Abstract
Autophagy is an intracellular lysosomal (vacuolar) degradation process that is characterized by the formation of double-membrane vesicles, known as autophagosomes, which sequester cytoplasm. As autophagy is involved in cell growth, survival, development and death, the levels of autophagy must be properly regulated, as indicated by the fact that dysregulated autophagy has been linked to many human pathophysiologies, such as cancer, myopathies, neurodegeneration, heart and liver diseases, and gastrointestinal disorders. Substantial progress has recently been made in understanding the molecular mechanisms of the autophagy machinery, and in the regulation of autophagy. However, many unanswered questions remain, such as how the Atg1 complex is activated and the function of PtdIns3K is regulated, how the ubiquitin-like conjugation systems participate in autophagy and the mechanisms of phagophore expansion and autophagosome formation, how the network of TOR signaling pathways regulating autophagy are controlled, and what the underlying mechanisms are for the pro-cell survival and the pro-cell death effects of autophagy. As several recent reviews have comprehensively summarized the recent progress in the regulation of autophagy, we focus in this Commentary on the main unresolved questions in this field.
Keywords: Apoptosis, Cell death, Lysosome, Vacuole, Yeast
Introduction
Autophagy is a stress-induced catabolic process involving the lysosome (or, in yeast, the analogous vacuole), which is conserved in all eukaryotes (Esclatine et al., 2009; Klionsky, 2005). According to the different pathways by which cargo is delivered to the lysosome or vacuole, autophagy is divided into three main types: chaperone-mediated autophagy (CMA), microautophagy and macroautophagy (Klionsky, 2005). CMA is a process that has been characterized in higher eukaryotes but not in yeast. In CMA, a chaperone protein binds first to its cytosolic target substrate and then to a receptor on the lysosomal membrane where the unfolding of the protein occurs. The unfolded cytosolic target protein is subsequently translocated directly into the lysosome for its degradation (Massey et al., 2004). Microautophagy translocates cytoplasmic materials into the lysosome or vacuole for degradation by either direct invagination, protrusion, or septation of the lysosomal or vacuolar membrane (Wang and Klionsky, 2004). Macroautophagy is characterized by the formation of a cytosolic double-membrane vesicle, the autophagosome. During macroautophagy, cytoplasmic proteins, organelles or other materials are surrounded by phagophores, which expand and close to form autophagosomes. These autophagosomes fuse with lysosomes (or vacuoles) to form autolysosomes, in which the cytoplasmic cargos are degraded by resident hydrolases. The resulting degradation products are then transported back into the cytosol through the activity of membrane permeases for reuse (Klionsky, 2007) (Fig. 1). In the yeast Saccharomyces cerevisiae, microautophagy engulfs cytosolic materials through an autophagic tube, which then scissions within the vacuole to release the contents into a vesicle within the vacuolar lumen for degradation (Uttenweiler and Mayer, 2008); microautophagy-like processes, such as one type of selective peroxisome degradation, are slightly different and involve targeted sequestration of the cargo (Dunn et al., 2005). The process and mechanism of microautophagy in mammalian cells are still not clear (Cuervo, 2004). Among the three main forms of autophagy, macroautophagy is the most widely studied and best characterized process. In this review, we will thus focus on macroautophagy, hereafter referred to as autophagy.
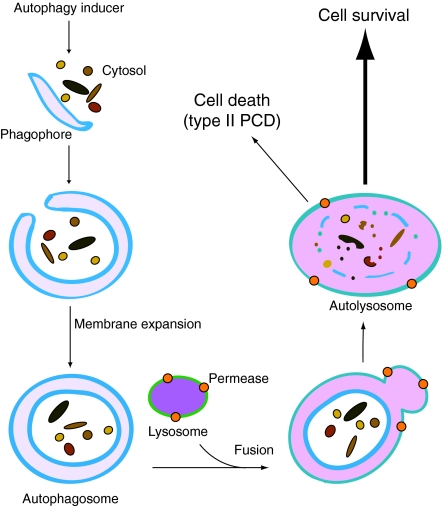
Fig. 1.
The autophagy pathway and its roles in cell survival and cell death. In the presence of an autophagy inducer, cytoplasmic materials, such as protein aggregates and organelles, are sequestered by a pre-autophagosomal membrane structure, the phagophore. The phagophore membrane then expands and encloses its cargo to form a double-membrane vesicle, the autophagosome. The autophagosome fuses with a lysosome (or a vacuole in yeast) to form an autolysosome, in which the enclosed cargo is degraded by acid hydrolases. After the resulting macromolecules are transported back into the cytosol through membrane permeases, they can either be used to synthesize proteins or can be oxidized by the mitochondria to generate ATP for cell survival. However, when autophagy occurs at excessive levels or under certain physiological conditions it can lead to type II programmed cell death (type II PCD). See the text for additional details.
Although autophagy is generally considered to be nonspecific, there are many examples of selective autophagy, including mitophagy (for mitochondria), ribophagy (for ribosomes), pexophagy (for peroxisomes) and reticulophagy (for the endoplasmic reticulum, ER) (He and Klionsky, 2009). By contrast, the yeast cytoplasm-to-vacuole targeting (Cvt) pathway is a biosynthetic pathway used to transport the vacuolar hydrolases α-mannosidase and aminopeptidase I (Ape1) from the cytosol to the vacuole under normal growth conditions. As the Cvt pathway shares the core autophagy machinery, which is composed of 17 autophagy-related (Atg) proteins found in all autophagy pathways, it is also defined as a type of selective autophagy (Inoue and Klionsky, 2010; Lynch-Day and Klionsky, 2010).
The primary role of autophagy is to protect cells under stress conditions, such as starvation. During periods of starvation, autophagy degrades cytoplasmic materials to produce amino acids and fatty acids that can be used to synthesize new proteins or are oxidized by mitochondria to produce ATP for cell survival (Levine and Yuan, 2005) (Fig. 1). However, when autophagy is excessively induced, it can result in autophagic cell death, so-called type II programmed cell death (PCD) (Fig. 1), which is distinct from type I PCD (apoptosis) and from necrosis (Chen et al., 2010; Levine and Yuan, 2005; Maiuri et al., 2007; Platini et al., 2010). In addition to stress management, autophagy is involved in normal development (Levine and Klionsky, 2004), senescence (Young et al., 2009), lifespan extension (Vellai et al., 2009), immunity and defense against microbial invasion (Deretic and Levine, 2009). Autophagy also has a role in many human pathophysiologies, such as cancer, myopathies, neurodegeneration, heart and liver diseases, and gastrointestinal disorders (Klionsky, 2005; Mizushima et al., 2008).
One fundamental conclusion that can be drawn from research on autophagy is that this process must be tightly regulated – too little or too much autophagy can be deleterious. In particular, if we hope to ever use autophagy for therapeutic purposes, it will be crucial to understand the details of its regulatory processes. The regulation of autophagy has been extensively studied in the past few years, and several reviews have thoroughly summarized the progress in this area (Bassham, 2009; Cebollero and Reggiori, 2009; Esclatine et al., 2009; Fimia and Piacentini, 2010; He and Klionsky, 2009; Meijer and Codogno, 2009; Yang and Klionsky, 2010). Therefore, in this review, we will focus on some of the main unresolved questions.
