Abstract
Purpose
To investigate the signal-to-noise-ratio (SNR) and data quality of time-reduced 1H 3D-MRSI techniques in the human brain at 3T.
Materials and Methods
Techniques that were investigated included ellipsoidal k-space sampling, parallel imaging, and EPSI. The SNR values for NAA, Cho, Cre, and lactate or lipid peaks were compared after correcting for effective spatial resolution and acquisition time in a phantom and in the brains of human volunteers. Other factors considered were linewidths, metabolite ratios, partial volume effects, and subcutaneous lipid contamination.
Results
In volunteers, the median normalized SNR for parallel imaging data decreased by 34–42%, but could be significantly improved using regularization. The normalized signal to noise loss in flyback EPSI data was 11–18%. The effective spatial resolutions of the traditional, ellipsoidal, SENSE, and EPSI data were 1.02, 2.43, 1.03, and 1.01cm3, respectively. As expected, lipid contamination was variable between subjects but was highest for the SENSE data. Patient data obtained using the flyback EPSI method were of excellent quality.
Conclusions
Data from all 1H 3D-MRSI techniques were qualitatively acceptable, based upon SNR, linewidths, and metabolite ratios. The larger FOV obtained with the EPSI methods showed negligible lipid aliasing with acceptable SNR values in less than 9.5 minutes without compromising the PSF.
Keywords: Spectroscopic Imaging, EPSI, Parallel Imaging, SNR
INTRODUCTION
Proton magnetic resonance spectroscopic imaging (1H MRSI) has become a routine clinical tool for non-invasively assessing the spatial distribution of different metabolites (1–3). This is of particular importance in the evaluation of neurological diseases. Many of the current studies that have applied point resolved (PRESS) 3D-MRSI are being performed at 1.5 Tesla (T) with a standard quadrature head coil and nominal spatial resolution of 1cm3 with acquisition times of 20 minutes or greater (4,5). The time required to obtain these data may limit the options for acquiring other valuable imaging sequences during a single patient visit.
By using a 3T scanner with an eight-channel phased array reception coil, it is possible to improve the signal to noise ratio (SNR) of the data by two-fold compared to 1.5T (6) without compromising the contrast between metabolite levels in normal appearing white matter (NAWM) and tumor (7). This improvement can either be used to maintain the spatial resolution and shorten the acquisition time or to improve the spatial resolution of the data with a similar acquisition time. The traditional method for localizing the signal relies on multiple phase encoding steps to gather the necessary spatial information. The time required to obtain these data for a given echo time (TE) depends linearly upon the repetition time (TR). Other than using the shortest possible TR, reducing the time needed to acquire the data requires the implementation of novel k-space sampling strategies (8–13).
Three well-known approaches that have been proposed to reduce the acquisition time for 1H 3D-MRSI data are ellipsoidal k-space encoding (9), parallel imaging reconstruction (10), and time varying readout gradient trajectories such as spiral spectroscopic imaging (11) and echo-planar spectroscopic imaging (EPSI) (12). Ellipsoidal k-space techniques are implemented by limiting the number of acquired phase encoding steps to a central ellipsoid of k-space, thus decreasing the number of samples obtained and reducing acquisition time. Parallel imaging techniques reduce the number of acquired k-space points by increasing the spacing between k-space samples. Additional spatial information from multiple receiver coils is then used to increase the spatial field-of-view (FOV) to the original size. EPSI and spiral SI techniques apply a time varying gradient waveform during data acquisition to simultaneously encode spectral and spatial information. This reduces the acquisition time by a factor determined by the number of phase encodes that would have been acquired in the EPSI or spiral dimensions. A variety of gradient trajectories may be considered based upon how they address waveform efficiency, robustness to gradient imperfections, and the tradeoff between spatial resolution and spectral bandwidth (13).
The purpose of this study was to investigate the relative benefits of time-reducing 1H 3D-MRSI techniques, and to determine which technique reduced acquisition time with the lowest cost in SNR and data quality. These issues were first investigated using 1H 3D-MRSI data from phantoms and healthy volunteers. Following this analysis, the most promising technique was applied to seventeen patients diagnosed with primary brain tumors in order to evaluate its robustness in a clinical setting.
MATERIALS AND METHODS
Images and 1H MRSI data were acquired using a 3T MR scanner with an eight-channel phased array head coil (GE Healthcare, Waukesha, WI, USA). Data were acquired on four separate occasions from an MR phantom (pH 7.2) that contained water, 3 mM choline (Cho), 10 mM creatine (Cre), 12.5 mM N-acetyl aspartate (NAA), 5 mM lactate (Lac), and 0.1% Magnevist (14). Comparison data were then acquired from the brains of ten healthy human volunteers. One of the ten volunteers did not receive a flyback EPSI scan and one other did not receive a symmetric EPSI scan because of limitations on scan time during these two studies. Seventeen EPSI datasets were also acquired in patients with primary brain tumors. All human subjects provided informed consent as required by our institutional review board.
