 | 0.36 | CCGGCGA/TCGCCGG | 0.00 | 0.40 | 0.00 | 0.00 | 0.00 | 0.00 | 0.07 | ||
| AATTACA/TGTAATT | 0.16 | 0.00 | 0.08 | AAATTTA/TAAATTT | 0.43 | 0.30 | 0.36 | CCGCCGA/TCGGCGG | 0.00 | 0.00 | 0.00 | 0.08 | 0.30 | 0.00 | 0.06 | ||
| ATGAGTG/CACTCAT | 0.16 | 0.00 | 0.08 | TCAACTA/TAGTTGA | 0.00 | 0.60 | 0.30 | CGGCAGG/CCTGCCG | 0.02 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| CAGCGAC/GTCGCTG | 0.16 | 0.00 | 0.08 | TTTTTTG/CAAAAAA | 0.00 | 0.60 | 0.30 | AAAATGA/TCATTTT | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| CATTCAA/TTGAATG | 0.16 | 0.00 | 0.08 | AATTGAG/CTCAATT | 0.43 | 0.00 | 0.21 | ACGCAAG/CTTGCGT | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| CCTCTCT/AGAGAGG | 0.16 | 0.00 | 0.08 | AGAAACA/TGTTTCT | 0.43 | 0.00 | 0.21 | AGCAGAG/CTCTGCT | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| CTCAACT/AGTTGAG | 0.16 | 0.00 | 0.08 | ATCGCCG/CGGCGAT | 0.43 | 0.00 | 0.21 | CACGCCG/CGGCGTG | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| TCTCAAA/TTTGAGA | 0.16 | 0.00 | 0.08 | ATGATTC/GAATCAT | 0.43 | 0.00 | 0.21 | CACTGCG/CGCAGTG | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| Octamers | |||||||||||||||||
| ATGTATGA/TCATACAT | 0.16 | 0.00 | 0.08 | AAGAAAAA/TTTTTCTT | 0.00 | 0.30 | 0.15 | GAAGTCAA/TTGACTTC | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 | 0.00 | 0.02 | ||
| CCCCTTCT/AGAAGGGG | 0.16 | 0.00 | 0.08 | TTTCTCTC/GAGAGAAA | 0.00 | 0.30 | 0.15 | GCGACCGA/TCGGTCGC | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 | 0.00 | 0.02 | ||
| CTTGTTCC/GGAACAAG | 0.16 | 0.00 | 0.08 | AAAAAAAC/GTTTTTTT | 0.00 | 0.00 | 0.00 | CCGCACGC/GCGTGCGG | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.11 | 0.02 | ||
| GAAGCAAG/CTTGCTTC | 0.16 | 0.00 | 0.08 | ACGGGCGA/TCGCCCGT | 0.00 | 0.00 | 0.00 | CCTATCTA/TAGATAGG | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.11 | 0.02 | ||
| AAAAAAAC/GTTTTTTT | 0.00 | 0.16 | 0.08 | AGAAAAAA/TTTTTTCT | 0.00 | 0.00 | 0.00 | CAAGAAGC/GCTTCTTG | 0.05 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | ||
| AGAAAAAA/TTTTTTCT | 0.00 | 0.16 | 0.08 | ATCAGGGA/TCCCTGAT | 0.00 | 0.00 | 0.00 | ACGGGCGA/TCGCCCGT | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| TCTTTGTG/CACAAAGA | 0.00 | 0.16 | 0.08 | ATGATGTA/TACATCAT | 0.00 | 0.00 | 0.00 | ATCAGGGA/TCCCTGAT | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| AAGAAAAA/TTTTTCTT | 0.00 | 0.00 | 0.00 | ATGTATGA/TCATACAT | 0.00 | 0.00 | 0.00 | ATGATGTA/TACATCAT | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| ACGGGCGA/TCGCCCGT | 0.00 | 0.00 | 0.00 | CAAGAAGC/GCTTCTTG | 0.00 | 0.00 | 0.00 | TCAAATTT/AAATTTGA | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| ATCAGGGA/TCCCTGAT | 0.00 | 0.00 | 0.00 | CCCCTTCT/AGAAGGGG | 0.00 | 0.00 | 0.00 | TGGGCTTG/CAAGCCCA | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| Nonamers | |||||||||||||||||