The role of the Atg1 complex in autophagy induction
The Atg1 complex is an essential component for autophagy induction
The Atg1 complex is involved in autophagy induction (see Box 1) and the target of rapamycin (TOR) – specifically, TOR complex 1 (TORC1) – is an upstream negative regulator of this complex (Chang and Neufeld, 2010; He and Klionsky, 2009). Differences exist in the regulation of autophagy by TORC1 among eukaryotes. For example, in yeast, TORC1 phosphorylation of Atg13 must be eliminated before it can interact with and activate Atg1 (Fig. 2A); however, mammalian Atg13 (ATG13) and Drosophila Atg13, the homologs of yeast Atg13, are always associated with ULK (ULK1 or ULK2, the mammalian homologs of yeast Atg1) and with Drosophila Atg1 (the Drosophila homolog of yeast Atg1), respectively, and Drosophila Atg13 becomes hyperphosphorylated during autophagy (Fig. 2B,C). The phosphorylation sites of the ULK complex in mammals have been mapped (Dorsey et al., 2009); mapping these sites and identifying the relevant kinases for the yeast proteins is underway (Kijanska et al., 2010; Yeh et al., 2010) and might provide additional information that resolves the apparent differences among these organisms. Another difference is that, in Drosophila, TORC1 binds to the Atg1 complex even when autophagy is induced (Fig. 2C), whereas in yeast and mammals TORC1 dissociates from their Atg1 complexes during autophagy (Fig. 2A,B). It is worthwhile pointing out that the Atg1 complex has not been fully characterized with regard to dynamic associations between its components, particularly in yeast.
Box 1. The core machinery of autophagy
In eukaryotes, the core machinery of autophagy includes the following four steps:
Induction
Induction is initiated by activation of the Atg1 complex. In yeast, the components of this complex include Atg1, Atg13 and the Atg17–Atg31–Atg29 subcomplex. The mammalian Atg1 complex (or ULK complex) is composed of the mammalian Atg1 homolog Unc-51-like kinases 1 or 2 (ULK1 or ULK2, respectively), mammalian autophagy related 13 homolog (ATG13), a putative counterpart of yeast Atg17, the RB1-inducible coiled-coil 1 (RB1CC1; also known as FIP200, focal adhesion kinase (FAK)-family-interacting protein of 200 kD) and Atg101 (an Atg13-binding protein).
Vesicle nucleation
This is the initial step that recruits proteins and lipids for autophagosome construction. Nucleation starts with the recruitment of Atg proteins to the phagophore assembly site (PAS) in yeast or to the PAS equivalent in mammals. The mechanism for nucleation is unclear, but activation of a phosphatidylinositol 3-kinase (PtdIns3K) complex is essential.
Vesicle expansion and completion
Autophagosome formation is a de novo process, in which membranes emerging at the PAS expand, either by direct flow from a source such as the ER or by vesicular addition, and then seal to enclose the cytosolic cargos. Two ubiquitin-like (Ubl) conjugation systems (Atg8 and Atg12) are involved in vesicle expansion and completion. Atg8 is conjugated to the lipid phosphatidylethanolamine (PE), whereas Atg12 is conjugated to Atg5.
Autolysosome formation
When the autophagosome is completed, it will fuse with the lysosome (the vacuole in yeast) to form an autolysosome, in which the cytosolic cargos will be degraded. The resulting products will be released back into the cytosol through permeases.
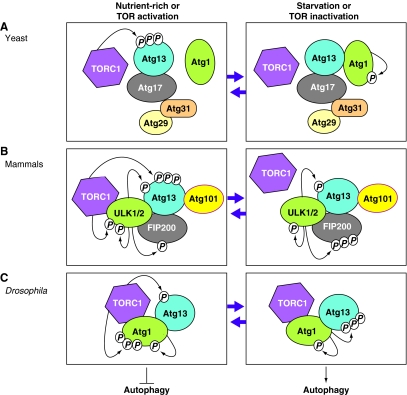
Fig. 2.
Dynamics of Atg1 complexes upon autophagy induction in different eukaryotes. (A) In yeast, under nutrient-rich conditions, the active TOR complex 1 (TORC1) hyperphosphorylates Atg13 (Kamada et al., 2010). This prevents the association of Atg1 with Atg13, which is bound to Atg17, Atg31 and Atg29, leading to inhibition of autophagy induction. Under starvation conditions when TORC1 is inactivated, Atg13 is no longer phosphorylated by TORC1, whereas Atg1 is autophosphorylated, leading to the association of Atg1 with the complex between Atg13, Atg17, Atg31 and Atg29, and subsequent autophagy induction (Cebollero and Reggiori, 2009; Chang and Neufeld, 2010; Kamada et al., 2010; Nakatogawa et al., 2009). (B) In contrast to yeast, mammalian ULK (ULK1 or ULK2, the homologs of yeast Atg1) forms a stable complex with mammalian Atg13, FIP200 (a putative counterpart of yeast Atg17) and Atg101 (an Atg13-binding protein), irrespective of TORC1 activation. Under nutrient-rich conditions, the active TORC1 associates with the ULK complex (ULK1 (or ULK2)–Atg13–FIP200-Atg101), phosphorylates ULK1 (or ULK2) and hyperphosphorylates Atg13, which inhibits the kinase activity of ULK1 (or ULK2) and thus blocks autophagy induction. Under starvation conditions when TORC1 is inactivated, TORC1 dissociates from the ULK complex, preventing phosphorylation of Atg13 and ULK1 (or ULK2) by TORC1 and leading to autophagy induction, whereas ULK1 (or ULK2) still phosphorylates Atg13 and itself, and hyperphosphorylates FIP200 (Chang and Neufeld, 2010; Mizushima, 2010; Yang and Klionsky, 2010). (C) Similar to the situation in mammals, in Drosophila Atg1 forms a complex with Atg13 irrespective of TORC1 activation (Chang and Neufeld, 2010). Under nutrient-rich conditions, the active TORC1 phosphorylates Atg13 and hyperphosphorylates Atg1, leading to the inhibition of autophagy induction. Under starvation conditions, when TORC1 is inactivated, Atg1 and Atg13 are no longer phosphorylated by TORC1, whereas Atg1 still phosphorylates itself and hyperphosphorylates Atg13, leading to autophagy induction. Figure modified from Chang and Neufeld (Chang and Neufeld, 2010) with permission.
In Caenorhabditis elegans, the Atg1 homolog uncoordinated-51 (UNC-51) (Meléndez and Levine, 2009), the Atg13 homolog ectopic PGL granules (EPG-1) (Tian et al., 2009), and possibly also other recently identified Atg proteins (e.g. EPG-7 or FIP200) (Tian et al., 2010) participate in autophagy. However, the functions of the Atg1 complex and its other components remain unclear in this organism (Meléndez and Levine, 2009).
How is the Atg1 complex activated?
One common feature of the activation of Atg1 complexes in different organisms is that the respective Atg13 proteins are no longer phosphorylated by TORC1 once autophagy has been induced (Fig. 2). A remaining issue, however, is whether this change in phosphorylation is essential for Atg1 complex activation and autophagy induction. Support for this idea is seen in yeast, as a non-phosphorylatable Atg13 mutant can bypass the TORC1 pathway and induce autophagy in growing cells (Kamada et al., 2010). In Drosophila, however, Atg13 is hyperphosphorylated under starvation conditions, which is dependent on Atg1 kinase activity (Chang and Neufeld, 2010). Furthermore, as ULK1 or ULK2 (in mammals) and Atg1 (in Drosophila) phosphorylate Atg13 during autophagy (Chang and Neufeld, 2010), does Atg1 also phosphorylate Atg13 in yeast? Under starvation conditions, ULK1, ULK2 (in mammals) and Atg1 (in Drosophila) are no longer phosphorylated by their respective TORC1s (Fig. 2B,C). Thus, similar to Atg13 in yeast, it is possible that activation of the ULK1, ULK2 and Atg1 complexes also involves the elimination of TORC1-dependent phosphorylation.
One additional issue that merits consideration is the physiological mechanism for downregulating autophagy. Most studies to date have focused on autophagy induction, but excessive autophagy is also deleterious. How do cells modulate the level of autophagy once it has been induced? A recent study suggests that death-associated protein 1 (DAP1) has a function in this process (Koren et al., 2010); however, it remains to be determined whether this protein targets the Atg1 complex, or other components of the autophagy pathway. The study of Atg1 effectors that bind to Atg1 and thereby regulate its activity, would greatly help in understanding the ways cells modulate the level of autophagy. However, no Atg1 effectors have been characterized outside of yeast (Kamada et al., 2000).
