Abstract
Lytic bacteriophages are hypothesized to contribute to the seasonality and duration of cholera epidemics in Bangladesh. However, the bacteriophages contributing to this phenomenon have yet to be characterized at a molecular genetic level. In this study, we isolated and sequenced the genomes of 15 bacteriophages from stool samples from cholera patients spanning a 10-year surveillance period in Dhaka, Bangladesh. Our results indicate that a single novel bacteriophage type, designated ICP1 (for the International Centre for Diarrhoeal Disease Research, Bangladesh cholera phage 1) is present in all stool samples from cholera patients, while two other bacteriophage types, one novel (ICP2) and one T7-like (ICP3), are transient. ICP1 is a member of the Myoviridae family and has a 126-kilobase genome comprising 230 open reading frames. Comparative sequence analysis of ICP1 and related isolates from this time period indicates a high level of genetic conservation. The ubiquitous presence of ICP1 in cholera patients and the finding that the O1 antigen of lipopolysaccharide (LPS) serves as the ICP1 receptor suggest that ICP1 is extremely well adapted to predation of human-pathogenic V. cholerae O1.
IMPORTANCE
The severe diarrheal disease cholera is caused by the bacterium Vibrio cholerae, which can be transmitted to humans from the aquatic environment. Factors that affect V. cholerae in the environment can impact the occurrence of cholera outbreaks; one of these factors is thought to be the presence of bacterial viruses, or bacteriophages. Bacteriophages that prey on V. cholerae in the environment, and potentially in humans, have not been extensively genetically characterized. Here, we isolated and sequenced the genomes of bacteriophages from cholera patient stool samples collected over a 10-year period in Dhaka, Bangladesh, a region that suffers from regular cholera outbreaks. We describe a unique bacteriophage present in all samples, infer its evolution by sequencing multiple isolates from different patients over time, and identify the host receptor that shows that the bacteriophage specifically predates the serogroup of V. cholerae responsible for the majority of disease occurrences.
INTRODUCTION
Cholera is a substantial health burden worldwide and is endemic in many parts of South Asia, Africa, South America, and Central America (1). Cholera epidemics can occur both in areas where cholera is endemic and areas where it is not endemic; the current outbreak in Haiti (2), a country that has not seen the disease in over a century, highlights the ongoing vulnerability of underdeveloped and tragedy-struck nations to explosive disease. Toxigenic Vibrio cholerae strains belonging to O1 and O139 serogroups are the causative agents of epidemic and pandemic cholera (3). V. cholerae O1 biotype El Tor, which can be distinguished by serotype (Ogawa and Inaba [4]) is the current leading cause of cholera both worldwide and regionally in Bangladesh (5, 6). V. cholerae O139, although it accounts for a much lower proportion of cholera, continues to circulate and cause disease with O1 El Tor. Cholera is characterized by acute secretory diarrhea that can rapidly lead to severe dehydration and death if left untreated. Toxigenic V. cholerae is present in the environment as a member of the aquatic ecosystem and can emerge to cause disease in humans (7, 8). Factors impacting the interepidemic persistence and ecology of V. cholerae in the aquatic environment can have great impact on the transmission and virulence of this organism (8–12). One environmental factor hypothesized to have an important role in shaping cholera outbreaks is the presence of V. cholerae lytic bacteriophages.
The dynamic between cholera incidence and lytic phages has recently been documented at the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B) located in Dhaka, Bangladesh. In this region of South Asia, where cholera has long been endemic, two distinct peaks of cholera occur each year, and each peak is inversely correlated with the presence of lytic phages in the aquatic environment (5). Intriguingly, the increase of lytic phage in the environment was observed to coincide with the increasing excretion of phage in stool samples from cholera patients (13). These observations led to the prediction that as V. cholerae increases in density in the environment as an outbreak proceeds, the level of predation by lytic phage reaches a height that promotes a decline in the outbreak (13, 14). In addition, experiments addressing the impact of lytic phage on growth of V. cholerae in the infant mouse infection model indicate that phage can dramatically reduce the load of V. cholerae in the small intestine (15, 16), which would contribute further to a decline in the outbreak if the phage behaves similarly in humans. However, the lytic phages hypothesized to play such a critical role in the dynamics of cholera outbreaks in Bangladesh have not been genetically characterized, and the diversity of phages involved in potentially modulating cholera outbreaks in this and other regions is unclear.
In this study, we isolated and sequenced the genomes of 15 phages from cholera patient stool samples collected at the ICDDR,B over a 10-year surveillance period. Our results indicate that a single novel phage type, designated ICP1 (for ICDDR,B cholera phage 1), is present in all cholera patient stool samples analyzed, while two unrelated phages (ICP2 and ICP3) have overlapped with ICP1 for distinct periods.
RESULTS
Isolation of phages.
Since 2000, there has been an ongoing NIH-funded prospective study of cholera at the ICDDR,B, in which stool samples are collected from patients for analysis and cryopreservation. These protocols have been reviewed and approved by both the ethical review committee at the ICDDR,B and the institutional review board at Massachusetts General Hospital. Thirty-one stool samples spanning a 10-year period between 2001 and 2010 were obtained and tested for the presence of phage capable of forming plaques on V. cholerae O1 El Tor. Of the 15 phages studied, three were previously isolated (15), while the remaining 12 were isolated in this study. Sequencing of the 15 phages revealed the presence of three distinct phage types designated ICP1, ICP2, and ICP3 (Table 1). The host range of ICP1 is limited to V. cholerae O1, while the host ranges for ICP2 and ICP3 also include non-O1 V. cholerae strains. Specifically, ICP2 is capable of forming plaques on V. cholerae O139 strain MO10, and ICP3 is capable of forming plaques on a non-O1, non-O139 strain CR034-24. Between one and seven additional isolates of each phage type were sequenced and based on the high-level DNA sequence homology were designated related isolates. The phage isolate from each type isolated at the earliest point in time was designated the prototype phage (ICP1, ICP2, and ICP3) and was analyzed in a detailed manner at the genomic level. The GenBank accession numbers for all phages are provided in Table S1 in the supplemental material.
TABLE 1 .