| AAGATGAAG/CTTCATCTT | 0.16 | 0.00 | 0.08 | ACTCCTTCA/TGAAGGAGT | 0.00 | 0.30 | 0.15 | ACGACTACG/CGTAGTCGT | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| AATGGGTGG/CCACCCATT | 0.16 | 0.00 | 0.08 | CAAATTACC/GGTAATTTG | 0.00 | 0.30 | 0.15 | AGCGAAGAA/TTCTTCGCT | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| AGAAGGAAG/CTTCCTTCT | 0.16 | 0.00 | 0.08 | CAGACTATT/AATAGTCTG | 0.00 | 0.30 | 0.15 | AGCACCAGC/GCTGGTGCT | 0.00 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | ||
| ATGGGTGAC/GTCACCCAT | 0.16 | 0.00 | 0.08 | CTTCTTATC/GATAAGAAG | 0.00 | 0.30 | 0.15 | GGTGGTATG/CATACCACC | 0.00 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | ||
| GAAGGAGAA/TTCTCCTTC | 0.16 | 0.00 | 0.08 | AAAAAAAAC/GTTTTTTTT | 0.00 | 0.00 | 0.00 | ACCCTCTCC/GGAGAGGGT | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 | 0.00 | 0.02 | ||
| GAGAAGAAG/CTTCTTCTC | 0.16 | 0.00 | 0.08 | AACAGGAGA/TCTCCTGTT | 0.00 | 0.00 | 0.00 | CCGCTGGAT/ATCCAGCGG | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 | 0.00 | 0.02 | ||
| GAGGAAGAA/TTCTTCCTC | 0.16 | 0.00 | 0.08 | AAGATGAAG/CTTCATCTT | 0.00 | 0.00 | 0.00 | GCTGTGACC/GGTCACAGC | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 | 0.00 | 0.02 | ||
| GAGGAAGAG/CTCTTCCTC | 0.16 | 0.00 | 0.08 | AATGGGTGG/CCACCCATT | 0.00 | 0.00 | 0.00 | ACCACCAGC/GCTGGTGGT | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.11 | 0.02 | ||
| TATAATTCG/CGAATTATA | 0.16 | 0.00 | 0.08 | ACAGCAACA/TGTTGCTGT | 0.00 | 0.00 | 0.00 | ACCACGGAC/GTCCGTGGT | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.11 | 0.02 | ||
| TCTTCGTCT/AGACGAAGA | 0.16 | 0.00 | 0.08 | ACCACCAGC/GCTGGTGGT | 0.00 | 0.00 | 0.00 | CCATCCTTA/TAAGGATGG | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.11 | 0.02 | ||
| Decamers | |||||||||||||||||
| ACTTTGAGTG/CACTCAAAGT | 0.16 | 0.00 | 0.08 | AAAAAGAAAA/TTTTCTTTTT | 0.00 | 0.00 | 0.00 | AAAAAGAAAA/TTTTCTTTTT | 0.00 | 0.00 | 0.00 | 0.00 | 0.30 | 0.00 | 0.05 | ||
| CAAAGTCACT/AGTGACTTTG | 0.16 | 0.00 | 0.08 | ACTTTGAGTG/CACTCAAAGT | 0.00 | 0.00 | 0.00 | CCACGCGTCG/CGACGCGTGG | 0.23 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.04 | ||
| TTTTTTTTCT/AGAAAAAAAA | 0.00 | 0.16 | 0.08 | AGCCCCAACG/CGTTGGGGCT | 0.00 | 0.00 | 0.00 | TTTTTTTTCT/AGAAAAAAAA | 0.00 | 0.00 | 0.13 | 0.00 | 0.00 | 0.00 | 0.02 | ||
| AAAAAGAAAA/TTTTCTTTTT | 0.00 | 0.00 | 0.00 | ATCTCCGCCG/CGGCGGAGAT | 0.00 | 0.00 | 0.00 | AGCCCCAACG/CGTTGGGGCT | 0.05 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | ||
Ara (Arabidopsis thaliana), Bra (Brassica napus), Lyc (Solanum lycopersicum), Sol (Solanum tuberosum), Ory (Oryza sativa), Sor(Sorghum bicolor), Tri (Triticum aestivum), Zea (Zea mays), Sac (Saccharum officinarum) and Hor (Hordeum vulgare).