Regulation of autophagy by the phosphatidylinositol 3-kinase complex
The PtdIns3K complex is involved in autophagy induction
The activation of a phosphatidylinositol 3-kinase (PtdIns3K) complex is an essential step in vesicle nucleation during autophagy (Box 1). In yeast, there are two PtdIns3K complexes. Complex I, which functions in autophagy, consists of Atg6 (Vps30), Atg14, Vps34 (the only PtdIns3K in yeast) and Vps15, whereas complex II contains Vps38 instead of Atg14 and functions in the vacuolar protein sorting (Vps) pathway (Cao and Klionsky, 2007; Nakatogawa et al., 2009).
In contrast to yeast, higher eukaryotes have three types of PtdIns3K (class I, II and III); the class I and III PtdIns3K complexes function as negative and positive regulators of autophagy, respectively (Baehrecke, 2005; Chang and Neufeld, 2010). The mammalian class III PtdIns3K complex is composed of beclin 1 (BECN1), phosphoinositide-3-kinase class 3 (PIK3C3, hereafter referred to as Vps34) and phosphoinositide-3-kinase, regulatory subunit 4 (PIK3R4, hereafter referred to as Vps15) (Funderburk et al., 2010; Yang and Klionsky, 2010). Atg14L (yeast Atg14-like) (Matsunaga et al., 2009; Zhong et al., 2009), activating molecule in beclin-1-regulated autophagy (AMBRA1) (Di Bartolomeo et al., 2010; Fimia et al., 2007), UV radiation resistance-associated gene (UVRAG) protein (Liang et al., 2008) and Bax-interacting factor 1 (Bif1) (Takahashi et al., 2007) positively regulate autophagy, whereas the RUN domain and cysteine-rich domain containing beclin-1-interacting protein (rubicon) (Matsunaga et al., 2009; Zhong et al., 2009) negatively regulates autophagy through interacting with the class III PtdIns3K. UVRAG (Liang et al., 2008) and rubicon (Matsunaga et al., 2009; Zhong et al., 2009) also regulate the endocytic pathway. The antiapoptotic Bcl-2 family members, such as Bcl-2 (BCL2), viral Bcl-2 homolog (vBcl-2) and Bcl-xL (BCL2L1), inhibit autophagy by binding to the BH3 domain of beclin 1 (He and Levine, 2010). Here, Bcl-2 originating from the ER rather than mitochodria is responsible for negatively regulating autophagy (He and Levine, 2010).
Similar to mammals, the class III PtdIns3K complex in Drosophila contains Atg6, Vps15 and Vps34; orthologs of Atg14L, UVRAG and Rubicon are also found in the Drosophila genome (Chang and Neufeld, 2010). In C. elegans, the class III PtdIns3K complex contains LET-512 (Vps34), ZK930.1 (Vps15) and BEC-1 (Atg6), and regulatory factors CED-9 (Bcl-2) and ERP-1 (Bif-1), but not an Atg14 equivalent, have been identified (Meléndez and Levine, 2009).
How is the function of PtdIns3K regulated?
The mammalian class III PtdIns3K has been divided into different types of complex based on the interaction of its core components (beclin 1, Vps34 and Vps15) with different interacting proteins (Funderburk et al., 2010; He and Levine, 2010). Proteins such as UVRAG, AMBRA1, Atg14L, and Bif-1 positively regulate autophagy, whereas other class III PtdIns3K-interacting proteins such as rubicon, Bcl-2 and Bcl-xL are negative regulators. Thus, what, if any, crosstalk occurs among these proteins, and how is this coordinated to regulate the function of class III PtdIns3K in autophagy induction? Further study of the roles of these class III PtdIns3K-interacting proteins under the same autophagy-inducing conditions might help answer these questions.
The role of vesicle expansion and completion in the regulation of autophagy
Atg8 and Atg12 are parts of unique Ubl conjugation systems
The Atg8 and Atg12 ubiquitin-like (Ubl) conjugation systems are involved in vesicle expansion and completion (see Box 1 and Fig. 3 for details). Conjugation reactions of Atg12 and Atg8 are catalyzed by the E1-like enzyme Atg7, and the E2-like enzymes Atg10 (for Atg12) and Atg3 (for Atg8). The cysteine protease Atg4 is required to remove an arginine residue (Arg117 in yeast) from nascent Atg8 for its maturation, and also to cleave Atg8 from phosphatidylethanolamine (PE) on the autophagosome membrane (referred to as deconjugation) after vesicle completion. Atg12 is conjugated to the Atg5 protein, and the Atg12–Atg5 complex might act as an E3 ubiquitin ligase homolog to catalyze the conjugation of Atg8 to PE. A number of observations suggest that there is crosstalk between the Atg8–PE and Atg12–Atg5 conjugation systems. First, the E1-like enzyme Atg7 activates both Atg12 and Atg8 (or its mammalian homolog LC3). Second, conjugation of Atg8 (LC3) to PE, and the subsequent autophagosome formation are dependent on the Atg12–Atg5-Atg16 complex (Geng and Klionsky, 2008).
Fig. 3.
The Atg8 and Atg12 ubiquitin-like conjugation systems in yeast. Atg8 (or the mammalian homolog LC3) is an ubiquitin-like (Ubl) protein that is cleaved by the cysteine protease Atg4 to expose a glycine residue that is then covalently conjugated with phosphatidylethanolamine (PE) (Ichimura et al., 2000). Atg12 is another Ubl protein that is covalently conjugated to Atg5 and the resulting Atg12–Atg5 conjugate is then attached to Atg16 to form a dimer of the trimeric Atg12–Atg5–Atg16 complex (Geng and Klionsky, 2008). Conjugation reactions of Atg12 and Atg8 are catalyzed by the E1-like enzyme Atg7 and the E2-like enzymes Atg10 (for conjugation of Atg12) and Atg3 (for Atg8) and result in the formation of either a multimeric complex with Atg5 (involving Atg12) or a lipid conjugate (with Atg8). In contrast to Atg5 conjugation to Atg12, Atg8 is conjugated to PE and not to a protein, thereby allowing membrane association. Furthermore, Atg4 cleaves the Atg8 (or LC3) precursor before conjugation and Atg8–PE (or LC3-II) during the subsequent deconjugation. Figure modified from Geng and Klionsky (Geng and Klionsky, 2008) with permission.
How do the ubiquitin-like conjugation systems participate in autophagy?
One of the main questions in this particular area is how the activity of Atg4 is controlled to prevent the premature deconjugation of Atg8 or LC3 from PE during autophagosome formation. One hypothesis that has been suggested is that the reactive oxygen species (ROS) H2O2 can inactivate Atg4 by oxidizing its catalytic cysteine residue to prevent cleavage of LC3–PE (or Atg8–PE) before autophagosome formation is complete (Scherz-Shouval et al., 2007). However, a pressing question in this model is how the levels of H2O2 can be temporally and spatially controlled to ensure that Atg4 remains active – as required in the following two situations: first, when it is used to cleave the LC3 (or Atg8) precursor and, second, when it is required to cleave the LC3–PE (or Atg8–PE) conjugate after autophagosome formation. A second hypothesis was put forward in a recent study in yeast (Nair et al., 2010), which suggests that Atg18 and Atg21, together with the Atg12–Atg5-Atg16 complex, form a barrier to block the access of Atg4 to Atg8–PE on the outer surface of the phagophore, thereby preventing the cleavage of Atg8–PE by Atg4. After autophagosome completion, Atg18, Atg21 and the Atg12–Atg5-Atg16 complex dissociate from autophagosomes, thus allowing Atg4 access to Atg8–PE.