General characteristics of the sequenced and annotated V. cholerae phages isolated from cholera patients at the ICDDR,B between 2001 and 2010
| Phage | Taxonomic family | Genome size (bp) | G+C content (%) | % of genome coding for proteins | No. of predicted CDSs | % CDSs similar to known proteins | % CDSs similar to conserved hypothetical proteins | % CDSs with homology to hypothetical proteins | No. of fully sequenced isolates |
|---|---|---|---|---|---|---|---|---|---|
| ICP1 | Myoviridae | 125,956 | 37.1 | 88.4 | 230 | 12 | 13 | 75 | 8 |
| ICP2 | Podoviridae | 49,675 | 42.7 | 93.7 | 73 | 19 | 18 | 63 | 2 |
| ICP3 | Podoviridae | 39,162 | 42.9 | 91.9 | 54 | 48 | 33 | 19 | 5 |
Prevalence of ICP1-, ICP2-, and ICP3-related phages in clinical specimens.
Primers (see Table S2 in the supplemental material) designed for the DNA polymerase in each phage were used to screen all stool samples by PCR; samples were confirmed positive for the presence of ICP2 or ICP3 with one additional primer set each. Three additional sets of primers designed to amplify polymorphic loci in ICP1-related phages were used to confirm the prevalence of these phages in all stool samples (Table 2).
TABLE 2 .
Detection of V. cholerae phages ICP1, ICP2, and ICP3 in cholera patient stool samples collected at the ICDDR,B between 2001 and 2010
| Phage | Presence of phage detected in individual stool samples collected in the following yra: |
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2001 |
2002 |
2003 |
2004 |
2005 |
2006 |
2007 |
2008 |
2009 |
2010 |
||||||||||||||||||||
| S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | S1 | S2 | S3 | |
| ICP1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| × | × | × | × | × | |||||||||||||||||||||||||
| ICP2 | + | + | + | + | + | + | |||||||||||||||||||||||
| × | × | ||||||||||||||||||||||||||||
| ICP3 | + | + | + | + | + | + | + | + | |||||||||||||||||||||
| × | × | × | × | ||||||||||||||||||||||||||
The presence of phage detected in individual patient stool samples S1, S2, and S3 collected between 2001 and 2010 is indicated as follows: +, phage detected by PCR; ×, phage isolated from stool sample.
ICP1 genome.
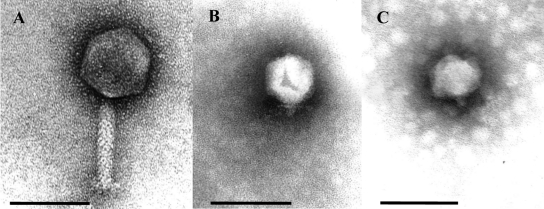
ICP1 is an unclassified member of the Myoviridae family, with an icosahedral head that is 86 nm long and a tail that is 106 nm long and 17 nm wide (Fig. 1A). ICP1 is morphologically unrelated to any Vibrio phages known (17), but it closely resembles a phage of Salinivibrio (18). The ICP1 genome is 125,956 bp with an average G+C content of 37% (Table 1), which is approximately 10% percent lower than that of its V. cholerae O1 host (19). The difference in G+C content between ICP1 and V. cholerae is apparent in their respective codon usage patterns (see Table S3 in the supplemental material). No tRNAs were found in ICP1 or in any of the seven additional ICP1-related isolates.
FIG 1 .
Electron micrograph images of V. cholerae phages isolated from stool samples from cholera patients. (A) ICP1, (B) ICP2, and (C) ICP3. Bars, 100 nm.
A total of 230 predicted coding sequences (CDSs) were identified, as well as 16 predicted promoters and seven rho-independent transcriptional terminators (Fig. 2). The majority of defined CDSs had no identifiable homologue (75%). In addition, 13% of CDSs showed homology to hypothetical proteins, leaving only 12% of the entire ICP1 genome that could be assigned putative function (Table 1). Of the CDSs with homology to proteins from phages, there were no majority with significant similarity to one particular phage, indicating that ICP1 is truly a unique phage of previously sequenced phages.
FIG 2 .
Genomic map of ICP1. The numbering below the ORF arrows is the numbering used in the text, tables, and GenBank. The small black arrows indicate the presence of predicted promoters, and stem loops are predicted transcriptional rho-independent terminators discovered using Mfold (for simplicity, not all promoters and terminators were shown). The short vertical marks along the genome show distance in kilobases, with a mark placed every 2 kb.
Half of the CDSs with predicted functions (14/28) are involved in nucleotide metabolism, replication, recombination, and repair. ICP1 carries genes encoding a DNA polymerase (DNAP) (open reading frame [ORF] 58) (Fig. 2), RNase H (ORF 55), primase/helicase (ORF 57), helicase (ORF 147), a recombination-associated protein (ORF 173), exonuclease (ORF 176), and DNA ligase (ORF 215). CDSs involved in nucleotide metabolism are nrdDG (ORF 113 and 114), nrdAB (ORF 203 and ORF 204), nadV (ORF 228) (Fig. 2), ribose-phosphate pyrophosphokinase (ORF 227), and a thymidylate synthase (ORF 193). NrdAB and NrdDG represent two distinct classes of ribonucleotide reductases (RNRs), which recycle ribonucleotides to supply deoxynucleoside triphosphates (dNTPs) for DNA replication (20). NrdAB belongs to the class Ia reductases that act under aerobic conditions. In contrast, NrdDG is a class III reductase that is inactivated by oxygen and is therefore required by cellular organisms under anaerobic growth conditions. Facultative anaerobes, like V. cholerae, rely on a class I enzyme during aerobiosis and a class III enzyme during anaerobiosis (20, 21). Homology of NrdAB and NrdD suggests that ICP1 RNRs are most closely related to RNRs from phages, namely, Escherichia phage rv5 (GenBank accession no. NC_011041), Erwinia phage φEa21-4 (GenBank accession no. NC_011811), and Enterobacteria phage Felix O1 (GenBank accession NC_005282). The NrdG of ICP1, on the other hand, shows the highest homology to V. cholerae O1 strain N16961 NrdG. In model systems, NrdG catalyzes the activation of NrdD; however, the FeS cluster of NrdG is first reduced by NADPH and flavodoxin reductase (22). It is possible that ICP1 NrdG requires cellular flavodoxin reductase for its activity, since no flavodoxin reductase homologue was detected in the ICP1 genome, and therefore, NrdG with similarity to V. cholerae NrdG has been selected for to permit this interaction during phage infection. However, the exact mechanism and function of these proteins during ICP1 infection of V. cholerae are not known. An intein was identified in NrdA of ICP1; this appears to be fairly commonplace, as many phage-carried RNR genes are interrupted by self-splicing introns or inteins (23, 24). Inteins are often found in proteins that are important for the survival of the organism, which in turn necessitates intein maintenance (25). Presumably nrdA contributes to the fitness of ICP1 as it does in phage T4; although nrdA is not essential in phage T4, nrdA mutants have a reduced burst size (26) and reduced DNA synthesis (27).