MR Data Acquisition Protocol
Data obtained from phantoms and healthy volunteers consisted of a scout sequence followed by an anatomic imaging sequence (axial T1-weighted 3D inversion recovery spoiled gradient recalled acquisition in steady-state (IRSPGR): FOV=24×24×18cm3, 256×256×120 voxels, 15° flip angle, TE=1.6ms, inversion time (TI) = 400ms, TR=7.3ms ); a set of 2D fast gradient echo proton density weighted coil sensitivity images (FOV=30×30×18cm3, 64×64×36 voxels, 20° flip angle, TE=2.1ms, TR=150ms); and three different 1H 3D-MRSI acquisitions (TE = 144 ms, TR=1100 ms). All 1H MRSI sequences applied PRESS volume localization with an 8cm × 8cm × 4cm selection from the center of the phantom or brain, with chemical shift selective (CHESS) (15) pulses for water suppression, and very selective suppression (VSS) (16) pulses to both sharpen the definition of the PRESS selected volume and to limit the degree of lipid contamination from the scalp.
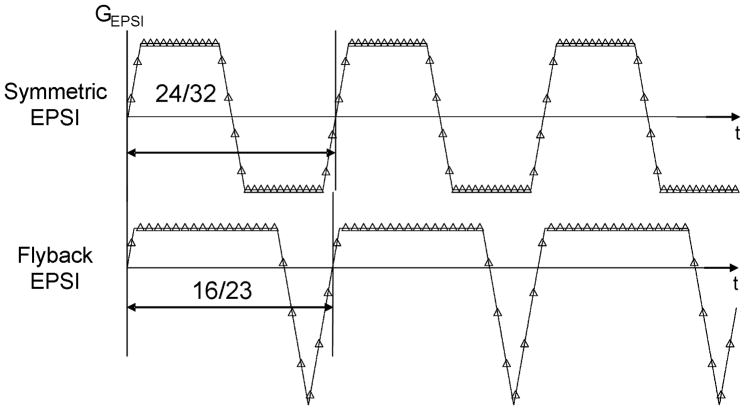
The acquisition parameters for the 1H 3D-MRSI data are summarized in Table 1 and the two EPSI trajectories are shown in Fig. 1. In all cases the echo-planar encoding was performed in the superior-inferior (SI) dimension. Ellipsoidal k-space sampling and parallel imaging data with a reduction factor of 2 in the right-left (RL) dimension (Rx=2) were simulated from the fully encoded MRSI data by eliminating the appropriate phase encoded k-space points. The MRSI data from patients were acquired using EPSI with the flyback gradient as described in Table 1.
Table 1.
List of all spectroscopic imaging techniques and their specifications, including: spatial field of view (FOV), spectral bandwidth (BW), number of scan repetitions (NEX), total acquisition time (tACQ), nominal resolution, and effective spatial resolution (as estimated via 3D-PSF simulation in MATLAB). The full-volume at half maximum PSF signal was used to represent effective spatial resolution. For traditional encoded 12×12×8 k-space sampling, ellipsoidal sampling, and SENSE sampling each point represented 9.04×10−6 cm3 (1 cm3 = 483 points). For all other 3D-MRSI techniques, each point represented 30.52×10−6 cm3 (1 cm3 = 323 points).
| Encoding Technique | FOV (cm3) | BW (Hz) | NEX | tACQ (min) | Nominal Resolution | Effective Spatial Resolution |
|---|---|---|---|---|---|---|
| Traditional MRSI | 12×12×8 | 2000 | 1 | 21.12 | 1cm3×1.95Hz | 1.017 cm3 |
| Ellipsoidal MRSI * | 12×12×8 | 2000 | 1 | 9.39 | 1cm3×1.95Hz | 2.473 cm3 |
| SENSE MRSI (Rx=2) * | 12×12×8 | 2000 | 1 | 10.56 | 1cm3×1.95Hz | 1.026 cm3 |
| SENSE w/REG * | 12×12×8 | 2000 | 1 | 10.56 | 1cm3×1.95Hz | 1.026 cm3 |
| Flyback EPSI | 16×16×16 | 988 | 2 | 9.46 | 1cm3×1.39Hz | 1.014 cm3 |
| Symmetric EPSI | 16×16×12 | 977 | 2 | 9.39 | 1cm3×1.91Hz | 1.015 cm3 |
| Symmetric w/ramps ‡ | 16×16×12 | 977 | 2 | 9.39 | 0.857cm3×1.91Hz | 0.868 cm3 |
These data sets were simulated from the traditionally encoded MRSI data.
From same dataset as symmetric EPSI data.
Figure 1.
Gradient waveforms used for two different echo-planar spectroscopic imaging (EPSI) encoding techniques. Fractions represent the number of samples on the plateau compared to the total number of samples per echo for each technique. Double-sided arrows represent the relative duration of each gradient echo.
Data Reconstruction
All data were reconstructed with custom software (6). The traditionally encoded 1H 3D-MRSI data were reconstructed by processing each of the 8 coil channels separately, apodizing with a 4Hz Lorentzian, applying phase and frequency corrections, and then removing residual water and baseline (17). The corrected spectroscopic images from each coil were then combined using coil sensitivity weights that were obtained from the low resolution proton density weighted images (18).
Ellipsoidal k-space sampling was simulated from the traditionally encoded MRSI data by reducing the number of phase encoding steps from 1152 (12×12×8) to 512 through zero filling the appropriate k-space points outside a normalized ellipsoidal radius of 1.2. This reduction would have resulted in an acquisition time of 9.39 minutes instead of the full 21.12 minutes. Data were then reconstructed in the same manner as the traditionally encoded data.