As was the case for dimers, in trimer frequencies motif patterns are different within as well as between families (Table 4). Among the Brassicaceae, GAA/TTC and AAG/CTT motifs were the most abundant, reaching frequencies of 8.36% and 6.73%, respectively. Contrasting values were verified for GAA/TTC between Arabidopsis (12.13%) and B. napus (4.59%), also the case for AAG/CTT between Arabidopsis (9.51%) and B. napus (3.96%). Some reports have claimed that AAG is the most frequent for Arabidopsis and B. rapa (Morgante et al., 2002; Hong et al., 2007). In the Solanaceae, GAA/TCC and AGA/TCT were the most frequent, with values of 4.75% and 4.60%, respectively. For both, frequency values were higher in S. tuberosum. Similar results were obtained in Arabidopsis, B. napus, B.rapa, S. Lycopersicum and S. tuberosum (Kumptla and Mukhopadhyay, 2005), as well as in Citrus (Jiang et al., 2006) where AAG/AGA/GAA motifs were the most frequent. In the Poaceae, the trimers CCG/CGG, CGC/GCG and GCC/GGC were the most frequent, corresponding to 5.89%, 5.85% and 5.06%, respectively, a total of 16.80% of all the microsatellites found. Within the family, different motifs were the most common, i.e., for O. sativa, S. bicolor and H. vulgare, CCG/CGG were predominant, for T. aestivum and S. officinarum GCC/GGC and for Z. mays CGC/GCG. Other studies have shown a predominance of CCG in the grass species Z. mays, H. vulgare, O. sativa, S. bicolor, T. aestivum, S. cereale and S. officinarum (Cordeiro et al., 2001; Kantety et al., 2002; Morgante et al., 2002; Varshney et al., 2002; Thiel et al., 2003; Nicot et al., 2004; Yu et al., 2004; La Rota et al., 2005; Peng and Lapitan, 2005). These motifs (CCG/CGG, CGC/GCG and GCC/GGC) seem to be less common in other families, where instead of values of around 16.8% (found for grasses), frequency was 0.56% in Brassicaceae and 0.36% in the Solanaceae.
Tetramers, pentamers and hexamers
For the loci formed by motifs longer than three nucleotides, only the ten highest average percentages for each family are shown (Tables 4 and 5).
In Brassicaceae, tetramer motifs occurring at higher frequencies were AAGA/TCTT, AAAC/GTTT or GAAA/TTTC adding to 1.04% of all motifs found. Other reports indicate that motifs AAAG/AAAT were predominant in Arabidopsis and AAAT in B. rapa (Cardle et al., 2000; Hong et al., 2007). For 5'UTR/CDS and 3'UTR Arabidopsis regions, the predominant motifs reported were AAAG/CTTT and AAAC/GTTT, respectively (Morgante et al., 2002; Zhang et al., 2004). For Solanaceae species, 1.96% of all motifs found were either TAAA/TTTA or TTAA/TTAA or AAGA/TCTT. These results agree with EST data from 20 dicot species (Kumptla and Mukhopadhyay, 2005). Among the grasses, 0.85% of all motifs were either CCTC/GAGG or AGGA/TCCT or CATC/GATG. Differences in predominant tetramer rates were found among the species (Table 4). Other reports have shown ACGT as the most abundant in barley (Varshney et al., 2002; Thiel et al., 2003), AAAG/CTTT and AAGG/CCTT in perennial ryegrass (Asp et al. 2007) and AAAG as the most frequent motif in rice BACs (McCouch et al., 2002).