Comparative sequence analysis of ICP1-related phages.
Seven additional isolates of ICP1 were isolated from stool samples between 2001 and 2006. Compared to ICP1, these seven isolates showed between 98 and 99.5% nucleotide identity over 95 to 100% of the length of their genomes. Of the 230 CDSs identified in ICP1, 201 were present in all isolates and therefore represent a set of core proteins found in ICP1-related phages. A nucleotide alignment of the annotated ICP1 genome with the seven ICP1-related isolates shows that synteny is conserved with no significant genome rearrangements observed (see Fig. S7 in the supplemental material). Phylogenetic analysis showed that relatedness was independent of their time of isolation (see Fig. S5 in the supplemental material). For example, two of the phages isolated in 2006 were more closely related to phages isolated in 2001 than to two other isolates recovered from samples in 2006.
The high conservation of genome synteny and sequence of ICP1-related phages indicates that this phage has been under conditions of stabilizing selection over the period sampled. To further investigate this, we identified synonymous substitutions (silent; dS) and nonsynonymous substitutions (amino acid altering; dN) for all 230 ICP1 CDSs within each of the ICP1-related phages. The average dN/dS ratio per CDS was then calculated to compare the strength of purifying selection acting on each CDS. Of these CDSs, 120 (52%) remained unchanged, and for 56 CDSs (24%), only synonymous changes were observed. The average dN/dS ratios for the 53 CDSs that had both synonymous and nonsynonymous changes were <1 for all except one (ORF 165, a conserved hypothetical). Thus, 52/53 CDSs that could be analyzed by dN/dS are undergoing purifying or stabilizing selection, in which changes to the coding sequence are being selected against (28).
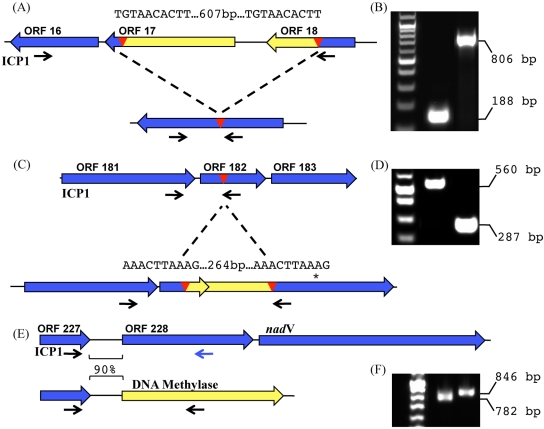
Genomic analysis of the seven ICP1-related isolates allowed us to identify several polymorphic loci that were further analyzed using PCR in all stool samples (Fig. 3). The first locus examined was a region of 607 bp flanked by 10-bp direct repeats encoding ORF 17 and ORF 18 in ICP1 (Fig. 3A and B). ICP1-related isolates from 2001 and 2004 (ICP1_2001-A and ICP1_2004-A) were identical to ICP1 at this locus; however, all phages recovered from 2005 and 2006 (phages ICP1_2005-A and ICP1_2006-A to ICP1_2006-D) showed a deletion of this region, resulting in the formation of one contiguous ORF representing an in-frame fusion of ORF 16 and ORF 18. Sequencing of PCR products from selected samples collected between 2008 and 2010 showed that the insertion observed in ICP1, which dominated before 2005, was present once again, suggesting a selective sweep at this locus. Unfortunately, like the majority of CDSs identified in ICP1, none of the coding sequences at this locus have homologues to suggest a reason for such a selective sweep. A similar polymorphism in ORF 182 was detected in ICP1-related phages (Fig. 3C and D). Here, a region of 264 bp flanked by 10-bp direct repeats was observed in three ICP1-related phages (ICP1_2004-A, ICP1_2006-C, and ICP1_2006-D). The deletion allele in ICP1 has a stop codon downstream, resulting in conversion of a large ORF into two small ORFs, ORF 182 and ORF 183. PCR to analyze this polymorphism was done on 29 stool samples, and in contrast to what was observed for the ORF 17/18 polymorphism (Fig. 3A), there appeared to be no temporal pattern associated with this polymorphism (data not shown). The nonessential region was detected in one-third of stool samples and was found throughout the 10-year period. Interestingly, single patient samples often showed both PCR products corresponding to the indel (insertion and deletion), indicating that a population of ICP1-related phages is mixed at the genomic level at this locus. No homologues were identified for any of the coding sequences involved in the polymorphism at this locus. A third locus corresponding to the replacement of ORF 228 and ORF 229 with a putative cytosine-specific DNA methylase was observed in ICP1-related phages (Fig. 3E and F). ORF 228 encodes a putative ribose-phosphate pyrophosphokinase, and ORF 229 encodes a putative nicotinamide phosphoribosyltransferase (nadV). Three of the seven sequenced ICP1-related phages (ICP1_2004-A, ICP1_2006-C, and ICP1_2006-D) have the methylase. When PCR was used to assay for the presence of either the methylase or ORF 228 from stool samples, only one additional stool sample from which no phage were recovered was positive for the methylase, while the remainder (27/29) were positive for ORF 228, indicating that these genes involved in nucleotide metabolism are likely selected for over the methylase.
FIG 3 .