Parallel imaging data were simulated from the traditional fully encoded MRSI data using a sensitivity encoding (SENSE) sampling scheme with a reduction factor of 2 along kx (Rx=2), which would have corresponded to an acquisition time of 10.56 minutes. This resulted in a 6×12×8 simulated spectral data acquisition matrix. The parallel imaging data were reconstructed using the SENSE algorithm according to the following formula (10,19,20):
| (1) |
where S is the coil sensitivity matrix, H is the complex conjugate transpose, ψ is the receiver noise matrix (19), λ is the regularization parameter (20), I is the identity matrix, A is the aliased spectrum, and v are the unaliased spectra. Tikhonov’s regularization parameter (20), λ, was set to 1 for regularized and to 0 for unregularized SENSE reconstruction.
The EPSI data were reconstructed one channel at a time and combined in the same manner as the traditionally encoded data, but with the application of an additional linear phase correction in the EPSI dimension of spectral k-space to prevent spatial chemical shift artifacts (21). For the initial analysis, only samples from the flat part (plateau) of the EPSI gradient waveforms were used. To reconstruct the data acquired with the symmetric EPSI waveform, the positive and negative gradient lobe data were processed separately and then combined in the spectral image domain. To recover the maximum possible information from the data, samples from the symmetric EPSI waveform transitions (ramps) were incorporated to also reconstruct the data at a nominal resolution of 0.86cm3 after applying a Gaussian-weighted average regridding algorithm (22).
Data Analysis
The spectroscopic imaging data were analyzed to quantify the SNR of the peak heights of Cho, Cre, NAA, Lac (in phantom only) and lipid (Lip) (in volunteers only). For the lipid peak, which was not always in phase, peak heights were assessed as the maximum positive or minimum negative peak height. The noise was calculated, using all voxels in the PRESS box, as the minimum standard deviation of all continuous 20Hz segments within a 250Hz region without metabolite signal. Metabolite SNR values were then normalized to acquisition time (tACQ) and effective spatial resolution (res) as follows:
| (2) |
The effective spatial resolution was estimated using a simulation of the 3D point-spread function (PSF) in MATLAB (The Mathworks, Inc.) using the method described by Maudsley et al. (9). By integrating the total number of simulated voxels greater than 50% of the maximum signal in the 3D-PSF, the full-volume at half maximum (FVHM) could be estimated to represent effective spatial resolution. In this simulation, for traditionally encoded 12×12×8 k-space sampling, ellipsoidal k-space sampling, and SENSE k-space sampling, 1 cm3 was represented by 110,592 (483) simulated voxels (1 simulated voxel = 9.04×10−6 cm3); however, for the larger FOV EPSI techniques, 1 cm3 was represented by 32,768 (323) simulated voxels (1 simulated voxel = 30.52×10−6 cm3) due to computer memory limitations for the simulation. The estimated effective spatial resolution for each technique as calculated from the FVHM of the 3D-PSF simulation is shown in the far right column of Table 1.
For phantom data, all voxels within the prescribed PRESS box were analyzed to compare nSNR of Cho, Cre, NAA, and Lac. Median nSNR values for each metabolite were compared along with the standard deviation (SD) of these medians across each acquisition technique. The median percent coefficient of variation (%CV = SD/Median) within the PRESS box was also compared across all techniques to assess the within acquisition spatial variance in metabolites. The linewidth of NAA was analyzed similarly by looking at the median values, the standard deviation of the medians, and the median %CV within each acquisition.
For the volunteer data, the T1-weighted 3D-IRSPGR images were automatically segmented into partial volumes of NAWM, normal appearing gray matter (NAGM), and cerebral spinal fluid (CSF) (23,24). Partial volume effects were qualitatively examined by looking at the voxels near the edge of the ventricles in the normal volunteers. Normalized SNR values for Cho, Cre, and NAA, and NAA linewidths were compared across all techniques in the volunteers (as described in phantoms) only in voxels within the press box that contained at least 90% NAWM. Cho:NAA ratios were assessed in all voxels containing at least 90% NAWM and a Wilcoxon signed rank test and Spearman rank correlation were used to compare with the traditionally encoded MRSI ratios. The number of voxels that were at least 90% outside the CSF and contained detectable metabolite peaks were also counted to determine the spectral quality across all 1H 3D-MRSI methods. Cho, Cre, and NAA Peaks were considered detectable if they had a raw SNR value greater than 5. Lipid peaks were considered detectable if they had a peak height greater than 25% of the median NAWM NAA peak. Voxels in the volunteers that contained a detectable lipid peak were considered to have lipid contamination because the PRESS box was prescribed completely within the brain parenchyma.
RESULTS
Phantom Data
Median nSNR values for NAA and Lac (± SD) for the phantom MRSI acquisitions are shown in Table 2, including median raw SNR values. The medians of the variation within each scan are also shown as a coefficient of variation (%CV). The nSNR values for the ellipsoidal data were corrected by a spatial resolution factor of 2.43 to account for the increased effective voxel size due to the PSF associated with the ellipsoidal sampling (9,24). The SNR values for SENSE data were not corrected for the geometry (or g) factor (18). It was estimated that the median phantom nSNR dropped by approximately 31–39% for NAA, Cho, Cre, and Lac relative to normalized values for fully phase encoded data which translates to a median g-factor between 1.45 and 1.63 for the unregularized SENSE data. The Tikhonov regularized SENSE data showed an nSNR reduction of 4–18%, and a lower median g-factor of 1.04–1.21. In regridded symmetric EPSI data, the median raw SNR decreased by 3.5% compared to plateau-only symmetric EPSI data, but the expected decrease due to finer effective spatial resolution was approximately 14%. After correcting the raw SNR values for effective spatial resolution, the regridding algorithm showed a median improvement in nSNR of approximately 13% compared to plateau-only data.
