Abstract
Objectives
To estimate the relative fitness differences between glycopeptide-resistant Enterococcus faecium (GREF) and glycopeptide-susceptible E. faecium (GSEF) from yearly surveillance data on the occurrence of GREF in Danish poultry farm environments.
Methods
A population genetic model was adapted to retrospectively estimate the biological fitness cost of acquired resistance. Maximization of a likelihood function was used to predict the longitudinal persistence of acquired resistance.
Results
Our analysis suggests strong selection against GREF following the 1995 ban on the glycopeptide growth promoter avoparcin. However, parameterizing the model with two selection coefficients suggesting a reduced negative effect of the acquired resistance on bacterial fitness over time significantly improved the fit of the model. Our analyses suggest that the acquired glycopeptide resistance will persist for >25 years.
Conclusions
Acquired resistance determinants in commensal E. faecium populations in Danish farm environments are likely to persist for decades, even in the absence of glycopeptide use.
Keywords: Enterococcus faecium, GREF, Danish
Introduction
Whereas a clear correlation exists between consumption of antibiotics and resistance development in microbial communities, the association between reduced usage and declining frequencies of resistance is less clear.1,2 Numerous experimental in vitro and in vivo studies have shown that newly acquired antibiotic resistance determinants impose a biological cost on their bacterial host.3 Theory predicts that after complete removal of antibiotic selective pressures, the frequency of resistance will decline as a function of the magnitude of these biological costs to the level where mutations or horizontal gene transfer eventually maintain it.4 However, validation of this prediction has been impeded by a lack of documented examples in which antibiotics have been completely removed from the environment. Long-term surveillance and analysis of appropriate empirical data sets are needed to better understand the fate of antibiotic resistance determinants after removal of positive directional selection.5
One example where a type of antibiotic has been completely removed from the environment concerns Danish and Norwegian poultry farms following the 1995 ban of the glycopeptide growth promoter avoparcin, which was used extensively in animal production in many European countries. Long-term use of avoparcin had selected for high relative proportions of vanA-type glycopeptide-resistant enterococci (GRE) in the farm environment.6 A ban was imposed because vanA conferred complete cross-resistance to clinically important glycopeptide antibiotics such as vancomycin and teicoplanin, considered to be antibiotics of last resort.
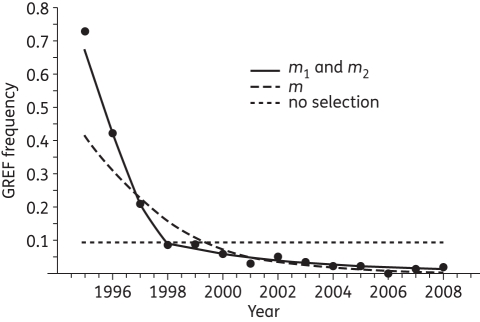
Surveillance data from Denmark demonstrated that the proportions of glycopeptide-resistant E. faecium (GREF) declined steeply in the first 3 years after the ban on avoparcin (Figure 1).7 However, the decline seems to have become more moderate in recent years. GREF were still isolated in 2% of the screened isolates in 2008.8 The Norwegian surveillance data are less comprehensive, but the existing data on GREF prevalence in poultry faeces are consistent with the Danish surveillance data.5 Here, we analyse the Danish surveillance data to retrospectively investigate this decline and estimate the difference in relative fitness between GREF and glycopeptide-susceptible E. faecium (GSEF). These estimates facilitate predictions of when the current surveillance programme will cease to detect GREF, and how long it will take for glycopeptide resistance determinants to be lost from the E. faecium population.
Figure 1.
Empirical observations of the frequency of GREF in Danish poultry samples 1995–2008 (filled circles). Lines show the fit of a maximum likelihood model to the surveillance data when there is no selection, a single selection coefficient for the entire time course (m) and two different selection coefficients (m1 and m2).
Methods
To quantify the biological cost of glycopeptide resistance, we applied a population genetic model to the available Danish surveillance data,7,8 under the assumption that the key difference between resistant and susceptible phenotypes is the presence or absence of horizontally acquired resistance determinants. The Danish surveillance data are based on non-selective isolations of E. faecium at slaughter and represent a stratified random sample of the respective populations. We assume, since GREF were obtained in direct non-selective plating assays, that the proportions of GREF reflect the actual proportions in poultry faeces.8 We estimate the cost of resistance based on a deterministic, continuous-time, haploid model of the frequency of resistance in response to selection:9,10
| (1) |
where pt is the frequency of resistance at time t, p0 is the frequency of resistance upon removal of the antibiotic from the environment (i.e. at the onset of the antibiotic-free period) and m is the continuous-time Malthusian selection coefficient. This model may be fitted to yearly observed count data on k GREF isolates detected in a sample of size n by a binomial maximum likelihood function:
| (2) |
Results and discussion
Maximizing Equation 2 with a directional selection parameter of zero (m = 0) yields stasis in GREF frequency and is highly unlikely (Figure 1). Estimation of a single selection parameter, m, representing a single Malthusian selection coefficient over the entire 13 year time course, yields a maximum likelihood estimate for m of −0.44. Converting from continuous to discrete time parameters via m = ln w yields a discrete-time relative fitness (w) of w = 0.64. To get an idea of the unit of discrete time (w is dimensionless) we can further calculate the discrete-time selection coefficient s of 0.36/year (w = 1 − s).11 These data suggest strong selection against GREF between 1995 and 2008.
