Abstract
ATTED-II (http://atted.jp) is a gene coexpression database for a wide variety of experimental designs, such as prioritizations of genes for functional identification and analyses of the regulatory relationships among genes. Here, we report updates of ATTED-II focusing on two new features: condition-specific coexpression and homologous coexpression with rice. To analyze a broad range of biological phenomena, it is important to collect data under many diverse experimental conditions, but the meaning of coexpression can become ambiguous under these conditions. One approach to overcome this difficulty is to calculate the coexpression for each set of conditions with a clear biological meaning. With this viewpoint, we prepared five sets of experimental conditions (tissue, abiotic stress, biotic stress, hormones and light conditions), and users can evaluate the coexpression by employing comparative gene lists and switchable gene networks. We also developed an interactive visualization system, using the Cytoscape web system, to improve the network representation. As the second update, rice coexpression is now available. The previous version of ATTED-II was specifically developed for Arabidopsis, and thus coexpression analyses for other useful plants have been difficult. To solve this problem, we extended ATTED-II by including comparison tables between Arabidopsis and rice. This representation will make it possible to analyze the conservation of coexpression among flowering plants. With the ability to investigate condition-specific coexpression and species conservation, ATTED-II can help researchers to clarify the functional and regulatory networks of genes in a broad array of plant species.
Keywords: Arabidopsis, Comparative transcriptomics, Database, Gene coexpression, Gene network, Rice
Introduction
Genes involved in related biological pathways are cooperatively expressed to establish their functions, and thus information on their coexpression is key to understanding the biological systems at the molecular level (Eisen et al. 1998). Coexpression data have been utilized in a wide variety of experimental designs, such as gene targeting, regulatory investigations and identification of potential partners in protein–protein interactions (Aoki et al. 2007, Usadel et al. 2009, Obayashi and Kinoshita 2010). The reliable estimation of coexpressed gene relationships requires large amounts of gene expression data obtained from DNA microarray experiments, which are available in public repositories (Craigon et al. 2004, Barrett et al. 2007, Swarbreck et al. 2008, Parkinson et al. 2009). Using these large public data sources, a number of coexpression databases have been constructed and are widely used.
One such coexpression database, ATTED-II, has the unique aspect of a network representation of gene coexpression, in addition to a simple gene list representation (Obayashi et al. 2007, Obayashi et al. 2009). The user can effectively find the functional relationships of genes and design experiments to confirm the gene functions by reverse genetics and general molecular biological techniques (Obayashi and Kinoshita 2010). To analyze a broad range of biological phenomena, most of the coexpression databases, including ATTED-II, provide a general and fundamental coexpression landscape called ‘condition-independent’ coexpression data, constructed from all available samples, and thus these coexpression data are valuable for many plant researchers (Usadel et al. 2009). Although the condition-independent coexpression is useful in many cases, assessing the biological implications can be difficult, because the coexpression is an average or static view of all potential gene relationships. Some gene relationships are enhanced under specific conditions, but their relationships can disappear under different conditions. Such dynamic gene relationships are difficult to analyze with the condition-independent coexpression data. To address this issue, we sought to investigate the dynamic gene relationships by comparing the coexpression under different sets of conditions, which we call ‘condition-specific’ coexpression.
Several condition-specific coexpression data sets are available in coexpression databases, such as BAR (Toufighi et al. 2005) and CSB.DB (Steinhauser et al. 2004). Although they are quite valuable, comparative views of the condition-specific coexpression data are not provided. To construct such a comparative view of condition-specific coexpression, we used a unique coexpression measure, MR, the mutual rank of the Pearson's correlation coefficient, which performs well to compare the coexpression strengths for various guide genes and different microarray conditions (Obayashi and Kinoshita 2009).
Another limitation of the previous ATTED-II database is the target species, since only Arabidopsis data are available, in spite of the societal demand for research on advantageous plants, such as crops, vegetables and trees. Although the Arabidopsis coexpression data could potentially be applied to such useful plants using the homologous gene relationships, the validity of the application is unknown. To overcome this limitation, we constructed the rice coexpression data, and provided a comparative view of Arabidopsis and rice coexpression using gene orthologs. Since Arabidopsis and rice are model dicot and monocot plants, respectively, the conservation of coexpression between these two species supports applications of the coexpression relationships to other flowering plants. The coexpression conservation has also been used to enhance the reliability of coexpression (Stuart et al. 2003, Oti et al. 2008, Obayashi and Kinoshita 2011), since if similar coexpression relationships are observed for orthologous genes in different species, then the possibility of experimental/technical artifacts is considered to be reduced.