Table 2.
Median raw SNR and normalized SNR (nSNR: cm−3 min−1/2) values for all spectroscopic imaging techniques for NAA and Lac in a phantom (N=4 scans). Standard deviation between the 4 scans is shown (for nSNR only) as median ± SD and the median variation within each scan is also presented as %CV. The Lac:NAA median ratio is displayed in the far right column.
| Encoding Technique | NAA raw SNR | NAA nSNR | Lac raw SNR | Lac nSNR | Lac:NAA |
|---|---|---|---|---|---|
| Traditional MRSI | 190.75 | 40.83 ± 1.53 (9.10%) | 43.43 | 9.30 ± 0.12 (15.20%) | 0.23 |
| Ellipsoidal MRSI | 289.92 | 38.26 ± 1.23 (9.09%) | 65.53 | 8.65 ± 0.21 (14.86%) | 0.23 |
| SENSE MRSI (Rx=2) | 83.86 | 25.15 ± 0.77 (16.64%) | 19.17 | 5.75 ± 0.23 (21.22%) | 0.23 |
| SENSE w/REG | 115.18 | 34.54 ± 2.41 (14.45%) | 22.81 | 6.84 ± 0.38 (24.06%) | 0.20 |
| Flyback EPSI | 116.03 | 37.20 ± 0.29 (9.10%) | 25.22 | 8.08 ± 0.17 (12.32%) | 0.22 |
| Symmetric EPSI | 109.87 | 35.32 ± 1.48 (9.52%) | 22.18 | 7.13 ± 0.53 (14.07%) | 0.21 |
| Symmetric w/ramps | 105.36 | 39.64 ± 1.39 (7.46%) | 20.95 | 7.88 ± 0.32 (14.12%) | 0.20 |
Volunteers
Table 3 shows median raw SNR and median nSNR (±SD) of NAA from NAWM in ten volunteers. Median Cho:NAA and Cre:NAA ratios are also shown, along with percentage of voxels containing aliased lipid. The plateau-only symmetric EPSI data showed a larger median nSNR loss (17–24%) than the flyback data (11–18%) for Cho, Cre, and NAA. Regridding the symmetric ramp data improved the median nSNR for Cho, Cre, and NAA by approximately 7.4, 3.1, and 3.7%, respectively. Both flyback and symmetric EPSI data had higher median nSNR values for all metabolites than unregularized parallel imaging data, which showed a median nSNR drop of approximately 34–42%. Regularization improved the parallel imaging data nSNR by a median of 34, 33, and 33%, respectively for Cho, Cre, and NAA, making the nSNR similar to that of the flyback and symmetric EPSI data.
Table 3.
Median raw SNR and normalized SNR (nSNR: cm−3 min−1/2) values for all spectroscopic imaging techniques for NAA in 10 volunteers. Standard deviation between the 10 scans is shown (for nSNR only) as median ± SD. Median metabolite ratios are also shown for each technique (Cho:NAA and Cre:NAA). Voxels were considered to have aliased lipid if Lip SNR was greater than 25% of median NAA SNR in NAWM. Flyback data were not acquired on one volunteer and symmetric data were not acquired on another.
| Encoding Technique | NAA raw SNR | NAA nSNR | Cho:NAA | Cre:NAA | % of voxels with aliased lipid |
|---|---|---|---|---|---|
| Traditional MRSI | 46.28 | 10.10 ± 1.09 (22.28%) | 0.48 | 0.39 | 0.0 ± 5 |
| Ellipsoidal MRSI | 68.75 | 9.22 ± 1.07 (23.48%) | 0.49 | 0.40 | 0.0 ± 7 |
| SENSE MRSI (Rx=2) | 20.54 | 6.16 ± 1.14 (23.19%) | 0.48 | 0.43 | 16.2 ± 27 |
| SENSE w/REG | 27.05 | 8.11 ± 1.74 (24.49%) | 0.49 | 0.42 | 12.3 ± 20 |
| Flyback EPSI | 25.87 | 8.29 ± 0.83 (21.12%) | 0.51 | 0.39 | 2.0 ± 4 |
| Symmetric EPSI | 24.73 | 8.05 ± 0.76 (26.18%) | 0.51 | 0.37 | 0.0 ± 2 |
| Symmetric w/ramps | 21.89 | 8.45 ± 1.14 (24.46%) | 0.50 | 0.37 | 2.2 ± 4 |
Linewidths, Metabolite Ratios, Lipid Contamination, and Qualitative Measures
The linewidths for NAA were similar across techniques, with median values of 5.5–6.5 Hz in phantom data and 9.2–11.1 Hz in volunteer data. Fully encoded MRSI data showed a median NAA linewidth of 6.2 Hz in the phantom and 10.1 Hz in volunteers. Table 4 shows median NAA linewidths for all of the techniques in phantoms and volunteers.
Table 4.