For pentamers, 0.80% (GAAAA/TTTTC, AAAAT/ ATTTT and AAAAC/GTTTT), 1.37% (AAAAT/ATTTT, AAAAG/CTTTT and AGAAG/CTTCT) and 0.83% (CTCTC/GAGAG, GAGGA/TCCTC and CTTCC/ GGAAG) were predominant in the Brassicaceae, Solanaceae and Poaceae, respectively. The major difference among plant families is the predominance of A/T in the Brassicaceae and Solanaceae. Also, reports on CDS regions in Arabidopsis, S. cerevisae and C.elegans, indicated the predominance of ACCCG and AAAAG (Toth et al. 2000). For eukaryotes in general, AAAAT, AAAAC and AAAAG are revealed as the most predominant (Li et al., 2004a). On the other hand, 5'UTR and 3'UTR regions of Arabidopsis were shown to be rich in AAGAG and AAAAC, respectively (Zhang et al., 2004). AAAAT (Hong et al., 2007) and AAAAT /AAAAG (Jiang et al., 2006) were described as being frequently found in the Rosaceae and Citrus, respectively. In transcripts from the TIGR database, the AGAGG motif was predominant in rice, AGGGG in barley and ACGAT in wheat (La Rota et al., 2005). Very little information was encountered on the preferential occurrence of pentamers in grasses, whereas that on eukaryotes (Toth et al., 2000; Li et al., 2004a), Citrus (Palmieri et al., 2007; Jiang et al., 2006), Arabidopsis (Zhang et al., 2004) and Rosaceae (Hong et al., 2007) offered variable results.
Hexamer patterns occurred among and within the three analyzed plant families (Table 5). To date, the predominance of AAGGAG hexamers in Arabidopsis, has been confirmed by only one other study (Toth et al., 2000). Other reports indicated the most encountered hexamers to be AAGATG, AAAGAG and AAAAAT in Arabidopsis (Zhang et al., 2004), AAAAAG in Citrus (Jiang et al., 2006), AACACG in S. cerevisae, ACCAGG in C. elegans, AAGGCC in mammals and CCCCGG in primates (Toth et al., 2000). The ten major occurrences for heptamers, octamers, nonamers and decamers are presented in Table 5. Occurrences are widely variable within and among families, making it difficult to establish either a pattern or discussion based on similarities.
Genome dynamics is very complex regarding microsatellite motifs in plants. The higher conservation of dimer motifs (AG/TC and GA/TC) seems to overcome evolutionary barriers distances such as those found between monocot and dicot plants. However, in the dicots, this conservation may not hold. Unexpectedly, Poaceae and Brassicaceae were closer when these motifs were analyzed. On the other hand, trimer microsatellites that are known to be predominant in coding regions followed the expected conservation pattern, with similar rates and predominant motifs (GAA/TTC) between the two dicot families. Trimers present at higher frequencies in the grasses tend to be formed by G/C arrangements, in contrast to dicot plants where G/A/T/C combinations are more frequent. The higher frequency of A/T- rich repeats is also found in pentamer motifs in the dicot families. Repeats of higher complexity did not reveal detectable conserved patterns in this study.
Conclusions
The occurrence of micro and minisatellites in rice sequences (11.28%) is higher than in other species, ranging from 2.5 to 5 times more sequences containing these repetitive DNA loci. The fact that species having larger genomes (T. aestivum, H. vulgare and S. officinarum) do not present a correspondingly higher frequency of repetitive loci implies there is no relationship between genome size and rates of tandem repeat occurrence in functional regions. However, the lower coverage of sequences present in databases for these species could also be a reason for the low rates found in some species. For Arabidopsis and rice, the results obtained are closer to reality, since both are considered model species and have been intensely studied.
The distribution of micro- and minisatellites was higher in CDS regions for all the studied species. Also, microsatellites (97%) were more common than mini-satellites (3%). Per family, the predominant dimer motifs were the same for Brassicaceae and Poaceae (AG/CT) and different for the Solanaceae (AT/AT). Trimers were the predominant repeats, ranging between 34.3% and 58.0%, with different rates depending on the family or species. For the Solanaceae, the predominant trimer motifs were not the same for S.lycopersicum (ATA/TAT and AAT/TTA) and S. tuberosum (GAA/TTC and AGA/TCT). This could be due to selection. Among the grasses, trimers formed by C/G were the most abundant. Nevertheless, specific motifs were variable between species.