PCR detection of ICP1-related phages in stool samples from cholera patients through analysis of polymorphic regions. Red triangles represent the indicated direct repeats, and the small black arrows indicate the positions of locus-specific primers. The sequence of the direct repeats is shown above the red triangles. (A) Schematic of the ORF 17 polymorphic locus. (B) An example of the ORF 17 polymorphism detected in stool samples from cholera patients by PCR. (C) Schematic of the ORF 182 polymorphic locus. The asterisk indicates the single nucleotide change found in phages carrying this insertion corresponding to an STOP → Gln mutation. (D) An example of the ORF 182 polymorphism. (E) Schematic of the ORF 228 polymorphic locus. ORF 228 encodes a putative ribose-phosphate pyrophosphokinase, while the downstream ORF encodes a putative nicotinate phosphoribosyltransferase. The observed polymorphism at this locus involves the complete replacement of ORF 228 and nadV with a putative site-specific DNA methylase. In addition, the nucleotide identity between the intergenic regions immediately upstream of methylase/ORF228 is indicated. (F) An example of the ORF 228 polymorphism.
Identification of V. cholerae O1 antigen as the ICP1 receptor.
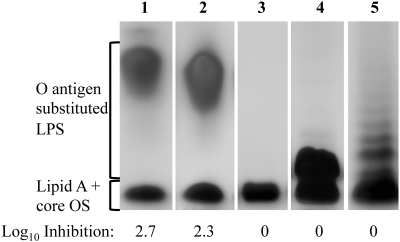
Previous experiments in which mice were challenged with a mixture of V. cholerae O1 El Tor and ICP1_2006-B resulted in the generation of phage-resistant V. cholerae isolates having an O-antigen-deficient phenotype (15). These results, along with the observation that ICP1 cannot produce plaques on other V. cholerae serogroups, indicate that an essential receptor for ICP1 is the lipopolysaccharide (LPS) O1 antigen. To test this hypothesis, we carried out a plaque inhibition assay using purified LPS extracted from strains representing different serogroups (O1 and O139), serotypes (O1 Inaba and Ogawa), and a rough mutant lacking the O1 antigen. Ten micrograms of purified LPS from O1 Inaba or Ogawa strains efficiently inhibited plaque formation by ICP1 (Fig. 4). No inhibition was observed when ICP1 was treated with LPS from V. cholerae O139, the rough mutant lacking the O1 antigen, or Escherichia coli. These results demonstrate that the O1 antigen is a receptor for ICP1 and not the core oligosaccharide. Furthermore, the results show that the terminal methyl group present in the O1 antigen of Ogawa serotype strains is not recognized, consistent with the observation that ICP1 forms plaques on both Inaba and Ogawa serotypes (data not shown).
FIG 4 .
SDS-PAGE pattern of purified LPS after silver staining and its ability to inactivate phage ICP1. Strains used for LPS preparation were as follows: V. cholerae strain AC53 (wild type [wt]; serotype Ogawa) (lane 1), AC304 (wt; serotype Inaba) (lane 2), AC304 rfbV::mTn5 (O-antigen mutant) (lane 3), AC2995 (O139) (lane 4), and E. coli O26:B6 (Sigma) (lane 5). The positions of the fully synthesized LPS with attached O antigen and lipid A core oligosaccharide (OS) precursor are indicated. The inhibition of ICP1 was determined by plaque inhibition assays using 10 µg of purified LPS and is indicated as the average log10 inhibition of two independent assays.
ICP2 genome.
ICP2-related phages were isolated from two stool samples from cholera patients in 2004 and 2006 and were detected in four additional stool samples between 2002 and 2007 (Table 2). ICP2 is morphologically a member of the Podoviridae family (Fig. 1B) with a capsid measuring 60 nm long and short tails measuring 13 nm long and 8 nm wide. ICP2 has a 49,675-bp genome predicted to encode 73 proteins (Table 1). The overall genome is organized into two transcriptional units in which ORFs 1 to 25 are transcribed in the orientation opposite that of ORFs 26 to 73 (see Fig. S1 in the supplemental material). A unique feature of the ICP2 genome is that it encodes two proteins (ORF 41 and ORF 42) to be involved in cobalamin (vitamin B12) biosynthesis. ORFs 41 and 42 encode putative CobT and CobS cobalt chelatase subunits, respectively. To our knowledge, CobT/CobS homologues have not been previously observed in phage genomes. Cobalamin is a structurally complex cofactor synthesized by bacteria and is required by enzymes from bacteria, protists, and animals (29). It is possible that these metabolic enzymes benefit ICP2 by enhancing the metabolism of the infected bacterial cell, which could in turn increase phage proliferation. Comparative analysis of ICP2 and the related phage, ICP2_2006-A, showed these two are 97.8% identical over 97.8% of their genomes at the nucleotide level. One interesting site of variation was seen between the predicted phage tail fiber proteins of ICP2 and ICP2_2006-A. These proteins are 54% identical and show a much higher level of amino acid conservation at the N terminus than at the C terminus. This indicates that there may be host range differences between the two phages, although we have not identified any thus far.
ICP3 genome.
ICP3 was recovered from a cholera patient stool sample from 2007, and four additional ICP3-related isolates were recovered from patient samples between 2007 and 2010, but not from patient samples before this (Table 2). ICP3 is morphologically a member of the Podoviridae (Fig. 1C) with a capsid measuring 60 nm long and short tails measuring 13 nm long by 8 nm wide. ICP3 is morphologically indistinguishable from coliphage T7 and Vibrio phage III of the typing set of Mukerjee (30). Phages of this morphological type have repeatedly been isolated and seem to be very common (31). The 39,162-bp genome has 54 predicted CDSs, all transcribed from the same strand (see Fig. S2 in the supplemental material). Genomic analysis using CoreGenes (32) demonstrated that ICP3 has 43% of its proteins in common with T7, placing ICP3 as a member of the “T7-like viruses” within the subfamily Autographivirinae (33). Functions could be assigned to almost half of the genes based on the similarity of the predicted products to known proteins (Table 1).
T7-like viruses utilize the host RNA polymerase (RNAP) for expression of early genes, which include a phage-borne RNAP (gene 1) (34). Middle and late regions are transcribed by the phage-borne RNAP, which recognizes conserved phage-specific promoters that are related to a 23-bp consensus sequence (35). Fourteen phage-specific promoters were identified in ICP3, and the consensus sequence shows sequence similarity to the T7 promoter consensus sequence (see Fig. S4 in the supplemental material). Specifically, the region from positions −7 to +6 is identical to the T7 consensus promoter sequence, while the region from positions −17 to −8 is unique to ICP3 compared to T7 and other T7-like viruses, consistent with the finding that this region is responsible for phage-specific promoter recognition (35).
Comparative sequence analysis of ICP3-related phages.