NAA linewidths for phantom and volunteers. Median values of the median linewidths are shown with standard deviation of the median linewidths as ±S.D. All voxels within the PRESS box were included for the phantom data, while only voxels containing at least 90% NAWM were used in the volunteer analysis.
| Encoding Technique | Phantom (N=4) NAA Linewidth (HZ) | Volunteer (N=10) NAA Linewidth (HZ) |
|---|---|---|
| Traditional MRSI | 6.2 ± 0.13 | 10.1 ± 0.64 |
| Ellipsoidal MRSI | 6.2 ± 0.15 | 10.3 ± 0.72 |
| SENSE MRSI (Rx=2) | 5.5 ± 0.22 | 9.2 ± 0.89 |
| Flyback EPSI | 5.9 ± 0.12 | 10.6 ± 0.50 |
| Symmetric EPSI | 6.5 ± 0.09 | 11.0 ± 0.52 |
| Symmetric EPSI w/ramps | 6.5 ± 0.11 | 11.1 ± 0.62 |
The relative metabolite ratios of Cho:NAA were similar across all MRSI techniques for volunteers compared with traditionally encoded MRSI (p<0.05 based upon a Spearman rank correlation). In the parallel imaging data set and both the symmetric and flyback datasets, the Wilcoxon signed-rank test showed only one volunteer had a significant difference in their Cho:NAA values when compared to fully encoded MRSI.
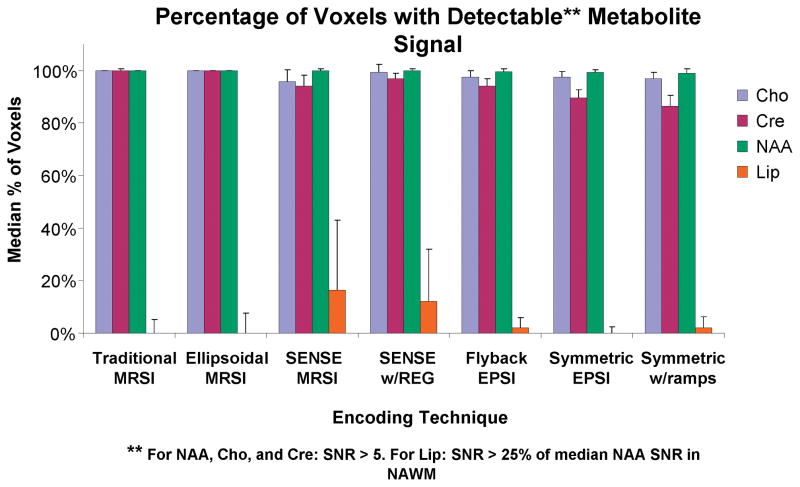
Analysis of spectral quality showed that between 96% and 100% of all voxels outside the ventricles had detectable NAA peaks (SNR > 5) for all 3D-MRSI techniques. For Cho and Cre the range was 85–100% and 82–100%, respectively. Figure 2 shows a bar graph of the median percentages of all detectable metabolites with error bars of one standard deviation for all techniques. Lipid aliasing was more noticeable in the parallel imaging techniques, as the median±SD percentage of voxels with lipid peaks greater than 25% of the median NAA signal from NAWM was 16.2±27% in the SENSE reconstructed data. With regularization, this number decreased to 12.3±20%. The EPSI acquisition techniques showed a lower median percentage of voxels with lipid aliasing (less than 2.2% for all techniques). Figure 3 shows two examples of the spectral quality for volunteers with traditional encoding, SENSE encoding, and flyback EPSI encoding.
Figure 2.
Median percentages of detectable voxels in each metabolite peak region are shown in the bar graph for each spectroscopic imaging technique. Error bars represent one standard deviation. Voxels were considered to have detectable NAA, Cho, and Cre signals if the metabolite raw SNR values were greater than 5. Voxels were considered to have aliased lipid if the lipid SNR was greater than 25% of the median SNR of NAA in NAWM.
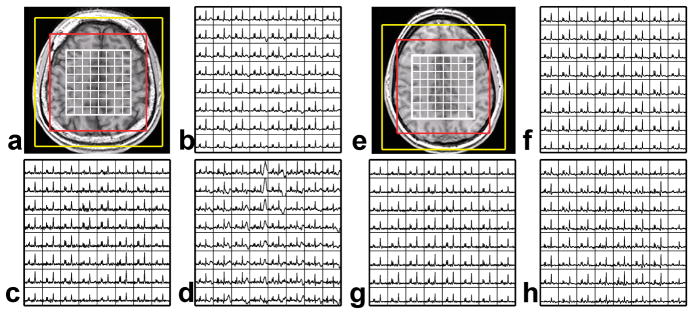
Figure 3.
Representative datasets from two volunteers. T1 weighted images are shown (a and e) with the spectral grid overlaid in white. The red box represents the FOV of the traditional, ellipsoidal, and SENSE encoded data, while the yellow box represents the FOV of the EPSI encoded data. Spectra acquired with a traditional phase encoding technique (b and f), a flyback EPSI technique (c and g), and with the SENSE technique (d and h) are shown for one slice with a 1cm3 nominal resolution. Lipid contamination is noticeable in d and slightly apparent in b.
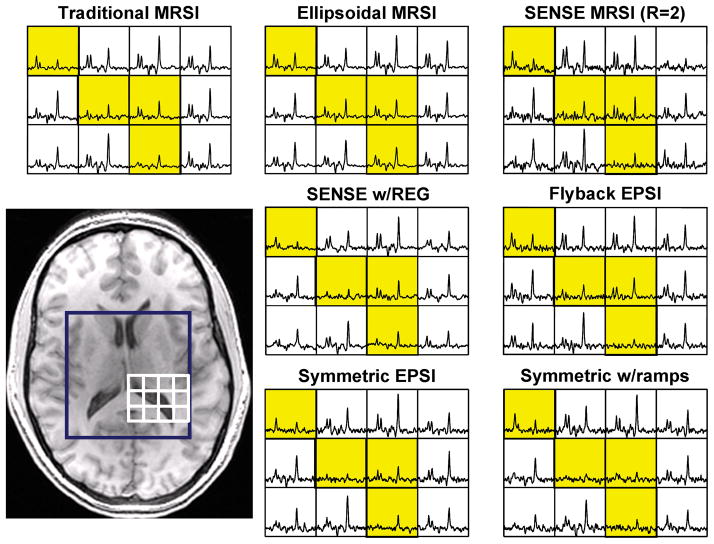
Figure 4 provides a representative dataset from a volunteer to show how partial voluming varies between techniques. The ellipsoidal encoded MRSI data are clearly inferior to the other data, as the definition of the ventricle is visibly reduced. As expected, the other techniques appear to have similar edge definition to the traditionally encoded MRSI.