However, a closer look at the empirical data in Figure 1 suggests that after a rapid decrease in the frequency of glycopeptide resistance between 1995 and 1998, the rate of decline appears to level out after 1998. The fit of the model is significantly improved by parameterizing Equation 1 with two different continuous-time fitnesses, m1 and m2, where m1 (−1.0) applies for pt from 1995 to 1998 and m2 (−0.16) applies for pt from 1998 to 2008 (log likelihood improved from −38.2 to −26.3). These values of m correspond to discrete-time fitnesses of w1 = 0.37 and w2 = 0.85. Because a more gradual change in population fitness is conceivable, we also fitted the model with a third selection parameter for the period from 1997 to 1999. Despite the resulting increase in the number of free parameters, addition of the third parameter worsened the fit of the model (log likelihood dropped from −26.3 to −27.4).
The best fit of the two-parameter model (Figure 1) supports a reduction in the rate of GREF decline after 1998. It also suggests that the biological cost of carrying glycopeptide resistance determinants was substantially reduced in the E. faecium populations after a few years. One potential explanation is that the initial strong selective disadvantage of being glycopeptide resistant was mitigated by compensatory evolution. Alternatively, it is possible that the reduced decline in GREF frequencies may have other causes, such as linked selection and possibly clonal shifts in the E. faecium populations.5 Revealing the proximate causes and factors resulting in this selection would require a detailed ecological/epidemiological model with extensive and poorly known ecological parameters. Since E. faecium is part of the normal flora of poultry it is reasonable to assume that the total population is relatively stable over time. As we assume a constant overall population size, many ecological factors may not be of direct relevance to our study as we are concerned with the effect of recently acquired glycopeptide resistance in previously susceptible phenotypes. Compensatory adaptation is not assured, and is not amenable to experimental tests in the genetically heterogeneous Enterococcus populations. Despite the inability to dissect additionally contributing factors, our likelihood analysis strongly suggests that a significant reduction in the cost of glycopeptide-resistant phenotypes has occurred over a short time period.
The difference in relative fitness between GREF and GSEF may be associated with the regulated expression of the Tn1546-encoded vanA operon. Foucault et al.12 recently demonstrated that the inactivated or inducible Tn1549-encoded vanB operon was not costly to the enterococcal host in the absence of vancomycin. However, while Tn1549 often has a chromosomal location, Tn1546 is far more frequently located on a genetically diverse set of large plasmids,5 and the initial cost of carrying acquired vancomycin resistance thus includes carrying the extrachromosomal elements.
The maximum likelihood model applied to the Danish surveillance data can further provide realistic estimates of how much time will pass before GREF will decline to frequencies at which the current surveillance programme in Denmark would not successfully isolate GREF by non-selective direct plating. Moreover, the time needed for complete loss of resistance in farm animals can be calculated (Table 1). Extrapolation from the model (assuming a single selection coefficient of m = −0.44) predicts sporadic detection until 2015, a low probability of detection until 2026 and complete reversal of glycopeptide resistance by 2036. However, extrapolation based on the better fitting model (with two selection coefficients and assuming no further drop in the second, smaller selection coefficient), predicts sporadic detection until 2027, increasingly reduced probability of detection until 2046 and complete loss of resistance by 2065. These estimates are helpful but are fairly crude predictions of a potentially complex outcome. The population genetic model applied here provides estimates for the waning of GREF relative to GSEF in response to selection only (i.e. the cost of acquired glycopeptide resistance). Clonal shifts in the heterogeneous E. faecium populations (unrelated to glycopeptide resistance carriage) could shorten or prolong the time. Moreover, ongoing compensatory evolution might fully restore the fitness of GREF strains and would in that case prolong the time until reversal of GREF.
Table 1.
Estimated time for loss of vancomycin resistance in E. faecium in the normal microbiota of poultry in Denmark
| Years (from 2010) to reach a frequency ofa |
|||
|---|---|---|---|
| Selection coefficients | 10−4 | 10−6 | 10−8 |
| m (−0.44) | 5 | 16 | 26 |
| m1 (−1.0) 1995–98 and m2 (−0.16) 1998–2008 | 17 | 36 | 55 |
aThe GREF detection limit varies from year to year in DANMAP (Danish Integrated Antimicrobial resistance Monitoring and Research Programme) (from 0.09 to 0.005).7,8 We assume that at a frequency of 10−4 GREF will only rarely be detected and at a frequency of 10−6 GREF will not be detected, and we consider resistance to be lost at a frequency of 10−8 (prevalence below the enterococcal population per individual farm animal).14
With these caveats in mind, it remains clear that whereas the prevalence of resistance is substantially reduced, and therefore the goal of lowering the exposure rate to humans of GREF infections from animal sources may have been met, the long-term persistence at the single farm animal level entails that reintroduction of glycopeptides will lead to a rapid population takeover by resistant phenotypes.