Four ICP3-related isolates recovered from stool samples between 2007 and 2010 were sequenced and compared to ICP3. These isolates were remarkably similar to ICP3, sharing between 99.6 and 99.8% nucleotide identity over 99.3 to 99.8% of their genome. A nucleotide alignment of the annotated ICP3 genome with the four ICP3-related isolates showed that the genomes are collinear, with no significant rearrangements observed (see Fig. S3 in the supplemental material).
We identified synonymous and nonsynonymous substitutions in ICP3-related phages for all 54 CDSs in ICP3. Seventeen of these CDSs (31%) had no mutations in any isolates relative to ICP3, and 18 CDSs (33%) had only synonymous changes. The dN/dS ratios for 18 of 19 CDSs that had both synonymous and nonsynonymous changes were <1 and thus are undergoing purifying or stabilizing selection. The one exception to this was gp12, which has a dN/dS value of >1, indicating positive or diversifying selection (28). In T7, the initial interaction with the bacterium is mediated by tail fibers (gp17) binding to LPS; subsequent interactions between one or both of the gp11 and gp12 tail proteins with unknown cell surface components may serve to situate the phage to a site where a successful infection can occur (36). The only CDS found to be undergoing positive selection in ICP3 was the T7 gp12 homologue, which may indicate that this protein is prone to evolutionary diversification to enhance the host range of this phage. In total, 50 of the 54 CDSs present in ICP3 were present in all ICP3-related isolates, and phylogenetic analysis of ICP3-related phages based on this shared core genome revealed that isolates recovered in 2007 and 2008 are more closely related to each other than isolates recovered in 2009 are (see Fig. S6 in the supplemental material).
DISCUSSION
The ecological factors supporting or promoting seasonal epidemics in areas where cholera is endemic, such as Bangladesh, are a topic of intense interest. A factor previously identified as having a role in the occurrence of V. cholerae outbreaks are lytic phages (5, 13). An interesting feature of lytic phages is that, similar to environmental factors, such as water temperature or rainfall amounts (37), phage may modulate the population of V. cholerae in the environment, thus impacting the occurrence of an outbreak; however, uniquely, these phages also travel with V. cholerae into the human host and are shed at appreciable amounts by infected patients, where like V. cholerae they could be spread to others via fecal-oral transmission. The impact of these phages on infectivity has only recently begun to be addressed (15, 16), and much more remains to be elucidated regarding the potential impact these phages have on V. cholerae both in the environment and in an infected host. We set out to characterize lytic phages collected from cholera patients and identified a unique phage type that is pervasive in all cholera patients from an area where cholera is endemic.
Phage genomes frequently contain a majority of novel genetic sequences of unknown function (23), and ICP1 is no exception, with 75% of its CDSs having no sequence similarity to any proteins seen thus far. Seven additional ICP1-related isolates from stool samples from cholera patients collected from 2001 to 2006 were sequenced and compared to ICP1, which revealed a stable ICP1 genome. Stability is suggested from the high level of nucleotide identity, lack of inversions and other major rearrangements, and the stabilizing selection inferred for virtually all genes harboring synonymous and nonsynonymous mutations. Indels were observed, and these included a small number of polymorphic loci that appear to frequently undergo alterations, two of them by recombination between 10-bp direct repeats. We were unable to identify any synonymous or nonsynonymous mutations that appeared in an isolate and were then carried forward in time, suggesting that these isolates represent a sampling of a large stable population of ICP1-related phages present in patients and presumably in the local environment in Dhaka, Bangladesh.
For several stool samples, we observed single-locus variants of ICP1 using PCR, in addition to the presence of the unrelated phages ICP2 and ICP3. In a previous study (16), it has similarly been observed that multiple phage types can be excreted by a single patient. These results parallel results for V. cholerae isolates from cholera patients (also in Dhaka, Bangladesh) in which intrapatient variation of V. cholerae dominated: 89% of stool samples had V. cholerae isolates with distinct genotypes (38). Taken together, these results suggest that both within and between cholera patients, the bacterial and phage burden during symptomatic disease is diverse. Selected isolates may represent only a fraction of the genomic diversity present during an outbreak. The relative stability of the ICP1 genome over time and its ubiquitous presence in cholera patients suggest that ICP1 is extremely well adapted to its host, specifically to pathogenic V. cholerae O1, and that there is significant selective pressure to maintain this particular genome structure.
Lytic phage from aquatic environments in Dhaka, Bangladesh, as well as from cholera patient stool samples at the ICDDR,B have been isolated and characterized by host range, electron microscopy, and restriction endonuclease cleavage patterns of purified phage genomes (5, 13). This work has identified the presence of multiple phage types, including phage JSF4 (5). Though sequence data are lacking, based on the KpnI restriction profile of phage JSF4 presented in supplemental data by Faruque et al. (13), we believe that JSF4 may be an ICP1-related isolate. JSF4 was recovered from cholera patient stool samples collected at the ICDDR,B during an epidemic that began in August 2004; during that study period, it was observed that as the epidemic progressed, an increasingly higher proportion of patients were observed to excrete JSF4 in their stool samples (13). JSF4 was also recovered from patient samples between 2004 and 2007, and again it was observed that this phage was more frequently isolated during the later stages of an epidemic (16). In both of these studies, patient samples were collected in which no phage were detected (for example, the percentage of patients excreting lytic phages ranged from ~5 to ~100% during the 12-week epidemic [13]). Assays for phages in stool samples in these studies were conducted by looking for plaque formation following inoculation of filter-sterilized stool samples with recipient V. cholerae strains. We also assayed for plaque formation, but in addition, we used PCR to more sensitively detect the presence of phages recovered in this study and observed that all of the stool samples had ICP1 and roughly half had at least one other phage.