Figure 4.
Qualitative examples of the partial volume effect in each spectroscopic imaging technique. Yellow boxes represent voxels containing ventricle.
Patients
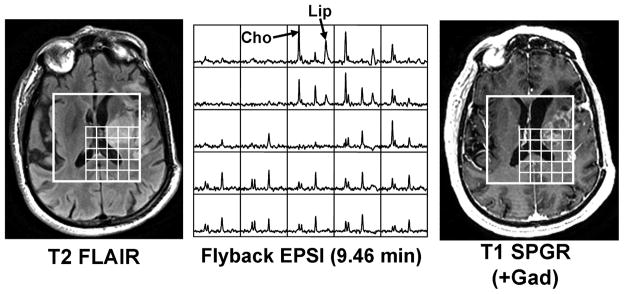
An example of data acquired on a glioma patient using a flyback EPSI sequence is shown in Figure 5. Differences in the pattern of metabolite levels were clearly visible in areas of T2 hyperintensity and contrast enhancement. Lipid, which is an important sign of necrosis, can be seen in multiple voxels in Figure 5. Based upon the analysis that we performed, these voxels would not have experienced aliasing from the scalp, and thus the lipid peaks may be attributed to the presence of necrosis. The median raw SNR values in NAWM for the seventeen patients were 21.15±4.52, 11.76±3.59 and 8.86±2.33 for NAA, Cho, and Cre, respectively. The median Cho:NAA metabolite ratios in NAWM were similar to the values from normal volunteers (not significantly different in a Mann-Whitney rank sum test (p > 0.10)). The median Cho:NAA ratio in the ten healthy volunteers’ NAWM was 0.53±0.04 for the traditionally encoded MRSI data, while the flyback data acquired in the seventeen patients had a median Cho:NAA ratio of 0.57±0.08 in NAWM. Lipid was detected within the tumors for 7 out of the 17 patient scans using the flyback EPSI technique. No lipid was detected outside of the T2 hyperintense or contrast enhancing lesions for any of the 17 scans.
Figure 5.
Typical data acquired on a glioma patient using a flyback EPSI sequence. High choline (Cho) and lipid (Lip) are markers of tumor pathology. Anatomical images (T2 FLAIR and T1 SPGR (+Gad)) are shown for reference.
DISCUSSION
The fast spectroscopic imaging techniques examined in this study produced data that would have been relevant for clinical applications. This is consistent with other studies in the literature (8–13). Each technique had its own advantages and limitations, and the choice of which is most appropriate for any specific application needs to take these factors into account. The ellipsoidal encoded MRSI data provided the highest absolute SNR, but had a broader PSF that caused a 2.43-fold increase in effective spatial resolution. This led to significant partial volume effects, which may not be important when looking at data with uniform concentrations of metabolites, but would make it extremely difficult to detect small lesions or heterogeneity in the case of tumors or other pathology. Showing a clear delineation between tumor and healthy tissue may be a crucial factor for surgeons and oncologists who wish to define the spatial extent of the lesion.
Spectroscopic imaging with parallel reconstruction methods can dramatically reduce acquisition time, but SNR may be lost due to the geometry of the coil array (18). The geometry or g-factor is the measurement used to assess such variations on a voxel by voxel basis. Our results showed a median nSNR loss in NAWM of volunteers of up to 42% in the unregularized data, which translated to a g-factor of approximately 1.7. After Tikhonov regularization was applied, the g-factor was reduced. For volunteers, the use of a regularization factor of 1.0 showed a median improvement in SENSE encoded nSNR of approximately 33–34%, reducing the g-factor to approximately 1.2. Tikhonov regularization has been shown here as an important technique for increasing the SNR for parallel imaging by aiding in the matrix inversion. Despite the improved SNR from regularization, SENSE techniques are more prone to spectral contamination from spatial aliasing than the other fast MRSI techniques because of the reduced FOV used for data acquisition. While this did not influence the quantification of NAA, it would have made it difficult to be certain whether the presence of lipid in tumor voxels was due to lipid contamination or to the tumor itself. Interpretation of patient data obtained via the SENSE methods will therefore be more dependent on the accuracy of coil calibration maps and the efficiency of VSS pulses in reducing the signal from subcutaneous lipid.