In conclusion, the presented results suggest that even when the biological cost of vancomycin resistance is initially high, the resistance determinants may persist for decades after a complete cessation of glycopeptide consumption in farm animals. From a clinical perspective, GREF of animal sources are generally not considered to be nosocomial pathogens at the strain level.13 However, the long-term persistence of GREF in food animals is worrying as a reservoir for horizontal transfer of the vanA gene cluster to clinically relevant enterococcal genetic lineages as well as to more pathogenic Gram-positive bacteria.
Funding
P. J. J. and K. M. N. were funded by the Norwegian Research Council (NFR), grant 172046. A. S. was supported by NFR grants 165997 and 183653, the European Commission (LSHE-CT-2007-037410 ‘ACE’) and the Medical Research Foundation, North Norway.
Transparency declarations
None to declare.
References
- 1.Seppala H, Klaukka T, Vuopio-Varkila J, et al. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. Finnish Study Group for Antimicrobial Resistance. N Engl J Med. 1997;337:441–6. doi: 10.1056/NEJM199708143370701. doi:10.1056/NEJM199708143370701. [DOI] [PubMed] [Google Scholar]
- 2.Enne VI, Livermore DM, Stephens P, et al. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet. 2001;357:1325–8. doi: 10.1016/S0140-6736(00)04519-0. doi:10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 3.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nature Rev Microbiol. 2010;8:260–71. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 4.Levin BR, Lipsitch M, Perrot V, et al. The population genetics of antibiotic resistance. Clin Infect Dis. 1997;24(Suppl 1):S9–16. doi: 10.1093/clinids/24.supplement_1.s9. [DOI] [PubMed] [Google Scholar]
- 5.Johnsen PJ, Townsend JP, Bohn T, et al. Factors affecting the reversal of antimicrobial-drug resistance. Lancet Infect Dis. 2009;9:357–64. doi: 10.1016/S1473-3099(09)70105-7. doi:10.1016/S1473-3099(09)70105-7. [DOI] [PubMed] [Google Scholar]
- 6.Wegener HC, Aarestrup FM, Jensen LB, et al. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerg Infect Dis. 1999;5:329–35. doi: 10.3201/eid0503.990303. doi:10.3201/eid0503.990303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aarestrup FM, Seyfarth AM, Emborg HD, et al. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob Agents Chemother. 2001;45:2054–9. doi: 10.1128/AAC.45.7.2054-2059.2001. doi:10.1128/AAC.45.7.2054-2059.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DANMAP. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria From Food Animals, Foods, and Humans in Denmark. ISSN 1600–2032. http://www.danmap.org/ [all DANMAP reports 2001–08, p. 38 (2001), p. 31 (2002), p. 32 (2003), p. 44 (2004), p. 51 (2005), p. 47 (2006), p. 59 (2007) and p. 68 (2008)] (6 December 2010, date last accessed) [Google Scholar]
- 9.Hartl D, Clark AG. Principles of Population Genetics. Sunderland, MA: Sinauer Associates, Inc.; 1997. [Google Scholar]
- 10.Nielsen KM, Townsend JP. Monitoring and modelling horizontal gene transfer. Nature Biotechnol. 2004;22:1110–14. doi: 10.1038/nbt1006. doi:10.1038/nbt1006. [DOI] [PubMed] [Google Scholar]
- 11.Lenski RE, Rose MR, Simpson SC, et al. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–41. doi:10.1086/285289. [Google Scholar]
- 12.Foucault ML, Depardieu F, Courvalin P, et al. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci USA. 2010;107:16964–9. doi: 10.1073/pnas.1006855107. doi:10.1073/pnas.1006855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willems RJ, Bonten MJ. Glycopeptide-resistant enterococci: deciphering virulence, resistance and epidemicity. Curr Opin Infect Dis. 2007;20:384–90. doi: 10.1097/QCO.0b013e32818be63d. doi:10.1097/QCO.0b013e32818be63d. [DOI] [PubMed] [Google Scholar]
- 14.Sorum M, Johnsen PJ, Aasnes B, et al. Prevalence, persistence, and molecular characterization of glycopeptide-resistant enterococci in Norwegian poultry and poultry farmers 3 to 8 years after the ban on avoparcin. Appl Environ Microbiol. 2006;72:516–21. doi: 10.1128/AEM.72.1.516-521.2006. doi:10.1128/AEM.72.1.516-521.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]