JSF4 titers in stool samples from cholera patients have been reported to be between 4.0 × 102 to 1.1 × 108 PFU/ml (13) and 1.8 × 103 to 5.7 × 107 PFU/ml (16). Despite seemingly high levels of lytic phage in the stool samples, V. cholerae O1 isolated from stool samples in which lytic phage were present are susceptible to lysis by the phage recovered from the same stool sample (this was observed with JSF4 [13, 16] and with ICP1_2006-B, ICP1_2006-C, and ICP_2006_D [15]). This is consistent with our finding that the receptor for ICP1 is the O1 antigen of V. cholerae. While LPS mutants lacking the O1 antigen dominate in in vitro cultures containing JSF4 (16) and can be isolated following in vivo coinfection of V. cholerae and ICP1_2006-B in mice (15), such phage-resistant O1 antigen-deficient bacteria would probably not dominate in human stool samples, as O1 antigen mutants are attenuated for virulence (16, 39, 40). This suggests that O1 antigen-deficient V. cholerae would be lost from the natural life cycle (41), because the fitness advantage to remaining O1 positive and virulent in humans is greater than the fitness advantage conferred by mutational escape from phages like ICP1.
The pervasiveness of ICP1-related phages in cholera patients treated at the ICDDR,B brings into question the life cycle of these phages; one could speculate that ICP1 may be capable of establishing lysogeny. Bioinformatics analysis of eight ICP1-related phages did not reveal anything at the genetic level that suggests establishment of classic lysogeny; for example, these phages appear to lack integrases and excisionases. Despite this, using PCR, we have been able to detect the transient presence of the ICP1 genomic DNA inside passaged bacterial colonies following ICP1 infection; however, the phage genome does not appear to be stably maintained in V. cholerae under the conditions tested (data not shown). A similar suggestion has been made for JSF4 (5, 14).
Finally, we also isolated and characterized two other unrelated types of phages from cholera patients in this study. ICP2-related phages were the least prevalent in clinical samples, and like ICP1, they were found to be genetically diverse and distinct from other sequenced phages in the databases. ICP3-related phages are T7-like and appeared in 2007 in patients at the ICDDR,B. In contrast to ICP1, both ICP2 and ICP3-related phages are not specific to V. cholerae O1, which is consistent with their less frequent isolation from stool samples from cholera patients, as they are not reliant on the most clinically dominant agent of cholera for their propagation.
MATERIALS AND METHODS
Strains and growth conditions.
V. cholerae strains used in this study are listed in Table S2 in the supplemental material. Strains were grown on Luria-Bertani (LB) agar or in LB broth at 37°C with 100 µg/ml streptomycin (SM) with the exceptions of strain AC304 rfbV::mTn5, which was grown in the presence of 25 µg/ml kanamycin and strain CR034-24, which was grown in the absence of antibiotics. To plate phages, strain AC53 was grown to mid-exponential phase (optical density at 600 nm [OD600] of ~0.3) and added to phage that were allowed to adsorb for 10 min at room temperature; the mixture was added to LB soft agarose (0.5%) and overlaid onto plates containing LB agar plus SM. The plates were incubated overnight at 37°C.
Phage isolation and detection in clinical specimens.
Phages ICP1_2006-B, ICP1_2006-C, and ICP1_2006-D were isolated previously and referred to as phages EN159, EN182, and EN191, respectively (15). ICP1 was isolated from a stool sample collected at the ICDDR,B in 2001 (S. Merrell and A. Camilli, unpublished data). Stool specimens were collected and stored from previous studies (42, 43). All other phages described in this study were isolated from stool samples as follows: ~100μl stool sample was added to 10 ml of LB plus strain AC53 (starting OD600 of 0.05), and the mixture was allowed to incubate overnight at 37°C. The filtrate was plated with strain AC53 using standard soft-agar overlay technique described above for detection of plaques. Phages were subjected to three rounds of plaque purification before being grown to a high titer for DNA isolation. For PCR, total DNA was extracted from 100-μl stool samples using the DNeasy blood and tissue kit (Qiagen). Primers for the amplification of V. cholerae 16S DNA (44) were used as a positive control. Select PCR products were confirmed by sequencing.
Phage DNA isolation and sequencing.
A modified version of the phage DNA extraction protocol described by Sambrook and Russell (45) was used. High-titer lysates were treated with benzonase endonuclease (Sigma) at 37°C; EDTA was omitted from the protocol. Paired-end Illumina libraries were created for each phage as previously described (46). Bar-coded adapters used (e.g., BC1a and BC1b) are listed in Table S2 in the supplemental material. Each purified, bar-coded sample was subjected to 15 cycles of PCR amplification using primers OLJ139 and OLJ140 (46). Sequencing was conducted at the Tufts University Core facility using an Illumina genome analyzer II.
Sequence analysis.
Genomes were assembled with Velvet (47). Per-base coverage values ranged from 1,000 to 7,000 times. Detailed manual analysis was conducted for ICP3, ICP2, and ICP1. Open reading frames were identified using Kodon (Applied Maths, Austin, TX). GeneMark.hmm (48) was used in conjunction with Kodon for ICP1. Batch BLAST (http://greengene.uml.edu/programs/NCBI_Blast.html) was used to identify homologues. Promoters were identified on the basis of sequence similarity to E. coli consensus σ70 promoter: TTGACA(N15-17)TATAAT, immediately upstream of an annotated gene. Rho-independent terminators were identified using Mfold (49). For phage ICP3, phage-specific promoters were discovered using PHIRE (50), and WebLogo (51) was used to generate a sequence logo for the consensus promoter. tRNAscan-SE v 1.21 (52) and Aragorn (53) were used to search the genomes for phage-encoded tRNA genes. Genomes were compared at the nucleotide level using the progressiveMauve algorithm (54). CoreGenes was used for comparisons at the proteomic level (32). ICP1-related, ICP2-related, and ICP3-related isolates were submitted for automated annotation as part of a pilot Phage Genomes Automatic Annotation by R. Brister (NCBI).
Genomic selection levels.
For every annotated CDS in ICP1 and ICP3, we used BLAST to identify homologous regions of at least 75% identity for every related phage isolate. Homologous regions were aligned using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). We determined the dN, dS, and dN/dS (55) for every pair and the average values for each ORF.
Phylogenetic analysis.
GRIMM (56) found no major rearrangements between the progenitor phage and the respective related isolates. Every aligned ORF found above that was present in every phage was concatenated into a whole-genome alignment of ORFs. The resulting contiguous sequence was provided as input to MrBayes (http://mrbayes.csit.fsu.edu/index.php), a program for the Bayesian estimation of phylogeny. Figures of each tree were generated using Figtree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Transmission electron microscopy.