In this study, the traditionally and ellipsoidal encoded data had a smaller FOV than the width of the brain. Despite this, only one of the ten volunteers showed significant lipid contamination. The in plane FOV for the EPSI data was more similar to the size of the brain and only one volunteer showed more than 4% contaminated voxels with these techniques. Six of the ten volunteers showed lipid contamination in greater than 15% of the voxels in the SENSE encoded data. Additionally, the variation in the signal within each phantom scan was slightly higher for the SENSE reconstruction with and without regularization, as seen in the larger %CV values in Table 2, compared to the other techniques. The SENSE unfolding technique attempts to correctly unravel the aliased signal, but this correction is not always perfect. This may be why the relatively uniform metabolite signal in the phantom appears to vary slightly more in the SENSE reconstructed data. The spectra in Figure 3 illustrate how relatively small lipid contamination in the traditionally encoded data can get magnified using the SENSE technique. Note that data acquired from the same subject with the FOV used for the EPSI acquisitions show no noticeable lipid contamination.
Echo-planar spectroscopic imaging techniques can have the most significant effect on reducing acquisition time, but these techniques require a trade off between spectral bandwidth and spatial resolution due to the limited gradient strength and slew rates available on clinical systems. Unless the gradient strength can be increased, faster spectral sampling would broaden the spatial resolution in the EPSI dimension. Gradient imperfections, asymmetries, inefficient sampling on the gradient waveform, and imperfect combination of data (in the case of the symmetric EPSI data) are also expected to provide a loss in nSNR for these techniques. These results show median nSNR losses in volunteers that range from 11–17% for NAA compared to the traditional phase encoded data.
Because of the more efficient spatial sampling, the FOV can be increased for the EPSI dimension at no cost until the maximum sampling frequency is reached. This is important when trying to determine if tissue contains lipid or lactate, which are markers of necrosis and/or hypoxia (25–27). The high level of lipid in the human scalp is of the order of 100 times the metabolites of interest, which makes lipid contamination a problem for small FOVs. This contamination can thus hinder the ability to assess real lipid or lactate, as they both resonate in the same region for 1H spectra. Another factor in our data acquisitions was that k-space was fully sampled with the EPSI techniques, which resulted in less partial voluming than the ellipsoidal techniques and thus maintained the ability to delineate the signals from adjacent tissues (see Figure 4). The flyback EPSI waveform was slightly less efficient than the symmetric EPSI waveform, but the data quality appeared superior. This is most likely due to the imperfect matching of the phases of the positive and negative gradient lobe data. After applying a Gaussian-weighted average regridding algorithm to the more efficient symmetric EPSI data, the nSNR did improve; but because the effective spatial resolution was reduced, the raw SNR was still lower than the flyback EPSI data.
Although the Cho:NAA ratios were not significantly different for NAWM, the raw median SNR of NAA in the patients was slightly lower than for the volunteers. The lower patient SNR may be attributed to the location of the selected volume for the patients, which was dictated by the tumor location that was sometimes below the selected volume in volunteers. This explains both the slightly lower median SNR and the larger variation in values that were observed. The flyback EPSI technique identified areas within the tumor that contained lipid for 7 of the 17 patients considered. This is important, as the presence of lipid is characteristic of high grade brain tumors, and the summed intensities of Lip/NAA within the region of the tumor having elevated Cho:NAA has been shown to be predictive of poor outcome (28). With the EPSI techniques, the larger FOV means that one can be confident that lipid signals within the tumor are coming from that spatial location rather than from subcutaneous fat.
In conclusion, this study compares the performance of a number of fast three dimensional spectroscopic imaging techniques. Ellipsoidal k-space encoding, parallel imaging with and without Tikhonov regularization, and echo-planar techniques all produce spectroscopic images that are qualitatively acceptable for clinical applications. Each technique has strengths and weaknesses, and it is possible to incorporate multiple strategies into a single fast spectroscopic imaging sequence and reconstruction package (29). The EPSI techniques seem to pose a slight advantage over the other techniques due to the increased FOV, which results in minimal lipid aliasing and preserves the PSF of larger phase encoded matrices. The flyback EPSI technique was shown to provide clinically relevant 3D-MRSI data in human glioma patients in less than 9.5 minutes with a 1cc spatial resolution.
Acknowledgments
This study was supported by the UC Discovery grants LSIT-01-10107 and ITL-BIO04-10148 which were funded in conjunction with GE Healthcare, and by NIH grants R01CA059880 and R01CA127612.