Another question regarding the role of Atg4 arises from structural studies of human Atg4B (ATG4B), which indicate that the free ATG4B is autoinhibited because its substrates (i.e. pro-LC3 and LC3–PE) cannot access its catalytic cysteine residue Cys74, which is covered by its regulatory loop Trp142 and the N-terminal tail (Noda et al., 2009; Satoo et al., 2009; Sugawara et al., 2005). When LC3 is bound to ATG4B, large conformational changes of these ATG4B domains take place, allowing LC3 access to the catalytic site (Satoo et al., 2009). However, the signals that trigger these conformational changes within Atg4 are unknown. One plausible hypothesis is that ROS, instead of inactivating Atg4, activates Atg4 by oxidizing its non-catalytic cysteine residues, triggering conformational changes of Atg4 when complete autophagosomes are formed and are ready for fusion with lysosomes – in which ROS are constitutively generated (Kubota et al., 2010). Indeed, multiple cysteine residues are found in the Atg4 proteins from different eukaryotes: 12 in yeast (S. cerevisiae), between 10 and 14 in human (ATG4A-C), 11 in D. melanogaster, and 9 cysteine residues in C. elegans. ROS might not be able to access Atg4 before the fusion of autophagosomes with lysosomes, because the eukaryotic cytosol is generally a reducing environment. However, two challenges of this hypothesis are to interpret first how oxidation of the catalytic cysteine residues of Atg4 proteins by ROS can be avoided and, second, how pre-LC3 (or Atg8) can be cleaved by Atg4 when it is not in close proximity to the lysosome (or vacuole).
Despite extensive studies, the exact functions of Atg8 or of LC3 are still unclear. In yeast, Atg8 can mediate the tethering and hemifusion of membranes in vitro (Nakatogawa et al., 2007), and the amount of Atg8 determines the size of autophagosomes (Xie et al., 2008). Atg8 and LC3 are also involved in cargo recognition (Kanki et al., 2009; Noda et al., 2008; Okamoto et al., 2009; Shintani et al., 2002). Mammalian cells have multiple Atg8 homologs including LC3A, LC3B, LC3C, γ-aminobutyric acid type A receptor associated protein (GABARAP), GABARAP-like 1, 2 and 3 (GABARAPL1, GABARAPL2, and GABARAPL3, respectively) (Geng and Klionsky, 2008; Weidberg et al., 2010). The question remains whether all mammalian Atg8 homologs participate in autophagy that is induced by the same stimulus. A recent study has demonstrated that LC3 is involved in phagophore membrane expansion, whereas the GABARAP family is required for autophagosome maturation (Weidberg et al., 2010). Thus, it is possible that the Atg8 homologs in mammals are involved in different stages of autophagosome formation. Finally, a recent report has demonstrated that Atg12 can be conjugated to Atg3 in mammals, and that this conjugation affects mitochondrial homeostasis and cell death, but not starvation-induced autophagy (Radoshevich et al., 2010). Other potential targets of Atg12 (or Atg8) have not yet been identified.
The source of autophagosomal membranes is an ongoing controversy
Although substantial progress has been achieved in understanding the molecular aspects of autophagy, the mechanism of formation of its most characteristic structure, the autophagosome, is still unclear. Various models have been proposed for autophagosome formation, but two predominate. In the first model, autophagosomes are formed by de novo synthesis of a nucleating structure, the phagophore, which expands by the addition of lipids that are transported by Atg9 from other sources (Kovács et al., 2007). In the second model, a subdomain of the ER is initially used as a cradle for the formation of the phagophore that expands between ER domains to which it is physically linked (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009). This latter model is further supported by three-dimensional electron tomography studies that demonstrate a direct connection between the phagophore and ER. However, none of these models can explain all the current data for autophagosome formation, but a mechanism involving de novo synthesis is probable because the phagophore appears to expand rather than to form in a single step from a pre-existing organelle (Mizushima et al., 2001).
What are the mechanisms of phagophore expansion and autophagosome formation?
As almost all intracellular organelles, such as the ER, Golgi, endosomes, mitochondria and the plasma membrane have been implicated in the formation of autophagosomes (Geng et al., 2010; Hailey et al., 2010; Luo et al., 2009; Lynch-Day et al., 2010; Ravikumar et al., 2010; Reggiori, 2006; Yen et al., 2010), it is possible that multiple sources, indeed, contribute to the autophagosomal membrane. If this is correct, a fundamental question is how the membrane is redirected from its normal itinerary within the cell to participate in autophagy. Another question is whether all of these sources contribute to autophagosome formation in the same autophagy process, or if there is a hierarchy and an order, in which different donor sources participate. If this is the case, the question arises of how this process is regulated.
A further issue is the location of phagophore formation. In yeast, most Atg proteins are recruited to the phagophore assembly site (PAS), which appears to function as the organizing center for autophagosome formation. However, an equivalent of the yeast PAS has not been identified in higher eukaryotes, and autophagosomes can form at multiple locations. Nonetheless, it has been proposed that mammalian cells might also construct their autophagosomes from a PAS equivalent. For example, under autophagy-inducing conditions, multiple green fluorescent protein (GFP)-LC3 containing vesicles can be observed in a mammalian cell, whereas typically only one or sometimes a few GFP-Atg8 dots and a single PAS are observed in a yeast cell. Thus, it is possible that, in contrast to yeast, mammalian cells have multiple PAS equivalents (Itakura and Mizushima, 2010).
Autophagy and TOR signaling
Known roles of TOR signaling pathways in autophagy regulation
TOR, specifically TORC1, can regulate autophagy in two ways; by directly phosphorylating Atg proteins, such as Atg13 and Atg1 as discussed above (Fig. 2), and by acting in a signal transduction cascade involving other proteins that can regulate autophagy (He and Klionsky, 2009). Many signaling pathways known to regulate autophagy merge at TORC1 (Fig. 4), including pathways that involve adenosine monophosphate-activated protein kinase (AMPK), which activates autophagy, and Akt (also known as protein kinase B) that downregulates autophagy (Esclatine et al., 2009; He and Klionsky, 2009; Klionsky, 2005). In mammalian cells, AMPK negatively regulates TORC1 by either directly inhibiting TORC1 (Gwinn et al., 2008; Yang and Klionsky, 2010) or activating tuberous sclerosis 2 (TSC2) protein, which is an upstream effector of TORC1 (Inoki et al., 2003). By contrast, the yeast AMPK homolog sucrose non-fermenting 1 (Snf1) is negatively regulated by TORC1 (Orlova et al., 2006). Both AMPK (Liang et al., 2007) and Snf1 (Wang et al., 2001) can positively regulate autophagy, but the mechanism used by Snf1 is not known.
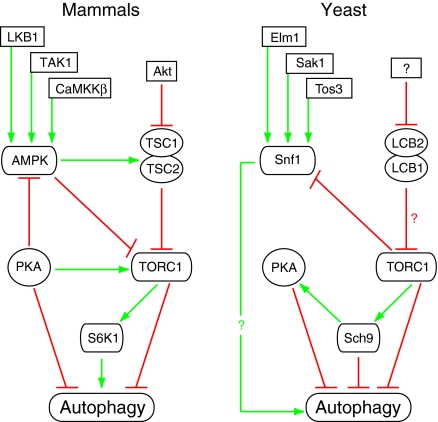
Fig. 4.