High-titer phage stocks were sedimented for 1 h at 25,000 × g using a Beckman (Palo Alto, CA) J2-21 high-speed centrifuge and a JA-18.1 fixed-angle rotor. Phages were washed twice in ammonium acetate buffer (0.1 M, pH 7.0). Purified phage particles were deposited on 3-mm carbon-coated copper grids and stained with 2% phosphotungstate at pH 7.2 (ICP2 and ICP3) or 2% uranyl acetate at pH 4.5 (ICP1) and examined with a Phillips EM 300 electron microscope. Magnification was monitored using T4 phage tails.
LPS extraction and plaque inhibition assays.
Cell surface polysaccharides were prepared and visualized as described recently (57). The ability of purified LPS to neutralize plaque formation was determined by mixing 104 PFU of ICP1 with 10 µg of purified LPS in 600 μl of LB plus 10 mM CaCl2 for 1 h at 37°C. Serial dilutions of this mixture were then plated using the soft-agar overlay method described earlier.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We thank D. Lazinski for critical evaluation and direction throughout this project.
This research was supported by National Institutes of Health grants AI055058 (A.C.), AI058935 (S.B.C.), and AI063079 (F.Q.). A.C. is a Howard Hughes Medical Institute investigator.
Footnotes
Citation Seed, K. D., K. L. Bodi, A. M. Kropinski, H.-W. Ackermann, S. B. Calderwood, et al. 2011. Evidence of a dominant lineage of Vibrio cholerae-specific lytic bacteriophages shed by cholera patients over a 10-year period in Dhaka, Bangladesh. mBio 2(1):e00334-10. doi:10.1128/mBio.00334-10.
REFERENCES
- 1. Zuckerman J. N., Rombo L., Fisch A. 2007. The true burden and risk of cholera: implications for prevention and control. Lancet Infect. Dis. 7:521–530 [DOI] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention 2010. Update: cholera outbreak—Haiti, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1473–1479 [PubMed] [Google Scholar]
- 3. Safa A., Nair G. B., Kong R. Y. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18:46–54 [DOI] [PubMed] [Google Scholar]
- 4. Stroeher U. H., Karageorgos L. E., Morona R., Manning P. A. 1992. Serotype conversion in Vibrio cholerae O1. Proc. Natl. Acad. Sci. U. S. A. 89:2566–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Faruque S. M., Naser I. B., Islam M. J., Faruque A. S., Ghosh A. N., Nair G. B., Sack D. A., Mekalanos J. J. 2005. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc. Natl. Acad. Sci. U. S. A. 102:1702–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sack R. B., Siddique A. K., Longini I. M., Jr., Nizam A., Yunus M., Islam M. S., Morris J. G., Jr., Ali A., Huq A., Nair G. B., Qadri F., Faruque S. M., Sack D. A., Colwell R. R. 2003. A 4-year study of the epidemiology of Vibrio cholerae in four rural areas of Bangladesh. J. Infect. Dis. 187:96–101 [DOI] [PubMed] [Google Scholar]
- 7. Colwell R. R. 2004. Infectious disease and environment: cholera as a paradigm for waterborne disease. Int. Microbiol. 7:285–289 [PubMed] [Google Scholar]
- 8. Reidl J., Klose K. E. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125–139 [DOI] [PubMed] [Google Scholar]
- 9. Vezzulli L., Guzman C. A., Colwell R. R., Pruzzo C. 2008. Dual role colonization factors connecting Vibrio cholerae’s lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr. Opin. Biotechnol. 19:254–259 [DOI] [PubMed] [Google Scholar]
- 10. Blokesch M., Schoolnik G. K. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:e81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bourassa L., Camilli A. 2009. Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae. Mol. Microbiol. 72:124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tamayo R., Patimalla B., Camilli A. 2010. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 78:3560–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Faruque S. M., Islam M. J., Ahmad Q. S., Faruque A. S., Sack D. A., Nair G. B., Mekalanos J. J. 2005. Self-limiting nature of seasonal cholera epidemics: role of host-mediated amplification of phage. Proc. Natl. Acad. Sci. U. S. A. 102:6119–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jensen M. A., Faruque S. M., Mekalanos J. J., Levin B. R. 2006. Modeling the role of bacteriophage in the control of cholera outbreaks. Proc. Natl. Acad. Sci. U. S. A. 103:4652–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nelson E. J., Chowdhury A., Flynn J., Schild S., Bourassa L., Shao Y., LaRocque R. C., Calderwood S. B., Qadri F., Camilli A. 2008. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 4:e1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zahid M. S., Udden S. M., Faruque A. S., Calderwood S. B., Mekalanos J. J., Faruque S. M. 2008. Effect of phage on the infectivity of Vibrio cholerae and emergence of genetic variants. Infect. Immun. 76:5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ackermann H. W., Kasatiya S. S., Kawata T., Koga T., Lee J. V., Mbiguino A., Newman F. S., Vieu J. F., Zachary A. 1984. Classification of Vibrio bacteriophages. Intervirology 22:61–71 [DOI] [PubMed] [Google Scholar]
- 18. Goel U., Kauri T., Ackermann H. W., Kushner D. J. 1996. A moderately halophilic Vibrio from a Spanish saltern and its lytic bacteriophage. Can. J. Microbiol. 42:1015–1023 [Google Scholar]
- 19. Heidelberg J., Eisen J., Nelson W., Clayton R., Gwinn M., Dodson R., Haft D. H., Hickey E. K., Peterson J. D., Umayam L., Gill S. R., Nelson K. E., Read T. D., Tettelin H., Richardson D., Ermolaeva M. D., Vamathevan J., Bass S., Qin H., Dragoi I., Sellers P., McDonald L., Utterback T., Fleishmann R. D., Nierman W. C., White O., Salzberg S. L., Smith H. O., Colwell R. R., Mekalanos J. J., Venter J. C., Fraser C. M. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nordlund P., Reichard P. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681–706 [DOI] [PubMed] [Google Scholar]
- 21. Torrents E., Aloy P., Gibert I., Rodriguez-Trelles F. 2002. Ribonucleotide reductases: divergent evolution of an ancient enzyme. J. Mol. Evol. 55:138–152 [DOI] [PubMed] [Google Scholar]
- 22. Torrents E., Eliasson R., Wolpher H., Graslund A., Reichard P. 2001. The anaerobic ribonucleotide reductase from Lactococcus lactis. Interactions between the two proteins NrdD and NrdG. J. Biol. Chem. 276:33488–33494 [DOI] [PubMed] [Google Scholar]
