Abstract
In this study, we used red cell glucose-6-phosphate dehydrogenase (G6PD) activity to screen for G6PD-deficient individuals in 373 unrelated asymptomatic adult men who were working with insecticides (organophosphorus and carbamate) in dengue prevention programs in 27 cities in São Paulo State, Brazil. Twenty-one unrelated male children suspected of having erythroenzymopathy who were attended at hospitals in São Paulo city were also studied. Fifteen of the 373 adults and 12 of the 21 children were G6PD deficient. G6PD gene mutations were investigated in these G6PD-deficient individuals by using PCR-RFLP, PCR-SSCP analysis and DNA sequencing. Twelve G6PD A-202A/376G and two G6PD Seattle844C, as well as a new variant identified as G6PD São Paulo, were detected among adults, and 11 G6PD A-202A/376G and one G6PD Seattle844C were found among children. The novel mutation c.660C > G caused the replacement of isoleucine by methionine (I220M) in a region near the dimer interface of the molecule. The conservative nature of this mutation (substitution of a nonpolar aliphatic amino acid for another one) could explain why there was no corresponding change in the loss of G6PD activity (64.5% of normal activity in both cases).
Keywords: glucose-6-phosphate dehydrogenase, mutations, polymorphism, variants
Over 440 glucose-6-phosphate dehydrogenase (G6PD) variants (Beutler and Vulliamy, 2002) have been grouped in five classes according to their enzymatic activity and clinical manifestations (World Health Organization-WHO criteria) (Beutler and Yoshida, 1988). The G6PD gene is highly polymorphic and so far more than 150 mutations resulting in distinct G6PD variants have been described (Beutler and Vulliamy, 2002; Hamel et al., 2002; Rodrigues et al., 2002; Vaca et al., 2003; Drousiotou et al., 2004; Grabowska et al., 2004; van Wijk et al., 2004; Yan et al., 2006; Maciag et al., 2007; Mason et al., 2007; Matsuoka et al., 2007; McDade et al., 2008; Minucci et al., 2008; Nuchprayoon et al., 2008). Most of these mutations show low enzyme activity and only five are class IV variants: G6PD São Borja337G > A (Weimer et al., 1993), G6PD A376A > G (Takizawa et al., 1987), G6PD San Luis Potosi376A > T (Vaca et al., 2003), G6PD Insuli989G > A (Sukumar et al., 2003), and G6PD Mira d'Aire1048G > C (Beutler and Vulliamy, 2002).
In this study, we screened for G6PD gene mutations in a series of G6PD-deficient adult males who were working with insecticides (organophosphorus and carbamate) in dengue prevention and in a group of unrelated male children suspected of having erythroenzymopathy.
Subjects: Blood samples from 373 unrelated asymptomatic adult males who had been exposed to toxic chemicals in different regions (27 cities) of São Paulo State, Brazil, and from 21 unrelated male children suspected of having erythroenzymopathy who were treated at hospitals in São Paulo city were screened for G6PD activity. This study was aproved by the Ethical Commitee of the Faculty of Pharmaceutical Sciences (protocol no. 214/2003) of the University of São Paulo, and formal consent was obtained from all participants prior to enrollment in the study.
Biochemical analysis: Red cell G6PD activity was assayed according to Beutler (1990). Individuals with G6PD activity lower than 70% of normal values were screened for G6PD gene mutations by PCR-RFLP (restriction fragment length polymorphism) or PCR-SSCP (single strand conformation polymorphism).
Fifteen of the 373 blood samples (4%) from adult males were G6PD deficient and screened for mutations. One sample with 64.5% of normal G6PD activity (7.8 IU.gHb-1.min-1 at 37 °C) compared to control levels (12.1 IU.gHb-1.min-1 at 37 °C) was also included in the mutation study since this value was considered to be borderline for normal activity (according to WHO criteria, normal activity is > 60% of control levels). Twelve of the 21 children were G6PD deficient and were also screened for mutations.