The TOR signaling pathways regulates autophagy in mammalian cells and yeast. In mammals, AMPK can be activated by its upstream factors LKB1, TAK1, and CaMKKβ (Yang and Klionsky, 2010). In yeast, Snf1 can be activated by its upstream factors Elm1, Sak1, and Tos3 (Hedbacker and Carlson, 2008). In yeast, the mammalian TSC1 and TSC2 homologs LCB2 and LCB1 (Gable et al., 2000) might form a complex that potentially inhibits TORC1, although this has not been experimentally demonstrated. A putative Akt homolog upstream of LCB2 and LCB1 also needs to be identified in yeast. Green arrows indicate interactions that induce autophagy, red bars indicate inhibition. See the text for details.
The protein kinase A (PKA) and TOR pathways are generally considered to be two parallel pathways that sense carbon and nitrogen sources, respectively (Stephan et al., 2010). However, studies in yeast and in mammals have indicated that there is crosstalk between these two pathways. In mammals PKA activates TORC1 (Mavrakis et al., 2006) by phosphorylating its subunit PRAS40 (Blancquaert et al., 2010). There, it can also phosphorylate and inactivate AMPK (Djouder et al., 2010), thus indirectly activating TORC1. By contrast, in yeast, TORC1 can indirectly activate PKA through activating the putative ribosomal protein S6 kinase (RPS6KB1, hereafter referred to as S6K1) homolog Sch9, which inhibits the mitogen activated protein kinase Slt2; Slt2 inactivates PKA by phosphorylating its regulatory subunit Bcy1 (Soulard et al., 2010). Mammalian PKA negatively regulates autophagy either by directly phosphorylating LC3 (Cherra et al., 2010) or by activating TORC1, which inhibits autophagy (Mavrakis et al., 2006). In yeast, PKA also negatively regulates autophagy (Yorimitsu et al., 2007) by phosphorylating Atg1 (Budovskaya et al., 2005) and Atg13 (Stephan et al., 2009).
In mammals, the TORC1 substrate S6K1 (Armour et al., 2009) is a positive regulator of autophagy, whereas yeast Sch9 is a negative regulator (Yorimitsu et al., 2007). Akt is an upstream negative regulator of the TSC1-TSC2 complex within the TOR signaling pathways that are involved in autophagy regulation in mammals (Yang and Klionsky, 2010) and Drosophila (Dutta and Baehrecke, 2008); however, the role of Akt in autophagy is not clear in C. elegans (Kang and Avery, 2010). In S. cerevisiae, Sch9 was originally proposed to be an Akt homolog (Sobko, 2006), but a later study demonstrated that Sch9 is more likely to be a S6K1 homolog (Urban et al., 2007), which means it would function as a substrate of TORC1 rather than as its regulator. Thus, a yeast Akt homolog still awaits identification or confirmation.
What controls the network of TOR signaling pathways that regulate autophagy?
Although TOR has been the most extensively studied regulator of autophagy, basic questions regarding its functions remain. For example, amino acids are main regulators of TOR and of autophagy; however, the factors upstream of TOR that sense amino acids are still unclear (Dann and Thomas, 2006; Meijer, 2008). Thus far, the networks of different TOR signaling pathways have been constructed in studies that focused primarily on one individual pathway. In the future, the relative importance of different pathways and their coordinated interplay needs to be addressed. For example, the AMPK pathway in mammals can activate autophagy either by directly inhibiting TORC1 or by activating TSC2. Yeast Snf1, however, which is also able to activate autophagy, but by a yet unknown mechanism, is a downstream target of TORC1 (Fig. 4). This raises the question whether AMPK is a downstream target of TORC1 in mammals, and Snf1 an upstream effector of TORC1 in yeast? A similar contrast in crosstalk applies to the interplay between PKA and TORC1 in mammals and yeast (Fig. 4). Furthermore, the roles of AMPK and PKA in Drosophila (Chang and Neufeld, 2010) and C. elegans (Kovács and Zhang, 2010; Meléndez and Levine, 2009) are not clear, and future studies are needed to delineate their functions with regard to autophagy. Along these lines, there are clear species-specific differences that need to be considered, such as why S6K1 is a positive regulator of autophagy in mammals, yet Sch9 negatively regulates it in yeast.
In recent years, studies have demonstrated that ROS can also regulate autophagy (Chen et al., 2009; Huang et al., 2009; Scherz-Shouval et al., 2007). ROS stimulate activation of AMPK (Jung et al., 2008) and of Snf1 (Hong and Carlson, 2007), indicating that ROS can induce autophagy through their effects on these two proteins. A recent study suggests that ROS activates AMPK by activating ataxia-telangiectasia mutated (ATM), which is an upstream activator of AMPK (Alexander et al., 2010). Future studies are required to determine whether ROS can also regulate other autophagic signaling pathways (He and Klionsky, 2009).
A final point to consider is that most of the regulatory components identified to date are kinases and that – presumably – there is a complementary phosphatase for each kinase. Future studies will be needed to identify these phosphatases and determine how their function is coordinated with that of the respective kinases.
Interplay between autophagy and other cellular processes
Crosstalk between autophagy and apoptosis
As both autophagy and apoptosis are important for the development and prevention of human diseases, the crosstalk between these two pathways has received increased attention, and several observations help to summarize this interplay in mammalian cells (Fig. 5). Under certain conditions, autophagy and apoptosis are two independent processes (Eisenberg-Lerner et al., 2009), whereas in other situations, the activation of autophagy inhibits apoptosis (Maiuri et al., 2007; Platini et al., 2010) or autophagy occurs upstream of apoptosis (Eisenberg-Lerner et al., 2009). Furthermore, regulators of apoptosis, such as Bcl-2 family members (Bcl-2 and Bcl-xL) (Levine et al., 2008) and CASP8 and FADD-like apoptosis regulator (CFLAR, also known as Flip) (Djavaheri-Mergny et al., 2010) can regulate autophagy, and proteins involved in autophagy, such as Atg5, beclin 1 and Atg4D, can also have a role in apoptosis (Fimia and Piacentini, 2010).
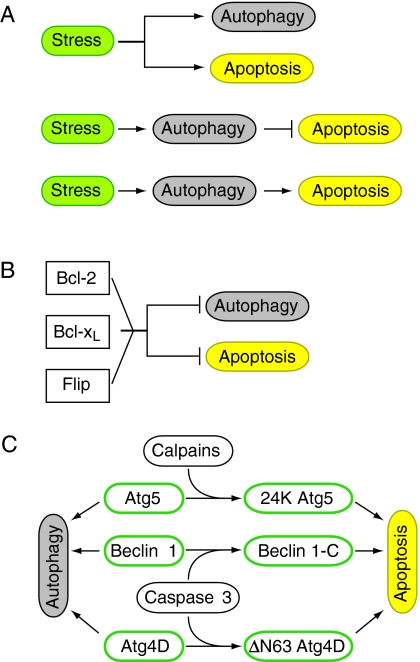
Fig. 5.
Crosstalk between autophagy and apoptosis in mammalian cells. (A) In a cell, the same stress induces both autophagy and apoptosis as independent processes (top). A stress can also induce autophagy, which then inhibits apoptosis (middle) or, alternatively, a stress induces autophagy, which can be the trigger of apoptosis (bottom). (B) Both autophagy and apoptosis are negatively regulated by Bcl-2, Bcl-xL and Flip. (C) Autophagy proteins Atg5, beclin 1 and Atg4D function in autophagy in their unmodified form, but also have a role in apoptosis after cleavage by either calpains, which target Atg5 and give rise to 24 K Atg5, or caspase 3, which cleaves beclin 1, resulting in a C-terminal fragment of beclin 1 (Beclin 1-C), and Atg4D, truncating it at the canonical caspase cleavage sequence (DEVD63K) to ΔN63 Atg4D.