Details of the new features are described in the following sections, along with examples of the comparative coexpressed gene list (Figs. 1, 3) and the switchable gene network (Fig. 2). The history of ATTED-II, as well as other miscellaneous updates, is summarized in Table 1.
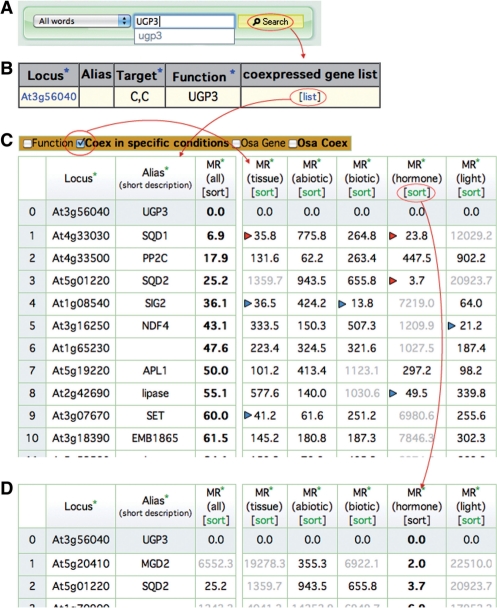
Fig. 1.
An example of condition-specific coexpression. (A) The search window on the top page of ATTED-II. The incremental search system provides a guess for the search query during the input. (B) Search results for UGP3. The link on the right is used for the coexpressed gene list. (C) Coexpressed gene list for UGP3 (At3g56040). The table on the left shows the condition-independent coexpressed gene list, sorted by MR (all) coexpression strength. When ‘Coex in specific conditions’ in the upper panel is checked, the left table appears for the five sets of condition-specific coexpression data. The colored triangles, which are not present on the database itself, are included to show strong coexpression with MR <50. (D) Same as C, but sorted by the MR (hormone) value.
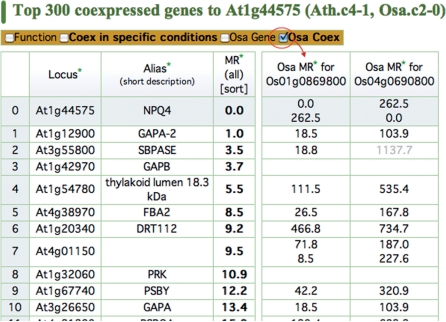
Fig. 3.
Coexpression comparison to investigate the functional orthologs, using the sequence orthologs. There are two rice orthologs for the Arabidopsis NPQ4 gene. Although both genes show similar coexpression to Arabidopsis NPQ4, Os01g0869800 shows stronger coexpression than Os04g0690800, suggesting that Os01g0869800 is the functional ortholog of Arabidopsis NPQ4.
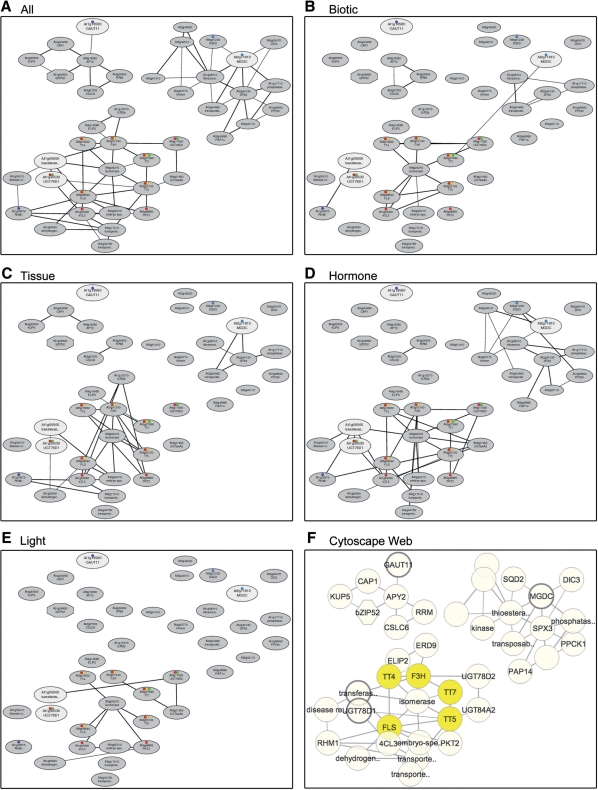
Fig. 2.