- 23. Hatfull G., Jacobs-Sera D., Lawrence J., Pope W., Russell D., Ko C., Weber R. J., Patel M. C., Germane K. L., Edgar R. H., Hoyte N. N., Bowman C. A., Tantoco A. T., Paladin E. C., Myers M. S., Smith A. L., Grace M. S., Pham T. T., O'Brien M. B., Vogelsberger A. M., Hryckowian A. J., Wynalek J. L., Donis-Keller H., Bogel M. W., Peebles C. L., Cresawn S. G., Hendrix R. W. 2010. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol. 397:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lazarevic V. 2001. Ribonucleotide reductase genes of Bacillus prophages: a refuge to introns and intein coding sequences. Nucleic Acids Res. 29:3212–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pietrokovski S. 2001. Intein spread and extinction in evolution. Trends Genet. 17:465–472 [DOI] [PubMed] [Google Scholar]
- 26. Yeh Y. C., Tessman I. 1972. Control of pyrimidine biosynthesis by phage T4. II. In vitro complementation between ribonucleotide reductase mutants. Virology 47:767–772 [DOI] [PubMed] [Google Scholar]
- 27. Chiu C. S., Cox S. M., Greenberg G. R. 1980. Effect of bacteriophage T4 nrd mutants on deoxyribonucleotide synthesis in vivo. J. Biol. Chem. 255:2747–2751 [PubMed] [Google Scholar]
- 28. Yang Z., Bielawski J. P. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278:41148–41159 [DOI] [PubMed] [Google Scholar]
- 30. Chatterjee S. N., Das J., Barua D. 1965. Electron microscopy of cholera phages. Indian J. Med. Res. 53:934–937 [PubMed] [Google Scholar]
- 31. Mitra K., Ghosh A. N. 2007. Characterization of Vibrio cholerae O1 El Tor typing phage S5. Arch. Virol. 152:1775–1786 [DOI] [PubMed] [Google Scholar]
- 32. Zafar N., Mazumder R., Seto D. 2002. CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinformatics 3:12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lavigne R., Seto D., Mahadevan P., Ackermann H. W., Kropinski A. M. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406–414 [DOI] [PubMed] [Google Scholar]
- 34. Chen Z., Schneider T. D. 2005. Information theory based T7-like promoter models: classification of bacteriophages and differential evolution of promoters and their polymerases. Nucleic Acids Res. 33:6172–6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imburgio D., Rong M., Ma K., McAllister W. 2000. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry 39:10419–10430 [DOI] [PubMed] [Google Scholar]
- 36. Kemp P., Garcia L. R., Molineux I. J. 2005. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 340:307–317 [DOI] [PubMed] [Google Scholar]
- 37. Constantin de Magny G., Murtugudde R., Sapiano M. R., Nizam A., Brown C. W., Busalacchi A. J., Yunus M., Nair G. B., Gil A. I., Lanata C. F., Calkins J., Manna B., Rajendran K., Bhattacharya M. K., Huq A., Sack R. B., Colwell R. R. 2008. Environmental signatures associated with cholera epidemics. Proc. Natl. Acad. Sci. U. S. A. 105:17676–17681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kendall E., Chowdhury F., Begum Y., Khan A., Li S., Thierer J., Bailey J., Kreisel K., Tacket C. O., LaRocque R. C., Harris J. B., Ryan E. T., Qadri F., Calderwood S. B., Stine O. C. 2010. Relatedness of Vibrio cholerae O1/O139 isolates from patients and their household contacts, determined by multilocus variable-number tandem-repeat analysis. J. Bacteriol. 192:4367–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nesper J., Lauriano C., Klose K., Kapfhammer D., Kraiss A., Reidl J. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Merrell D., Hava D., Camilli A. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471–1491 [DOI] [PubMed] [Google Scholar]
- 41. Nelson E. J., Harris J. B., Morris J. G., Jr., Calderwood S. B., Camilli A. 2009. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7:693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weil A. A., Arifuzzaman M., Bhuiyan T. R., LaRocque R. C., Harris A. M., Kendall E. A., Hossain A., Tarique A. A., Sheikh A., Chowdhury F., Khan A. I., Murshed F., Parker K. C., Banerjee K. K., Ryan E. T., Harris J. B., Qadri F., Calderwood S. B. 2009. Memory T-cell responses to Vibrio cholerae O1 infection. Infect. Immun. 77:5090–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harris J. B., Khan A. I., LaRocque R. C., Dorer D. J., Chowdhury F., Faruque A. S., Sack D. A., Ryan E. T., Qadri F., Calderwood S. B. 2005. Blood group, immunity, and risk of infection with Vibrio cholerae in an area of endemicity. Infect. Immun. 73:7422–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gonzalez-Escalona N., Fey A., Hofle M. G., Espejo R. T., Guzman C. A. 2006. Quantitative reverse transcription polymerase chain reaction analysis of Vibrio cholerae cells entering the viable but non-culturable state and starvation in response to cold shock. Environ. Microbiol. 8:658–666 [DOI] [PubMed] [Google Scholar]
- 45. Sambrook J., Russell D. W. 2001. Extraction of bacteriophage λ DNA from large-scale cultures using proteinase K and SDS. In Sambrook J., Russell D. W., Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Majerczyk C. D., Dunman P. M., Luong T. T., Lee C. Y., Sadykov M. R., Somerville G. A., Bodi K., Sonenshein A. L. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zerbino D. R., Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Besemer J., Lomsadze A., Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lavigne R., Sun W. D., Volckaert G. 2004. PHIRE, a deterministic approach to reveal regulatory elements in bacteriophage genomes. Bioinformatics 20:629–635 [DOI] [PubMed] [Google Scholar]
- 51. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Laslett D., Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Darling A. E., Mau B., Perna N. T. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nei M., Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 56. Tesler G. 2002. GRIMM: genome rearrangements web server. Bioinformatics 18:492–493 [DOI] [PubMed] [Google Scholar]
- 57. Bishop A. L., Schild S., Patimalla B., Klein B., Camilli A. 2010. Mucosal immunization with Vibrio cholerae outer membrane vesicles provides maternal protection mediated by antilipopolysaccharide antibodies that inhibit bacterial motility. Infect. Immun. 78:4402–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.