What is the crosstalk between autophagy and apoptosis
One of the most important questions in this field is what are the underlying mechanisms of the pro-cell survival effect of autophagy versus its pro-cell death effects? Autophagic degradation of active caspase-8, a positive effector of apoptosis, is responsible for the inhibition of apoptotic cell death in mammalian cells (Hou et al., 2010). The pro-cell death effect of autophagy could be related to the activation of apoptosis, which would imply that autophagy is an upstream event of apoptosis (Nezis et al., 2010) or, alternatively, autophagy could be independent of apoptosis, such as in situations, in which autophagy-induced cell death does not exhibit any of the characteristic apoptotic features, such as caspase activation and DNA fragmentation (Voss et al., 2010). Thus, it has been controversial as to how ‘autophagic cell death’ should be defined and, for consistency, we here define it as ‘cell death that is triggered by autophagy’ (pro-cell death effect). In contrast to the above-mentioned mechanism for the cytoprotective effect of autophagy against stress, one possible mechanism for autophagic cell death could involve the autophagic degradation of a negative effector of apoptosis. This is supported by a recent demonstration that autophagic degradation of the Drosophila inhibitor of apoptosis (IAP) dBruce controls apoptotic cell death in nurse cells during late Drosophila melanogaster oogenesis (Nezis et al., 2010). Other possible mechanisms underlying the dual functions of autophagy (in cell survival and cell death) need to be explored in the future.
Factors that make it possible to differentiate the pro-cell survival and pro-cell death effects of autophagy have been reported recently. For instance, Draper (Drpr), the D. melanogaster ortholog of the C. elegans engulfment receptor CED-1 (McPhee et al., 2010) and Jun N-terminal-kinase (JNK) (Shimizu et al., 2010) are involved in autophagic cell death, but not in autophagy-induced cell survival. Furthermore, death-associated protein kinase (DAPK) has been proposed to convert autophagy from a cell survival mechanism to one of the initiation of cell death (Bialik and Kimchi, 2010). It remains to be investigated how exactly these factors (Drpr, JNK, DAPK, etc.) are able to direct autophagy from a survival to a death pathway.
Several Atg proteins have been implicated in apoptosis. For example, caspase 3 cleaves the human Atg4 family member Atg4D to generate a truncated product, ΔN63 Atg4D that, when overexpressed, induces autophagy-independent apoptosis (Betin and Lane, 2009). Similarly, under conditions that induce apoptosis, Atg5 is cleaved by calpains, generating a truncated product 24K Atg5 that, when overexpressed by itself, induces apoptosis, but not autophagy (Yousefi et al., 2006). Caspases also cleave beclin 1 at Asp149 during apoptosis resulting in the inhibition of autophagy (Djavaheri-Mergny et al., 2010; Luo and Rubinsztein, 2010; Wirawan et al., 2010). It remains to be investigated whether other Atg proteins are also cleaved during apoptosis.
However, apoptosis-inducing factors might also have a role in autophagy, as for example, caspases are involved in autophagy in Drosophila (Hou et al., 2008; Kim et al., 2010). In mammalian cells, the expression of caspase 9 is mediated by the expression of beclin 1, but the role of caspase 9 in autophagy has not been investigated (Wang et al., 2007).
Regulation of autophagy within the context of other cellular networks
Autophagy is regulated by a complex network that consists of different signaling pathways. A recent yeast study highlights the importance of investigating the regulation of autophagy in the context of a network of multiple signaling pathways (Yang et al., 2010); in S. cerevisiae, Pho85, which exists in different cyclin-dependent kinase (Cdk)-containing Pho85-cyclin complexes, can have both positive and negative effects on autophagy. Pho85 in complex with the cyclins Pho80 and Pc15 negatively regulates autophagy, whereas it has a positive effect on autophagy when it forms a complex with the cyclins Clg1 or Pcl1. This indicates that autophagy regulators can have opposing roles (i.e. positive or negative) depending on their interactions with different components of a signaling network. In mammalian cells, Cdk1 and Cdk5 negatively regulate autophagy by affecting the interaction of Vps34 with beclin 1 (Furuya et al., 2010); however, it is not known if they can also have a positive role in autophagy.
Remaining questions regarding the cellular network regulating autophagy
The most obvious question is: what is the complete regulatory network that controls autophagy? Even if we limit this consideration to starvation-induced autophagy, this process is regulated in response to a wide range of nutrients, including amino acids, phosphate, nitrogen and glucose, and questions remain with regard to which signaling pathway(s) dominate, and how are they interconnected to modulate the level of autophagy. Compared with yeast, genes in higher eukaryotes cannot be that easily mutated, knocked out or overexpressed, making it more difficult to study the entire autophagy network. However, the approach of gene silencing techniques using small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) is now widely used and should start to provide detailed information in higher eukaryotes soon. For example, in a recent study applying RNA interference (RNAi) coupled with proteomic analysis, a network of basal autophagy in human cells was constructed that consists of 751 interactions and 409 candidate interacting proteins (Behrends et al., 2010). It is expected that the networks of autophagy induced by different stress conditions could also be explored following up on this pioneer study.
Conclusions
Although we have learned much in the past decade regarding the molecular aspects of autophagy, many questions remain – as outlined here. Further characterization of the known 35 Atg genes (Kanki and Klionsky, 2010; Suzuki et al., 2010; Nazarko et al., 2011), and the discovery of additional genes involved in autophagy regulation will help to answer these questions in the future. Along with mechanistic studies of autophagy regulation, the identification of genetic and pharmacological regulators of autophagy could have practical health benefits. Currently, two main approaches have been used to develop new autophagy regulators (inhibitors or activators). The first approach is the screening of small molecules targeting autophagy (Farkas et al., 2009; Zhang et al., 2007). The second approach is to design and develop autophagy regulators according to the structural information of Atg proteins (Miller et al., 2010). Recently, additional autophagy regulators have been discovered through such a screening approach (Balgi et al., 2009; Choi et al., 2010; Renna et al., 2010). More studies using these and other approaches are expected to help us in understanding the regulatory network of autophagy and in making the therapeutic modulation of autophagy a reality.
Acknowledgments
We apologize to authors whose work has not been cited owing to space limitations. This work was supported by Public Health Service Grant GM53396 to D.J.K. Deposited in PMC for release after 12 months.