An example of NetworkDrawer to represent condition-specific coexpression (http://atted.jp/top_draw.shtml#NetworkDrawer). Four genes with UDP-glycosyltransferase activity were used. (A) Condition-independent coexpressed gene network, as a default network. (B–E) Condition-specific coexpressed gene network using biotic stress samples (B), tissue and development samples (C), hormone treatment samples (D) and light treatment samples (E). (F) The same view as A, using the Cytoscape web system, which is also available on NetworkDrawer in ATTED-II. Yellow color in a network indicates a common KEGG pathway, ‘ath00941: Flavonoid Biosynthesis’. Annotation on the Cytoscape web system can be changed interactively.
Table 1.
Evolution of ATTED-II versions over the last 3 years
| Version | Date | Arabidopsis | Rice | Gene model | Other annotations |
|---|---|---|---|---|---|
| ver.6.0 | 2010/09/28 | ↑ | c2.0 | ↑ | Updated |
| ver.5.5 | 2009/07/08 | ↑ | ↑ | TAIR9 | Updated |
| ver.5.4 | 2009/07/01 | ↑ | c1.0 | ↑ | ↑ |
| ver.5.3 | 2009/05/27 | ↑ | ↑ | Updated | |
| ver.5.2 | 2008/07/19 | ↑ | TAIR8 | Updated | |
| ver.5.1 | 2008/04/08 | c4.1 | ↑ | ↑ | |
| ver.5.0 | 2008/03/18 | c4.0 | TAIR7 | Updated |
↑The same condition as the previous version. Details of the updates of ‘Other annotations’ are available from http://atted.jp/versions.shtml
Results and Discussion
Condition-specific coexpression
We prepared condition-specific coexpression data sets for the following five conditions: tissues, abiotic stresses, biotic stresses, hormones and light conditions, which were obtained from the AtGenExpress international collaboration (Schmid et al. 2005, Kilian et al. 2007, Goda et al. 2008). Fig. 1 shows how to use the condition-specific coexpression views in the coexpressed gene list. The UGP3 gene is used as an example, since Okazaki et al. (2009) recently identified the enzyme it encodes as being involved in sulfolipid biosynthesis. They analyzed and discussed condition-specific coexpression, and thus this gene represents a good example to evaluate the condition-specific coexpression view on the new ATTED-II. On the top page of ATTED-II, the gene name can be searched on the main search window, using either the AGI code, the gene name or a keyword (Fig. 1A). As a result, one locus, At3g56040, was found as a UGP3 gene (Fig. 1B). The coexpressed gene list can be viewed by clicking the link on the right side, which will yield a simple list of the top 300 coexpressed genes for the query (the table on the left in Fig. 1C). In the list, the locus ID, the alias name and the coexpression MR value are shown as a default. The top line is the guide gene itself, i.e. UGP3 in this case, and the next lines show the most strongly coexpressed genes. The product of UGP3 was originally identified as an enzyme involved in sulfolipid biosynthesis, based on the gene coexpression relationships with the sulfolipid (SQD1 and SQD2) and galactolipid biosynthesis genes (MGD2 and MGD3) (Okazaki et al. 2009). This coexpressed gene list shows that two sulfolipid genes (SQD1 and SQD2) are strongly coexpressed with UGP3, in ranking positions 1 and 3 (a smaller MR means a stronger coexpression).
Additional information is available by clicking the checkbox in the operation panel above the table. When‘Coex in specific conditions’ in the panel is checked, the five sets of condition-specific coexpression data will appear (the table on the right in Fig. 1C), where weak coexpression data with MR >1,000 are shown in gray, and the data in the column used for sorting are shown in bold face. The condition-specific coexpression patterns are different for each coexpressed gene. For example, SQD1 is strongly coexpressed under the tissue and hormone conditions, while SQD2 is specifically coexpressed under the hormone condition, as indicated by the red triangles in the figure. Please note that the colored triangles shown in Fig. 1C to show strong coexpression with MR <50 are not on the database but are for ease of viewing. In a similar way (see blue triangles in the figure), the pattern of condition-specific coexpression is diverse. These complicated coexpression patterns may suggest the multiple aspects of UGP3 functions, or simply reflect the evolutionary history of this gene, as described by Okazaki et al. (2009). In addition, Okazaki et al. discussed the coordinated regulation between sulfolipid and galactolipid biosynthesis under hormone control, including the auxin/cytokinin cross-talk system (Okazaki et al. 2009). This hormone regulation can be analyzed by sorting the coexpression table for hormone condition (Fig. 1D), which revealed strong coexpression of UGP3 with SQD1 (MR = 23.8), SQD2 (MR = 3.7), MGD2 (MR = 2.0) and MDG3 (MR = 13.1).