References
- 1.Bertolino A, Callicott JH, Nawroz S, et al. Reproducibility of proton magnetic resonance spectroscopic imaging in patients with schizophrenia. Neuropsychopharmacology. 1998 Jan;18(1):1–9. doi: 10.1016/S0893-133X(97)00090-0. [DOI] [PubMed] [Google Scholar]
- 2.Tedeschi G, Bonavita S, Barton NW, et al. Proton magnetic resonance spectroscopic imaging in the clinical evaluation of patients with Niemann-Pick type C disease. J Neurol Neurosurg Psychiatry. 1998 Jul;65(1):72–9. doi: 10.1136/jnnp.65.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurhanewicz J, Vigneron DB, Nelson SJ. Three-dimensional magnetic resonance spectroscopic imaging of brain and prostate cancer. Neoplasia. 2000 Jan-Apr;2(1–2):166–89. doi: 10.1038/sj.neo.7900081. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govindaraju V, Gauger GE, Manley GT, Ebel A, Meeker M, Maudsley AA. Volumetric proton spectroscopic imaging of mild traumatic brain injury. AJNR Am J Neuroradiol. 2004 May;25(5):730–7. [PMC free article] [PubMed] [Google Scholar]
- 5.Laprie A, Pirzkall A, Haas-Kogan DA, et al. Longitudinal multivoxel MR spectroscopy study of pediatric diffuse brainstem gliomas treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2005 May 1;62(1):20–31. doi: 10.1016/j.ijrobp.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Osorio JA, Ozturk-Isik E, et al. Considerations in applying 3D PRESS H-1 brain MRSI with an eight-channel phased-array coil at 3 T. Magn Reson Imaging. 2006 Dec;24(10):1295–302. doi: 10.1016/j.mri.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Osorio JA, Ozturk-Isik E, Xu D, et al. 3D 1H MRSI of brain tumors at 3.0 Tesla using an eight-channel phased-array head coil. J Magn Reson Imaging. 2007 Jul;26(1):23–30. doi: 10.1002/jmri.20970. [DOI] [PubMed] [Google Scholar]
- 8.Pohmann R, von Kienlin M, Haase A. Theoretical evaluation and comparison of fast chemical shift imaging methods. J Magn Reson. 1997 Dec;129(2):145–60. doi: 10.1006/jmre.1997.1245. [DOI] [PubMed] [Google Scholar]
- 9.Maudsley AA, Matson GB, Hugg JW, Weiner MW. Reduced phase encoding in spectroscopic imaging. Magn Reson Med. 1994 Jun;31(6):645–51. doi: 10.1002/mrm.1910310610. [DOI] [PubMed] [Google Scholar]
- 10.Dydak U, Weiger M, Pruessmann KP, Meier D, Boesiger P. Sensitivity-encoded spectroscopic imaging. Magn Reson Med. 2001 Oct;46(4):713–22. doi: 10.1002/mrm.1250. [DOI] [PubMed] [Google Scholar]
- 11.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn Reson Med. 1998 Jun;39(6):889–98. doi: 10.1002/mrm.1910390606. [DOI] [PubMed] [Google Scholar]
- 12.Posse S, Tedeschi G, Risinger R, Ogg R, Le Bihan D. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn Reson Med. 1995 Jan;33(1):34–40. doi: 10.1002/mrm.1910330106. [DOI] [PubMed] [Google Scholar]
- 13.Chen AP, Cunningham CH, Ozturk-Isik E, et al. High-speed 3T MR spectroscopic imaging of prostate with flyback echo-planar encoding. J Magn Reson Imaging. 2007 Jun;25(6):1288–92. doi: 10.1002/jmri.20916. [DOI] [PubMed] [Google Scholar]
- 14.Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. 2004 Mar;51(3):435–40. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 15.Haase A, Frahm J, Hänicke W, Matthaei D. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol. 1985 Apr;30(4):341–4. doi: 10.1088/0031-9155/30/4/008. [DOI] [PubMed] [Google Scholar]
- 16.Tran TK, Vigneron DB, Sailasuta N, et al. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000 Jan;43(1):23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 17.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001 Aug;46(2):228–39. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 18.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999 Nov;42(5):952–62. [PubMed] [Google Scholar]
- 19.Ozturk-Isik E, Crane JC, Cha S, Chang SM, Berger MS, Nelson SJ. Unaliasing lipid contamination for MR spectroscopic imaging of gliomas at 3T using sensitivity encoding (SENSE) Magn Reson Med. 2006 May;55(5):1164–9. doi: 10.1002/mrm.20860. [DOI] [PubMed] [Google Scholar]
- 20.Lin FH, Kwong KK, Belliveau JW, Wald LL. Parallel imaging reconstruction using automatic regularization. Magn Reson Med. 2004 Mar;51(3):559–67. doi: 10.1002/mrm.10718. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham CH, Vigneron DB, Chen AP, et al. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005 Nov;54(5):1286–9. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 22.Thompson AR, Bracewell RN. Interpolation and Fourier transformation of fringe visibilities. Astron J. 1974;79(1):11–24. [Google Scholar]
- 23.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Vigneron DB, Cha S, et al. Relationship of MR-derived lactate, mobile lipids, and relative blood volume for gliomas in vivo. AJNR Am J Neuroradiol. 2005 Apr;26(4):760–9. [PMC free article] [PubMed] [Google Scholar]
- 25.Alger JR, Frank JA, Bizzi A, et al. Metabolism of human gliomas: assessment with H-1 MR spectroscopy and F-18 fluorodeoxyglucose PET. Radiology. 1990;177:633–641. doi: 10.1148/radiology.177.3.2243962. [DOI] [PubMed] [Google Scholar]
- 26.Herholz K, Heindel W, Luyten PR, et al. In vivo imaging of glucose consumption and lactate concentration in human gliomas. Ann Neurol. 1992;31:319–327. doi: 10.1002/ana.410310315. [DOI] [PubMed] [Google Scholar]
- 27.Remy C, Fouilhe N, Barba I, et al. Evidence that mobile lipids detected in rat brain glioma by 1H nuclear magnetic resonance correspond to lipid droplets. Cancer Res. 1997;57:407–414. [PubMed] [Google Scholar]
- 28.Crawford FW, Khayal IS, McGue C, Saraswathy S, Pirzkall A, Cha S, Lamborn KR, Chang SM, Berger MS, Nelson SJ. Relationship of pre-surgery metabolic and physiological MR imaging parameters to survival for patients with untreated GBM. J Neurooncol. 2009 Feb;91(3):337–51. doi: 10.1007/s11060-008-9719-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin FH, Tsai SY, Otazo R, et al. Sensitivity-encoded (SENSE) proton echo-planar spectroscopic imaging (PEPSI) in the human brain. Magn Reson Med. 2007 Feb;57(2):249–57. doi: 10.1002/mrm.21119. [DOI] [PubMed] [Google Scholar]