References
- Alexander A., Cai S. L., Kim J., Nanez A., Sahin M., MacLean K. H., Inoki K., Guan K. L., Shen J., Person M. D., et al. (2010). ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. USA 107, 4153-4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour S. M., Baur J. A., Hsieh S. N., Land-Bracha A., Thomas S. M., Sinclair D. A. (2009). Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging 1, 515-528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke E. H. (2005). Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6, 505-510 [DOI] [PubMed] [Google Scholar]
- Balgi A. D., Fonseca B. D., Donohue E., Tsang T. C., Lajoie P., Proud C. G., Nabi I. R., Roberge M. (2009). Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS ONE 4, e7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D. C. (2009). Function and regulation of macroautophagy in plants. Biochim. Biophys. Acta 1793, 1397-1403 [DOI] [PubMed] [Google Scholar]
- Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010). Network organization of the human autophagy system. Nature 466, 68-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betin V. M., Lane J. D. (2009). Caspase cleavage of Atg4D stimulates GABARAPL1 processing and triggers mitochondrial targeting and apoptosis. J. Cell. Sci. 122, 2554-2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialik S., Kimchi A. (2010). Lethal weapons: DAP-kinase, autophagy and cell death: DAP-kinase regulates autophagy. Curr. Opin. Cell Biol. 22, 199-205 [DOI] [PubMed] [Google Scholar]
- Blancquaert S., Wang L., Paternot S., Coulonval K., Dumont J. E., Harris T. E., Roger P. P. (2010). cAMP-dependent activation of mammalian target of rapamycin (mTOR) in thyroid cells. Implication in mitogenesis and activation of CDK4. Mol. Endocrinol. 24, 1453-1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Deminoff S. J., Herman P. K. (2005). An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 102, 13933-13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Klionsky D. J. (2007). Physiological functions of Atg6/Beclin 1, a unique autophagy-related protein. Cell Res. 17, 839-849 [DOI] [PubMed] [Google Scholar]
- Cebollero E., Reggiori F. (2009). Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793, 1413-1421 [DOI] [PubMed] [Google Scholar]
- Chang Y.-Y., Neufeld T. P. (2010). Autophagy takes flight in Drosophila. FEBS Lett. 584, 1342-1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Azad M. B., Gibson S. B. (2009). Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 16, 1040-1052 [DOI] [PubMed] [Google Scholar]
- Chen Y., Azad M. B., Gibson S. B. (2010). Methods for detecting autophagy and determining autophagy-induced cell death. Can. J. Physiol. Pharmacol. 88, 285-295 [DOI] [PubMed] [Google Scholar]
- Cherra S. J., III, Kulich S. M., Uechi G., Balasubramani M., Mountzouris J., Day B. W., Chu C. T. (2010). Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol. 190, 533-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I. K., Cho Y. S., Jung H. J., Kwon H. J. (2010). Autophagonizer, a novel synthetic small molecule, induces autophagic cell death. Biochem. Biophys. Res. Commun. 393, 849-854 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. (2004). Autophagy: in sickness and in health. Trends Cell Biol. 14, 70-77 [DOI] [PubMed] [Google Scholar]
- Dann S. G., Thomas G. (2006). The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett. 580, 2821-2829 [DOI] [PubMed] [Google Scholar]
- Deretic V., Levine B. (2009). Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5, 527-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo S., Corazzari M., Nazio F., Oliverio S., Lisi G., Antonioli M., Pagliarini V., Matteoni S., Fuoco C., Giunta L., et al. (2010). The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191, 155-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M., Maiuri M. C., Kroemer G. (2010). Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 29, 1717-1719 [DOI] [PubMed] [Google Scholar]
- Djouder N., Tuerk R. D., Suter M., Salvioni P., Thali R. F., Scholz R., Vaahtomeri K., Auchli Y., Rechsteiner H., Brunisholz R. A., et al. (2010). PKA phosphorylates and inactivates AMPKα to promote efficient lipolysis. EMBO J. 29, 469-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey F. C., Rose K. L., Coenen S., Prater S. M., Cavett V., Cleveland J. L., Caldwell-Busby J. (2009). Mapping the phosphorylation sites of Ulk1. J. Proteome. Res. 8, 5253-5263 [DOI] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J. A. K. W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. (2005). Pexophagy: the selective autophagy of peroxisomes. Autophagy 1, 75-83 [DOI] [PubMed] [Google Scholar]
- Dutta S., Baehrecke E. H. (2008). Warts is required for PI3K-regulated growth arrest, autophagy, and autophagic cell death in Drosophila. Curr. Biol. 18, 1466-1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A., Bialik S., Simon H. U., Kimchi A. (2009). Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 16, 966-975 [DOI] [PubMed] [Google Scholar]
- Esclatine A., Chaumorcel M., Codogno P. (2009). Macroautophagy signaling and regulation. Curr. Top. Microbiol. Immunol. 335, 33-70 [DOI] [PubMed] [Google Scholar]
- Farkas T., Hoyer-Hansen M., Jäättelä M. (2009). Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy 5, 1018-1025 [DOI] [PubMed] [Google Scholar]
- Fimia G. M., Piacentini M. (2010). Regulation of autophagy in mammals and its interplay with apoptosis. Cell. Mol. Life Sci. 67, 1581-1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia G. M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., et al. (2007). Ambra1 regulates autophagy and development of the nervous system. Nature 447, 1121-1125 [DOI] [PubMed] [Google Scholar]
- Funderburk S. F., Wang Q. J., Yue Z. (2010). The Beclin 1-VPS34 complex-at the crossroads of autophagy and beyond. Trends Cell Biol. 20, 355-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T., Kim M., Lipinski M., Li J., Kim D., Lu T., Shen Y., Rameh L., Yankner B., Tsai L. H., et al. (2010). Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol. Cell 38, 500-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K., Slife H., Bacikova D., Monaghan E., Dunn T. M. (2000). Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275, 7597-7603 [DOI] [PubMed] [Google Scholar]
- Geng J., Klionsky D. J. (2008). The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 9, 859-864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Nair U., Yasumura-Yorimitsu K., Klionsky D. J. (2010). Post-golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 21, 2257-2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D. W., Rambold A. S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P. K., Lippincott-Schwartz J. (2010). Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656-667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433-1437 [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Levine B. (2010). The Beclin 1 interactome. Curr. Opin. Cell Biol. 22, 140-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M. (2008). SNF1/AMPK pathways in yeast. Front. Biosci. 13, 2408-2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. P., Carlson M. (2007). Regulation of Snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282, 16838-16845 [DOI] [PubMed] [Google Scholar]
- Hou W., Han J., Lu C., Goldstein L. A., Rabinowich H. (2010). Autophagic degradation of active caspase-8: A crosstalk mechanism between autophagy and apoptosis. Autophagy 6, 891-900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. C., Chittaranjan S., Barbosa S. G., McCall K., Gorski S. M. (2008). Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182, 1127-1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Canadien V., Lam G. Y., Steinberg B. E., Dinauer M. C., Magalhaes M. A., Glogauer M., Grinstein S., Brumell J. H. (2009). Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA 106, 6226-6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488-492 [DOI] [PubMed] [Google Scholar]
- Inoki K., Zhu T., Guan K. L. (2003). TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577-590 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Klionsky D. J. (2010). Regulation of macroautophagy in Saccharomyces cerevisiae. Semin. Cell Dev. Biol. 21, 664-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Mizushima N. (2010). Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.-N., Yang W. K., Kim J., Kim H. S., Kim E. J., Yun H., Park H., Kim S. S., Choe W., Kang I., et al. (2008). Reactive oxygen species stabilize hypoxia-inducible factor-1 alpha protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis 29, 713-721 [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507-1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., Yonezawa K., Ohsumi Y. (2010). Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30, 1049-1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C., Avery L. (2010). Death-associated protein kinase (DAPK) and signal transduction: fine-tuning of autophagy in Caenorhabditis elegans homeostasis. FEBS J. 277, 66-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Klionsky D. J. (2010). The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 75, 795-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009). Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M., Dohnal I., Reiter W., Kaspar S., Stoffel I., Ammerer G., Kraft C., Peter M. (2010). Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 6, 1168-1178 [DOI] [PubMed] [Google Scholar]
- Kim Y.-I., Ryu T., Lee J., Heo Y.-S., Ahnn J., Lee S.-J., Yoo O. (2010). A genetic screen for modifiers of Drosophila caspase Dcp-1 reveals caspase involvement in autophagy and novel caspase-related genes. BMC Cell Biol. 11, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. (2005). The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931-937 [DOI] [PubMed] [Google Scholar]
- Koren I., Reem E., Kimchi A. (2010). DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20, 1093-1098 [DOI] [PubMed] [Google Scholar]
- Kovács A. L., Zhang H. (2010). Role of autophagy in Caenorhabditis elegans. FEBS Lett. 584, 1335-1341 [DOI] [PubMed] [Google Scholar]
- Kovács A. L., Palfia Z., Rez G., Vellai T., Kovács J. (2007). Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy 3, 655-662 [DOI] [PubMed] [Google Scholar]
- Kubota C., Torii S., Hou N., Saito N., Yoshimoto Y., Imai H., Takeuchi T. (2010). Constitutive reactive oxygen species generation from autophagosome/lysosome in neuronal oxidative toxicity. J. Biol. Chem. 285, 667-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463-477 [DOI] [PubMed] [Google Scholar]