Condition-specific coexpression is a powerful method to investigate regulatory mechanisms, as shown in the above example. However, the calculation of the condition-specific coexpression requires the experimental sets to be carefully defined, which is difficult to accomplish with the rapidly growing amount of microarray data that are publicly available. One possible means to find appropriate groups of conditions automatically is to perform a principal component analysis of the conditions. The coexpression trend under the iterative subtraction of major contributing conditions is one way to summarize multiple condition-specific coexpression data. We provide this coexpression trend as the coexpression stability in the CoexViewer tool (Kinoshita and Obayashi 2009). A link icon to the CoexViewer accompanies each gene on the coexpressed gene list (not shown in Fig. 1).
Improvement of NetworkDrawer to represent condition-specific coexpression
Condition-specific coexpression data are also available on gene networks. ATTED-II provides a tool called NetworkDrawer, in Draw box, to draw gene networks. Fig. 2A is the original view of the coexpressed gene network generated by NetworkDrawer, which uses condition-independent coexpression, when we used four genes with UDP-glycosyltransferase activity (At1g06000, At1g18580, At1g30530 and At2g11810). In the gene networks, the bigger, white nodes (circles) indicate the query genes, the smaller, gray nodes are additional genes to draw the coexpressed gene networks, and the black edges (lines) show the gene coexpression. As seen in the network, these four query genes are involved in three distinct coexpression networks (Fig. 2A). By following the color of the nodes, the user can check the common KEGG annotations among the genes on the networks. For example, the yellow dots in the networks correspond to ‘ath00941: Flavonoid Biosynthesis’ in the KEGG pathway. The detailed KEGG map view in GenomeNet is available from the summary table in NetworkDrawer. By using this functionality, the biological meaning of each network can easily be interpreted. Fig. 2B–E shows the condition-specific coexpression relationships. The coexpression under biotic stress conditions (Fig. 2B) reveals a weak link between the bottom and upper-right clusters, suggesting conditional relationships between the two functional clusters. The gene coexpression under tissue (Fig. 2C), hormone (Fig. 2D) and light (Fig. 2E) conditions is similar to the condition-independent coexpression (Fig. 2A), but the coexpression under the hormone conditions is the tightest, which suggests that these coexpression networks mainly reflect hormone-driven conditions.
In addition to these network pictures, we prepared an interactive network drawer, using the Cytoscape Web system (Lopes et al. 2010), which is a web implementation of Cytoscape (Shannon et al. 2003). Fig. 2F shows the same coexpression network with Cytoscape Web on ATTED-II, where the user can zoom in and out, edit node positions and obtain detailed information about the node by placing the cursor over each node or clicking on it. Coexpressed gene networks marked with several genome-wide annotations, such as predicted protein subcellular localizations, are available on the Cytoscape Web system. Moreover, we prepared a scheme to use stand-alone Cytoscape for further analyses, where any genome-wide information can be overlaid on the coexpressed gene network. The GraphML file provided in the NetworkDrawer can be imported in Cytoscape using our graphmlreader plug-in, by simply dragging and dropping the GraphML file link in NetworkDrawer to the Cytoscape window. Details about the use of Cytoscape are described on our web page (http://atted.jp/help/cytoscape.shtml).
Gene coexpression for different species
To expand the coexpression analyses with other useful plant species, homologous gene information is used. If a gene pair in one species is coexpressed, then the homologous gene pairs in the other species can be hypothesized to be coexpressed to a similar extent. This assumption may be true for some closely related species, but is less reliable for distantly related species. Some changes in coexpression patterns during evolution have been observed in animal coexpression (Obayashi and Kinoshita 2011). To check the feasibility of coexpression conservation, we prepared coexpression data from rice and provided comparative views in the coexpressed gene list. If a coexpression relationship is conserved in the two species, then the conservation of the gene coexpression in all flowering plants can be expected with more confidence. The details for the raw data used to calculate the rice coexpression are shown in Table 2. Please note that although condition-independent coexpression data are used for the Arabidopsis and rice comparison, some important experimental conditions may be absent in one of the species, which may reduce the conserved coexpression.
Table 2.