Disagreements between earlier reports and the results obtained in the present work, where dimers were also frequent in CDS regions, could be due to the fact that the Unigene database contains predominantly EST clusters. Therefore, there is a tendency for under-representing the UTR regions in the annotated sequences. This is true for all species, except Arabidopsis. This could be solved by manually curating the genes, thereby defining the different regions. Achievement, however, would require a community effort.
The obtained results shed light on the patterns of tandem repeat occurrence within and between different plant families, thereby facilitating the use of plant-breeding strategies based on the transfer of markers from model to orphan species.
Acknowledgments
The authors thank CNPq for fellowships and grants, as well as Dr. Dario Abel Palmieri (UNESP/Assis-SP) and Dr. Olivier Panaud (University of Perpignan) for fruitful discussions.
Footnotes
Associate Editor: Márcio de Castro Silva Filho
References
- Asp T., Frei U.K., Didion T., Nielsen K.K., Lübberstedt T. Frequency, type, and distribution of EST-SSRs from three genotypes of Lolium perenne, and their conservation across orthologous sequences of Festuca arundinacea, Brachypodium distachyon, and Oryza sativa. BMC Plant Biol. 2007;7:36. doi: 10.1186/1471-2229-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G.I. Evolution of simple sequence repeats. Comput Chem. 1996;20:41–48. doi: 10.1016/s0097-8485(96)80006-4. [DOI] [PubMed] [Google Scholar]
- Cardle L., Ramsay L., Milbourne D., Macaulay M., Marshall D., Waugh R. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics. 2000;156:847–854. doi: 10.1093/genetics/156.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conne B., Stutz A., Vassalli J.D. The 3' untranslated region of messenger RNA: A molecular `hotspot' for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- Cordeiro G.M., Casu R., McIntyre C.L., Manners J.M., Henry R.J. Microsatellite markers from sugarcane (Saccharum spp. ) ESTs cross transferable to erianthus and sorghum. Plant Sci. 2001;160:1115–1123. doi: 10.1016/s0168-9452(01)00365-x. [DOI] [PubMed] [Google Scholar]
- Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. Expansion of a CUG trinucleotide repeat in the 3'untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C.P., Piao Z.Y., Kang T.W., Batley J., Yang T.J., Hur Y.K., Bhak J., Park B.S., Edwards D., Lim Y.P. Genomic distribution of simple sequence repeats in Brassica rapa. Mol Cells. 2007;23:349–356. [PubMed] [Google Scholar]
- Iyer R.R., Pluciennik A., Rosche W.A., Sinden R.R., Wells R.D. DNA polymerase III proofreading mutants enhance the expansion and deletion of triplet repeat sequences in Escherichia coli. J Biol Chem. 2000;275:2174–2184. doi: 10.1074/jbc.275.3.2174. [DOI] [PubMed] [Google Scholar]
- Jiang D., Zhong G.Y., Hong Q.B. Analysis of microsatellites in citrus unigenes. Acta Genetica Sinica. 2006;33:345–353. doi: 10.1016/S0379-4172(06)60060-7. [DOI] [PubMed] [Google Scholar]
- Jung S., Abbott A., Jesudurai C., Tomkins J., Main D. Frequency, type, distribution and annotation of simple sequence repeats in Rosaceae ESTs. Funct Integr Genomics. 2005;5:136–143. doi: 10.1007/s10142-005-0139-0. [DOI] [PubMed] [Google Scholar]
- Kantety R.V., La Rota M., Matthews D.E., Sorrells M.E. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol. 2002;48:501–510. doi: 10.1023/a:1014875206165. [DOI] [PubMed] [Google Scholar]
- Kashi Y., King D.G. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Kumpatla S.P., Mukhopadhyay S. Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome. 2005;48:985–998. doi: 10.1139/g05-060. [DOI] [PubMed] [Google Scholar]