- Levine B., Yuan J. (2005). Autophagy in cell death: an innocent convict? J. Clin. Invest. 115, 2679-2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Sinha S., Kroemer G. (2008). Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 4, 600-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., et al. (2008). Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776-787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Shao S. H., Xu Z. X., Hennessy B., Ding Z., Larrea M., Kondo S., Dumont D. J., Gutterman J. U., Walker C. L., et al. (2007). The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 9, 218-224 [DOI] [PubMed] [Google Scholar]
- Luo S., Rubinsztein D. C. (2010). Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 17, 268-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Chen Q., Cebollero E., Xing D. (2009). Mitochondria: one of the origins for autophagosomal membranes? Mitochondrion 9, 227-231 [DOI] [PubMed] [Google Scholar]
- Lynch-Day M. A., Klionsky D. J. (2010). The Cvt pathway as a model for selective autophagy. FEBS Lett. 584, 1359-1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day M. A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C. R., Brumell J. H., Ferro-Novick S., Klionsky D. J. (2010). Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107, 7811-7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007). Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8, 741-752 [DOI] [PubMed] [Google Scholar]
- Massey A., Kiffin R., Cuervo A. M. (2004). Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36, 2420-2434 [DOI] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385-396 [DOI] [PubMed] [Google Scholar]
- Mavrakis M., Lippincott-Schwartz J., Stratakis C. A., Bossis I. (2006). Depletion of type IA regulatory subunit (RIα) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Hum. Mol. Genet. 15, 2962-2971 [DOI] [PubMed] [Google Scholar]
- McPhee C. K., Logan M. A., Freeman M. R., Baehrecke E. H. (2010). Activation of autophagy during cell death requires the engulfment receptor Draper. Nature 465, 1093-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer A. J. (2008). Amino acid regulation of autophagosome formation. Methods Mol. Biol. 445, 89-109 [DOI] [PubMed] [Google Scholar]
- Meijer A. J., Codogno P. (2009). Autophagy: regulation and role in disease. Crit. Rev. Clin. Lab. Sci. 46, 210-240 [DOI] [PubMed] [Google Scholar]
- Meléndez A., Levine B. (2009). Autophagy in C. elegans. WormBook, 1-26 [DOI] [PubMed] [Google Scholar]
- Miller S., Tavshanjian B., Oleksy A., Perisic O., Houseman B. T., Shokat K. M., Williams R. L. (2010). Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science 327, 1638-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. (2010). The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132-139 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. (2001). Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657-668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U., Cao Y., Xie Z., Klionsky D. J. (2010). Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 285, 11476-11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165-178 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458-467 [DOI] [PubMed] [Google Scholar]
- Nazarko V. Y., Nazarko T. Y., Farre J.-C., Stasyk O. V., Warnecke D., Ulaszewski S., Cregg J. M., Sibirny A. A., Subramani S. (2011). Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris. Autophagy 7, (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis I. P., Shravage B. V., Sagona A. P., Lamark T., Bjørkøy G., Johansen T., Rusten T. E., Brech A., Baehrecke E. H., Stenmark H. (2010). Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell Biol. 190, 523-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008). Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211-1218 [DOI] [PubMed] [Google Scholar]
- Noda N. N., Ohsumi Y., Inagaki F. (2009). ATG systems from the protein structural point of view. Chem. Rev. 109, 1587-1598 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87-97 [DOI] [PubMed] [Google Scholar]
- Orlova M., Kanter E., Krakovich D., Kuchin S. (2006). Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryotic Cell 5, 1831-1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platini F., Perez-Tomas R., Ambrosio S., Tessitore L. (2010). Understanding autophagy in cell death control. Curr. Pharm. Des. 16, 101-113 [DOI] [PubMed] [Google Scholar]
- Radoshevich L., Murrow L., Chen N., Fernandez E., Roy S., Fung C., Debnath J. (2010). ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142, 590-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D. C. (2010). Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12, 747-757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F. (2006). 1. Membrane origin for autophagy. Curr. Top. Dev. Biol. 74, 1-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna M., Jimenez-Sanchez M., Sarkar S., Rubinsztein D. C. (2010). Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J. Biol. Chem. 285, 11061-11067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoo K., Noda N. N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2009). The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341-1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749-1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Konishi A., Nishida Y., Mizuta T., Nishina H., Yamamoto A., Tsujimoto Y. (2010). Involvement of JNK in the regulation of autophagic cell death. Oncogene 29, 2070-2082 [DOI] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E., Klionsky D. J. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobko A. (2006). Systems biology of AGC kinases in fungi. Sci. STKE 2006, re9 [DOI] [PubMed] [Google Scholar]
- Soulard A., Cremonesi A., Moes S., Schutz F., Jeno P., Hall M. N. (2010). The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21, 3475-3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K. (2009). The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 106, 17049-17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K. (2010). The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy 6, 294-295 [DOI] [PubMed] [Google Scholar]
- Sugawara K., Suzuki N. N., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2005). Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 280, 40058-40065 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kondo C., Morimoto M., Ohsumi Y. (2010). Selective transport of α-mannosidase by autophagic pathways. Identification of a novel receptor, Atg34p. J. Biol. Chem. 285, 30019-30025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Coppola D., Matsushita N., Cualing H. D., Sun M., Sato Y., Liang C., Jung J. U., Cheng J. Q., Mule J. J., et al. (2007). Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 9, 1142-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E., Wang F., Han J., Zhang H. (2009). epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy 5, 608-615 [DOI] [PubMed] [Google Scholar]
- Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., et al. (2010). C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042-1055 [DOI] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., et al. (2007). Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663-674 [DOI] [PubMed] [Google Scholar]
- Uttenweiler A., Mayer A. (2008). Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol. Biol. 445, 245-259 [DOI] [PubMed] [Google Scholar]
- Vellai T., Takacs-Vellai K., Sass M., Klionsky D. J. (2009). The regulation of aging: does autophagy underlie longevity? Trends Cell. Biol. 19, 487-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss V., Senft C., Lang V., Ronellenfitsch M. W., Steinbach J. P., Seifert V., Kogel D. (2010). The pan-Bcl-2 inhibitor (-)-gossypol triggers autophagic cell death in malignant glioma. Mol. Cancer Res. 8, 1002-1016 [DOI] [PubMed] [Google Scholar]
- Wang C.-W., Klionsky D. J. (2004). Microautophagy. In Autophagy (ed. Klionsky D. J.), pp. 107-114 Georgetown: Landes Bioscience; [Google Scholar]
- Wang Z., Wilson W. A., Fujino M. A., Roach P. J. (2001). Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 21, 5742-5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. H., Xu L., Duan Z. L., Zeng L. Q., Yan N. H., Peng Z. L. (2007). Beclin 1-mediated macroautophagy involves regulation of caspase-9 expression in cervical cancer HeLa cells. Gynecol. Oncol. 107, 107-113 [DOI] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E., Vande Walle L., Kersse K., Cornelis S., Claerhout S., Vanoverberghe I., Roelandt R., De Rycke R., Verspurten J., Declercq W., et al. (2010). Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 1, e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D. J. (2008). Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290-3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Geng J., Yen W.-L., Wang K., Klionsky D. J. (2010). Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol. Cell 38, 250-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y. Y., Wrasman K., Herman P. K. (2010). Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 185, 871-882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.-L., Shintani T., Nair U., Cao Y., Richardson B. C., Li Z., Hughson F. M., Baba M., Klionsky D. J. (2010). The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188, 101-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.-L. (2009). 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180-1185 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Zaman S., Broach J. R., Klionsky D. J. (2007). Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 4180-4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. R. J., Narita M., Ferreira M., Kirschner K., Sadaie M., Darot J. F. J., Tavare S., Arakawa S., Shimizu S., Watt F. M., et al. (2009). Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798-803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. (2006). Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8, 1124-1132 [DOI] [PubMed] [Google Scholar]
- Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A. D., Xie X., Ma D., et al. (2007). Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. USA 104, 19023-19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009). Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468-476 [DOI] [PMC free article] [PubMed] [Google Scholar]