Coexpression data information in ATTED-II
| Species | Sample type | Version | Number of genes | Number of TAIR-ID | Number of microarray slides | Microarray platform | Data source | Last update |
|---|---|---|---|---|---|---|---|---|
| Arabidopsis | All | Ath.c4.1 | 20969 | 58 | 1388 | GPL198 | TAIR | 2008/04/08 |
| Arabidopsis | Tissue | Ath.c4.1_tis | 20969 | 1 | 236 | GPL198 | TAIR | 2010/09/28 |
| Arabidopsis | Stress | Ath.c4.1_str | 20969 | 9 | 534 | GPL198 | TAIR | 2010/09/28 |
| Arabidopsis | Hormone | Ath.c4.1_hor | 20969 | 13 | 200 | GPL198 | TAIR | 2010/09/28 |
| Arabidopsis | Biotic | Ath.c4.1_bio | 20969 | 6 | 200 | GPL198 | TAIR | 2010/09/28 |
| Arabidopsis | Light | Ath.c4.1_lig | 20969 | 1 | 48 | GPL198 | TAIR | 2010/09/28 |
| Rice | All | Osa.c2.0 | 20204 | 20 | 310 | GPL2025 | ArrayExpress | 2010/09/28 |
In Fig. 3, the table on the left shows the Arabidopsis coexpressed gene list from NPQ4, which functions in non-photochemical quenching in PSII. As in Fig. 1C, the top line is the guide gene itself, and the following lines indicate the most strongly coexpressed genes ordered by their coexpression strength, as measured by MR. Checking ‘Osa Coex’ in the upper checkbox will show the orthologous coexpression in rice, in the columns on the right. There are two rice orthologs for NPQ4, according to the homolog information in HomoloGene (Sayers et al. 2010), and thus two columns for rice are shown in the table on the right.
In the rice coexpression table (the ‘MR for Os01g0869800’ column in the table), the gene on the left in the second line shows the coexpression between the rice ortholog to Arabidopsis NPQ4 (i.e. Os01g0869800) and that to GAPA-2, with a coexpression strength of 18.5. This value indicates very strong coexpression, because the MR value ranges from 1 to the total number of genes in the species for the coexpression analysis (about 20,000 for both Arabidopsis and rice). In the same way, most of the coexpression in this list is conserved between Arabidopsis and rice, and, therefore, these coexpressed relationships are expected to be conserved in other flowering plant species. It is noteworthy that this conservation of the coexpressed gene list supports not only each coexpression relationship but also the reliability of the original expression pattern of NPQ4 measured by microarray, because such conserved coexpression would not be expected from pseudo expression data produced by microarray technical artifacts. Note that in some cases, such as the seventh line for At4g01150 in the rice orthologs (‘MR for Os01g0869800’ columns), there are multiple orthologous genes in a species, and the coexpression values are shown in parallel in a single cell (71.8 and 8.5).
In addition to evaluating the reliabilities of coexpression between a pair of genes and the expression pattern of the guide gene, a coexpression comparison can be used to identify ‘functional orthologs’ among genes with similar sequences. In Fig. 3, there are two rice ‘orthologs’ or genes with similar sequences to the Arabidopsis guide gene (NPQ4). Both of the rice genes show strong coexpression with the orthologous genes of the coexpressed genes in Arabidopsis. However, for almost all of the coexpressed gene relationships, the gene on the left side (Os01g0869800) shows stronger coexpression, suggesting that Os01g0869800 is more likely to be a functional ortholog, rather than the gene on the right (Os040690800). The gene on the right might have acquired some other cellular functions after gene duplication. In addition to the analysis for an Arabidopsis coexpressed gene list, the same analysis can be performed for the rice coexpressed gene lists, which are available from a link that will appear by turning on the ‘Osa gene’ checkbox in the operation panel.
Other improvements
The top page of ATTED-II was revised to intuitively show the four major functions of our database. ATTED-II has nine tools, in addition to the pre-calculated pages. To access these tools easily, we classified them into two categories, ‘Search box’ and ‘Draw box’. The Search box is composed of tools to search genes by some aspects, such as coexpression and protein–protein interactions, and their outputs are gene tables. On the other hand, the tools in Draw box can draw various pictures, such as a gene network, a hierarchical gene cluster and a detailed view of gene coexpression. There are two additional boxes, ‘Browse’ and ‘Bulk download’. The Browse box has a link to the gene networks for six subcellular locations. These gene networks are huge, and, therefore, they are provided as a Google Map interface. The Bulk download box is used for downloading the coexpression data of Arabidopsis and rice, as well as other miscellaneous tables, to construct ATTED-II. These data are now available under the Creative Commons Attribution license.