DNA analysis: Genomic DNA was extracted from leukocytes (Salazar et al., 1998) and RFLP analysis for G6PD A+376G, G6PD A-202A/376G and G6PD Mediterranean 563C polymorphic mutations was done using a procedure modified from Saad et al. (1997). The polymerase chain reaction (PCR) for RFLP or SSCP was done in a total volume of 50 μL using 100 ng of genomic DNA, 15 pmol of each oligonucleotide, 0.2 mM of each dNTP, 1 U of DNA polymerase (Biotools), and 1x reaction buffer (10x PCR buffer; Biotools). The primers used were previously described by Beutler et al. (1989) and Poggi et al. (1990). For RFLP analysis, the PCR products of exons 4, 5 and 6 were digested with NlaIII, FokI and MboII restriction endonucleases, respectively. Negative samples for A+376G, A-202A/376G and Mediterranean563C mutations were analyzed by SSCP, according to Beutler et al. (1989) and Poggi et al. (1990). The PCR products were denatured in 95% formamide, 0.005% bromophenol blue, 20 mM EDTA and 0.005% xylenecyanol in 10 mM Tris-HCl, pH 8.0, at 95 °C for 8 min followed by rapid cooling on ice. Polyacrylamide gel electrophoresis was done at 600 V, 25 mA and 15 W using GeneGel Excel 12.5/24 in the GenePhor System (Pharmacia Biosciences, Uppsala, Sweden for 90 min at two temperatures (5 °C and 15 °C). The gels were stained with silver nitrate (Sambrook et al., 1989). The resulting bands were compared with a normal control and fragments with any alteration were purified (QIAquick PCR purification Kit, Qiagen Inc., Chastworth, CA, USA) and directly sequenced with sense and antisense primers in a model 3100 capillary automatic sequencer (Applied Biosystem, Foster City, CA, USA). For haplotype determination, six intragenic polymorphic sites were analyzed, as described by Rodrigues et al. (2002): exon 5, 376A > G, FokI; intron 5, 611C > G, PvuII; intron 8, 163C > T, BspHI; exon 10, 116G > A, PstI; exon 11, 1311 C > T, Bcl I and intron 11, 93T > C, NlaIII. Restriction enzyme digestions were done according to the manufacturer's instructions (New England Biolabs, Ipswich, MA, USA). The polymorphic haplotypes were recorded as (+) or (-) if the restriction site was present or absent, respectively.
PCR-RFLP analysis showed that 12 of the 15 adult males had G6PD A-202A/376G mutation (202G > A and 376G > A mutations). All but one of the G6PD-deficient children (11/12) also had this mutation. All of the mutations had the haplotype VIa (+ + - + - +).
Two G6PD-deficient adults and the child without the G6PD A-202A/376G mutation had a different SSCP pattern for exon 8 compared to the controls. This exon was subsequently sequenced and the variant identified as G6PD Seattle (844G > C). These variants had the haplotype I (- - + + - -).
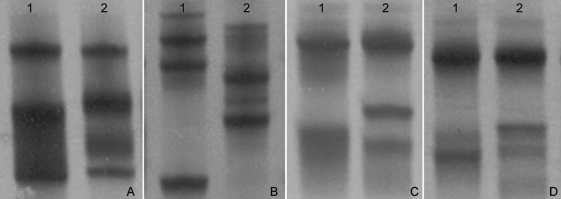
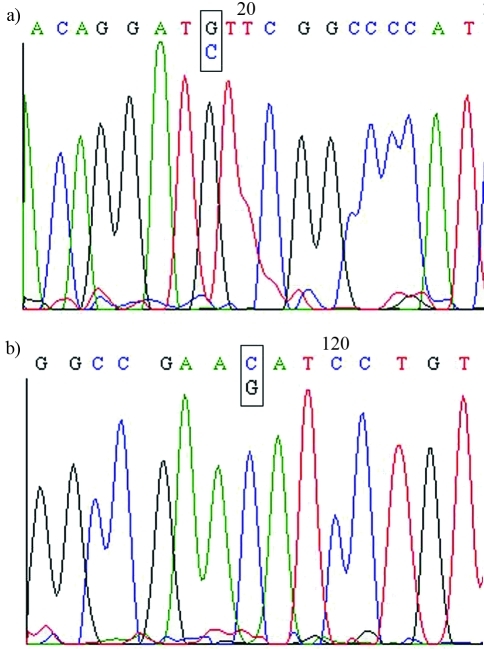
The sample with 64.5% G6PD activity showed no mutations in RFLP analysis, but SSCP analysis revealed a different profile in exon 7 compared to the normal control (Figure 1). DNA sequencing of the PCR product of exon 7 revealed a novel point mutation, c.660C > G (Figure 2), that led to the replacement of isoleucin by methionine (I220M). This mutation was associated with haplotype I (- - + + - -) and the new variant was designated as G6PD São Paulo.