- La Rota M., Kantety R.V., Yu J.K., Sorrells M.E. Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genomics. 2005;6:23. doi: 10.1186/1471-2164-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson M.J., Zhang L. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol. 2006;7:R14. doi: 10.1186/gb-2006-7-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Xia Q., Lu C., Zhou Z., Xiang Z. Analysis on frequency and density of microsatellites in coding sequences of several eukaryotic genomes. Genom Proteom Bioinf. 2004b;2:24–31. doi: 10.1016/S1672-0229(04)02004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Korol A.B., Fahima T., Beiles A., Nevo E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- Li Y.C., Korol A.B., Fahima T., Nevo E. Microsatellites within genes: Structure, function, and evolution. Mol Biol Evol. 2004a;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Maia L.C. da, Palmieri D.A., de Souza V.Q., Kopp M.M., de Carvalho F.I., Costa de Oliveira A. SSR Locator: Tool for simple sequence repeat discovery integrated with primer design and PCR simulation. Int J Plant Genomics. 2008:412696. doi: 10.1155/2008/412696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch S.R., Teytelman L., Xu Y., Lobos K.B., Clare K., Walton M., Fu B., Maghirang R., Li Z., Xing Y., et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L. ) DNA Res. 2002;9:199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- Morgante M., Olivieri A.M. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993;3:175–182. [PubMed] [Google Scholar]
- Morgante M., Hanafey M., Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Nicot N., Chiquet V., Gandon B., Amilhat L., Legeai F., Leroy P., Bernard M., Sourdille P. Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs) Theor Appl Genet. 2004;109:800–805. doi: 10.1007/s00122-004-1685-x. [DOI] [PubMed] [Google Scholar]
- Palmieri D.A., Novelli V.M., Bastianel M., Cristofani M., Monge G.A., Carlos E.F., Oliveira A.C., Machado M.A. Frequency and distribution of microsatellites from ESTs of citrus. Genet Mol Biol. 2007;30:1009–1018. [Google Scholar]
- Parida S.K., Anand Raj Kumar K., Dalal V., Singh N.K., Mohapatra T. Unigene derived microsatellite markers for the cereal genomes. Theor Appl Genet. 2006;112:808–817. doi: 10.1007/s00122-005-0182-1. [DOI] [PubMed] [Google Scholar]
- Peng J.H., Lapitan N.L. Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct Integr Genomics. 2005;5:80–96. doi: 10.1007/s10142-004-0128-8. [DOI] [PubMed] [Google Scholar]
- Philips A.V., Timchenko L.T., Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in yotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Mishra R.K., Singh L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S., DeClerck G., Lukashova A., Lipovich L., Cartinhour S., McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L. ): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T., Michalek W., Varshney W., Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L. ) Theor Appl Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- Thornton C.A., Wymer J.P., Simmons Z., McClain C., Moxley R.T. Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat Genet. 1997;16:407–409. doi: 10.1038/ng0897-407. [DOI] [PubMed] [Google Scholar]
- Tóth G., Gáspári Z., Jurka J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R.K., Graner A., Sorrells M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Varshney R.K., Thiel T., Stein N., Langridge P., Graner A. In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett. 2002;7:537–546. [PubMed] [Google Scholar]
- Varshney R.K., Hoisington D.A., Tyagi A.K. Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 2006;24:490–499. doi: 10.1016/j.tibtech.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Yu J.K., Dake T.M., Singh S., Benscher D., Li W., Gill B., Sorrells M.E. Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome. 2004;47:805–818. doi: 10.1139/g04-057. [DOI] [PubMed] [Google Scholar]
- Zhang L., Yuan D., Yu S., Li Z., Cao Y., Miao Z., Qian H., Tang K. Preference of simple sequence repeats in coding and non coding regions of Arabidopsis thaliana. Bioinformatics. 2004;20:1081–1086. doi: 10.1093/bioinformatics/bth043. [DOI] [PubMed] [Google Scholar]
- Zhang L., Zuo K., Zhang F., Cao Y., Wang J., Zhang Y., Sun X., Tang K. Conservation of noncoding microsatellites in plants: Implication for gene regulation. BMC Genomics. 2006a;7:323. doi: 10.1186/1471-2164-7-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yu S., Cao Y., Wang J., Zuo K., Qin J., Tang K. Distributional gradient of amino acid repeats in plant proteins. Genome. 2006b;49:900–905. doi: 10.1139/g06-054. [DOI] [PubMed] [Google Scholar]