API is available to retrieve gene coexpression at the following URL: http://atted.jp/API/coex/(AGIcode)/(coexpression_measure)/(cutoff). Details of API are described at http://atted.jp/help/API.shtml.
Materials and Methods
Data sources
To calculate the coexpression data, we used the gene expression data measured by Affymetrix GeneChip platforms. For the condition-specific coexpression data, the AtGenExpress data were used, which are downloadable from TAIR (Swarbreck et al. 2008). Some microarray data were derived from diverse sample backgrounds, such as different tissues and genotypes. When the sample background differences are distinct, variations in the sample background can easily hide the responses to the treatments when calculating the gene-to-gene Pearson's correlation coefficient. Therefore, we independently applied per-gene normalization to the microarray data for each sample background, to ignore the background differences. Details of the calculations are available at http://atted.jp/help/coex_cal.shtml. The GeneChip data for rice were obtained from ArrayExpress. The details of the ATTED-II coexpression data are provided in Table 2 and in our database (http://atted.jp/top_statistics.shtml).
Measuring gene coexpression
To evaluate the strength of coexpression, the Pearson and Spearman correlation coefficients are widely used, but we found that these measures were not suitable for direct comparisons among different species. However, the correlation rank-based measure, MR (which is calculated as the geometric mean of the correlation rank of gene A to gene B and of gene B to gene A), is more suitable to compare the coexpression data in multiple species, on average (Obayashi and Kinoshita 2009). In addition, we have performed several successful case studies, using MR coexpression data, to identify new gene functions in applications for Arabidopsis (Obayashi and Kinoshita 2010). Therefore, we adopted MR as the coexpression measure in ATTED-II, to compare the coexpression strengths among multiple species.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology, Japan [Grants-in-Aid for Scientific Research (No. 21770035) and for Publication of Scientific Research Results (No. 228063) to T.O., and a Grant-in-Aid for Innovative Areas ‘HD physiology’ (No. 22136005) to K.K.].
Acknowledgments
We thank Dr. Keiichiro Ono (UCSD Medical School) for valuable discussions about the utilization of Cytoscape. Super-computing resources were provided by the Human Genome Center, Institute of Medical Science, The University of Tokyo.
Glossary
Abbreviations
- MR
mutual rank.
References
- Aoki K., Ogata Y., Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48:381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., Sasaki E., Akiyama K., Maruyama-Nakashita A., Nakabayashi K., et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Obayashi T. Multi-dimensional correlations for gene coexpression and application to the large-scale data of Arabidopsis. Bioinformatics. 2009;25:2677–2684. doi: 10.1093/bioinformatics/btp442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 2009;37:D987–D991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009;16:249–260. doi: 10.1093/dnares/dsp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K. Coexpression landscape in ATTED-II: usage of gene list and gene network for various types of pathways. J. Plant Res. 2010;123:311–319. doi: 10.1007/s10265-010-0333-6. [DOI] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K. COXPRESdb: a database to compare gene coexpression in seven model animals. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1147. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., et al. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Shimojima M., Sawada Y., Toyooka K., Narisawa T., Mochida K., et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell. 2009;21:892–909. doi: 10.1105/tpc.108.063925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oti M., van Reeuwijk J., Huynen M.A., Brunner H.G. Conserved co-expression for candidate disease gene prioritization. BMC Bioinformatics. 2008;9:208. doi: 10.1186/1471-2105-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H., Kapushesky M., Kolesnikov N., Rustici G., Shojatalab M., Abeygunawardea N., et al. ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., et al. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser D., Usadel B., Luedemann A., Thimm O., Kopka J. CSB.DB: a comprehensive systems-biology database. Bioinformatics. 2004;20:3647–3651. doi: 10.1093/bioinformatics/bth398. [DOI] [PubMed] [Google Scholar]
- Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Swarbreck D., Wilks C., Lamesch P., Berardini T.Z., Garcia-Hernandez M., Foerster H., et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. The Botany Array Resource: e-Northerns, expression angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Usadel B., Obayashi T., Mutwil M., Giorgi F.M., Bassel G.W., Tanimoto M., et al. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009;32:1633–1651. doi: 10.1111/j.1365-3040.2009.02040.x. [DOI] [PubMed] [Google Scholar]