Figure 1.
SSCP patterns of G6PD exon 7. Electrophoresis was done at 5 °C (A), 8 °C (B), 12 °C (C) and 15 °C (D). Lane 1: G6PD B control. Lane 2: G6PD São Paulo variant.
Figure 2.
DNA sequencing of G6PD exon 7. The use of sense (A) and antisense (B) primers revealed the novel mutation c.660C > G of the G6PD São Paulo variant.
Polymorphic variants: Most of the individuals included in the DNA analysis had a G6PD A- 202G > A mutation that is frequently found in African descendents from Central Africa, thus confirming the results obtained by Saad et al. (1997). In their study of 150 G6PD-deficient blood donors, these authors observed the A- 202G > A mutation in 146 individuals (97.3%), indicating that this variant is the most frequent G6PD deficiency in the population of southeastern Brazil. G6PD Mediterranean but did not detect any G6PD Seattle. In contrast, in a sample from the same population, we found G6PD Seattle but not G6PD Mediterranean. This variation may reflect the ethnic heterogeneity of the population in the state of São Paulo.
Haplotype analysis: All individuals with G6PD A- 202G > A had the characteristic haplotype VIa (+ + - + - +), which is very common in Brazilian (Saad et al., 1997) and Portuguese (Rodrigues et al., 2002) populations. Seattle variant carriers had the haplotype I (- - + + - -) that was also present in the G6PD São Paulo variant described here.
G6PD São Paulo variant: The large number of G6PD mutations that cause low enzymatic activity (classes I, II and III) indicates that the tertiary structure of the protein probably has many more critical sites that affect the enzymes stability and activity than non-critical sites. However, this conclusion may reflect the fact that most molecular studies of G6PD have been done only in patients with hemolytic crises. Class IV variants have rarely been detected because carriers have near normal G6PD activity, and only five of them have been studied molecularly (Weimer et al., 1993; Takizawa et al., 1987; Beutler and Vulliamy, 2002; Vaca et al., 2003; Sukumar et al., 2003). As shown here we have identified a novel class IV G6PD variant in a male with borderline G6PD activity. A novel mutation c.660 (C > G) was detected in exon 7, surprisingly close to the dimer interface. According to Naylor et al. (1996) and Au et al. (2000), exon 10, the first half of exon 11, and the second half of exon 6 up to the first half of exon 7 are involved in the dimer interface and are crucial for G6PD stability and activity.
All of the mutations described so far in this region (Harilaou 648T > G, class I; Radlowo 679C > T, class I; “Mexico City” 680G > A, class III; A- 680G > T, class III) significantly reduce G6PD activity (Hirono and Beutler, 1988; Poggi et al., 1990; Beutler et al., 1992; Jablonska-Skwiecinska et al., 1999). In the Harilaou, Radlowo and “Mexico City” A- mutations, the changes occurred between residues with distinct characteristics: 216Phe (nonpolar aromatic) > Leu (nonpolar aliphatic), 227Arg (polar alkaline) > Trp (apolar aromatic), 227Arg (polar alkaline) > Gln (polar neutral), and Arg (polar alkaline) > Leu (apolar aliphatic), respectively (Hirono and Beutler, 1988; Poggi et al., 1990; Beutler et al., 1992; Jablonska-Skwiecinska et al., 1999). It is possible that the substitution of a nonpolar aliphatic amino acid for another amino acid with similar properties (I220L) was crucial for maintaining fairly normal catalytic activity in the novel G6PD variant identified here.
Acknowledgments
This work was sponsored by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, grant n. 03/008766-6).
Footnotes
Associate Editor: Francisco Mauro Salzano
References
- Au S.W., Gover S., Lam V.M.S., Adams M.J. Human glucose-6-phosphate dehydrogenase: The crystal structure reveals a structural NADP+ molecule and provides insights into enzyme deficiency. Structure. 2000;8:293–303. doi: 10.1016/s0969-2126(00)00104-0. [DOI] [PubMed] [Google Scholar]
- Beutler E. Erythrocyte enzyme assays. In: Williams W.J., Beutler E., Erslev A.J., Lichtman M.A., editors. Hematology. New York: McGraw-Hill; 1990. pp. 1723–1726. [Google Scholar]
- Beutler E., Vulliamy T.J. Hematologically important mutations: Glucose-6-phosphate dehydrogenase. Blood Cells Mol Dis. 2002;28:93–103. doi: 10.1006/bcmd.2002.0490. [DOI] [PubMed] [Google Scholar]
- Beutler E., Yoshida A. Genetic variation of glucose-6-phosphate dehydrogenase: A catalog and future prospects. Medicine. 1988;67:311–334. doi: 10.1097/00005792-198809000-00003. [DOI] [PubMed] [Google Scholar]
- Beutler E., Kuhl W., Vives-Corrons J.L., Prchal J.T. Molecular heterogeneity of glucose-6-phosphate dehydrogenase A- Blood. 1989;74:2550–2555. [PubMed] [Google Scholar]
- Beutler E., Westwood B., Prchal J., Vaca G., Bartsocas C.S., Baronciani L. New glucose-6-phosphate dehydrogenase mutations from various ethnic groups. Blood. 1992;80:255–256. [PubMed] [Google Scholar]
- Drousiotou A., Touma E.H., Andreou N., Loiselet J., Angastiniotis M., Verrelli B.C., Tishkoff S.A. Molecular characterization of G6PD deficiency in Cyprus. Blood Cells Mol Dis. 2004;33:25–30. doi: 10.1016/j.bcmd.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Grabowska D., Jablonska-Skwiecinska E., Plochocka D., Chelstowska A., Lewandowska I., Witos I., Majewska Z., Rokicka-Milewska R., Burzynska B. A novel mutation in the glucose-6-phosphate dehydrogenase gene in a subject with chronic nonspherocytic hemolytic anemia - characterization of enzyme using yeast expression system and molecular modeling. Blood Cells Mol Dis. 2004;32:124–130. doi: 10.1016/j.bcmd.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Hamel A.R., Cabral I.R., Sales T.S.I., Costa F.F., Saad S.T.O. Molecular heterogeneity of G6PD deficiency in an Amazonian population and description of four new variants. Blood Cells Mol Dis. 2002;28:399–406. doi: 10.1006/bcmd.2002.0524. [DOI] [PubMed] [Google Scholar]
- Hirono A., Beutler E. Molecular cloning and nucleotide sequence of cDNA for human glucose-6-phosphate dyhydrogenase variant A- Proc Natl Acad Sci USA. 1988;85:3951–3954. doi: 10.1073/pnas.85.11.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska-Skwiecinska E., Lewandowska I., Plochocka D., Topczewski J., Zimowski J.G., Klopocka J., Burzynska B. Several mutations including two novel mutations of the glucose-6-phosphate dehydrogenase gene in Polish G6PD deficient subjects with chronic nonspherocytic hemolytic anemia, acute hemolytic anemia, and favism. Hum Mutat. 1999;14:477–484. doi: 10.1002/(SICI)1098-1004(199912)14:6<477::AID-HUMU6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Maciag M., Plochocka D., Jablonska-Skwiecinska E., Mendek-Czajkowska E., Gomaszewska E., Strojny W., Balwierz W., Zdebska E., Burzynska B. Molecular analysis of three novel G6PD variants: G6PD Pedoplis-Ckaro, G6PD Piotrkow and G6PD Krakow. Acta Biochim Pol. 2007;54:877–881. [PubMed] [Google Scholar]
- McDade J., Abramova T., Mortier N., Howard T., Ware R.E. A novel G6PD mutation leading to chronic hemolytic anemia. Pediatr Blood Cancer. 2008;51:816–819. doi: 10.1002/pbc.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.J., Bautista J.M., Gilsanz F. G6PD deficiency: The genotype-phenotype association. Blood Rev. 2007;21:267–283. doi: 10.1016/j.blre.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Matsuoka H., Thuan D.T.V., van Thien H., Kanbe T., Jalloh A., Hirai M., Arai M., Dung N.T., Kawamoto F. Seven different glucose-6-phosphate dehydrogenase variants including a new variant distributed in Lam Dong Province in southern Vietnam. Acta Med Okayama. 2007;61:213–219. doi: 10.18926/AMO/32873. [DOI] [PubMed] [Google Scholar]
- Minucci A., Concolino P., Vendittelli F., Giardina B., Zuppi C., Capoluongo E. Glucose-6-phosphate dehydrogenase Buenos Aires: A novel de novo missense mutation associated with severe enzyme deficiency. Clin Biochem. 2008;41:742–745. doi: 10.1016/j.clinbiochem.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Naylor C.E., Rowland P., Basak A.K., Gover S., Mason P.J., Bautista J.M., Vulliamy T.J., Luzzatto L., Adams M.J. Glucose-6-phosphate dehydrogenase mutations causing enzyme deficiency in a model of the tertiary structure of the human enzyme. Blood. 1996;87:2974–2982. [PubMed] [Google Scholar]
- Nuchprayoon I., Louicharoen C., Charoenvej W. Glucose-6-phosphate dehydrogenase mutations in Mon and Burmese of southern Myanmar. J Hum Genet. 2008;53:48–54. doi: 10.1007/s10038-007-0217-3. [DOI] [PubMed] [Google Scholar]
- Poggi V., Town M., Foulkes N.S., Luzzatto L. Identification of a single base in a new human mutant glucose-6-phosphate dehydrogenase gene by polymerase chain reaction amplification of the entire coding region genomic DNA. Biochem J. 1990;271:157–160. doi: 10.1042/bj2710157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M.O., Freire A.P., Martins G., Pereira J., Martins M.C., Monteiro C. Glucose-6-phosphate dehydrogenase deficiency in Portugal: Biochemical and mutational profiles, heterogeneity, and haplotype association. Blood Cells Mol Dis. 2002;28:249–259. doi: 10.1006/bcmd.2002.0505. [DOI] [PubMed] [Google Scholar]
- Saad S.T.O., Salles T.S.I., Carvalho M.H., Costa F.F. Molecular characterization of glucose-6-phosphate dehydrogenase deficiency in blood donors from Brazil. Hum Hered. 1997;47:17–21. doi: 10.1159/000154383. [DOI] [PubMed] [Google Scholar]
- Salazar L.A., Hirata M.H., Cavalli S.A., Machado M.O., Hirata R.D. Optimized procedure for DNA isolation from fresh and cryopreserved clotted human blood useful in clinical molecular testing. Clin Chem. 1998;44:1748–1750. [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sukumar S., Mukherjee M.B., Colah R.B., Mohanty D. Molecular characterization of G6PD Insuli - A novel 989 CGC → CAC (330 Arg → His) mutation in the Indian population. Blood Cells Mol Dis. 2003;30:246–247. doi: 10.1016/s1079-9796(03)00018-4. [DOI] [PubMed] [Google Scholar]
- Takizawa T., Yoneyama Y., Miwa S., Yoshida A. A single nucleotide base transition is the basis of the common human glucose-6-phosphate dehydrogenase variant (A+) Genomics. 1987;1:228–231. doi: 10.1016/0888-7543(87)90048-6. [DOI] [PubMed] [Google Scholar]
- Yan T., Cai R., Mo O., Zhu D., Ouyang H., Huang L., Zhao M., Huang F., Li L., Liang X., et al. Incidence and complete molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Guangxi Zhuang autonomous region of southern China: Description of four novel mutations. Haematologica. 2006;91:1321–1328. [PubMed] [Google Scholar]
- Vaca G., Arámbula E., Monsalvo A., Medina C., Nuñez C., Sandoval L., López-Guido B. Glucose-6-phosphate dehydrogenase (G6PD) mutations in Mexico: Four new G-6-PD variants. Blood Cells Mol Dis. 2003;31:112–120. doi: 10.1016/s1079-9796(03)00119-0. [DOI] [PubMed] [Google Scholar]
- van Wijk R., Huizinga E.G., Prins I., Kors A., Rijksen G., Bierings M., van Solinge W. Distinct phenotypic expression of two de novo missense mutations affecting the dimer interface of glucose-6-phosphate dehydrogenase. Blood Cells Mol Dis. 2004;32:112–117. doi: 10.1016/j.bcmd.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Weimer T.A., Salzano F.M., Westwood B., Beutler E. Molecular characterization of glucose-6-phosphate dehydrogenase variants from Brazil. Hum Biol. 1993;65:41–47. [PubMed] [Google Scholar]