Abstract
This study examined molecular (DNA hypermethylation), clinical, histopathological, demographical, smoking, and alcohol variables to assess diagnosis (early versus late stage) and prognosis (survival) outcomes in a retrospective primary laryngeal squamous cell carcinoma (LSCC) cohort. The study cohort of 79 primary LSCC was drawn from a multi-ethnic (37% African American), primary care patient population, diagnosed by surgical biopsies in the Henry Ford Health System from 1991 to 2004 and followed from 5 to 18 years (through 2009). Of the 41 variables, univariate risk factors of p < 0.10 were tested in multivariate models (logistic regression (diagnosis) and Cox (survival) models (p < 0.05)). Aberrant methylation of estrogen receptor 1 (ESR1; p = 0.01), race as African American (p = 0.04), and tumor necrosis (extensive; p = 0.02) were independent predictors of late stage LSCC. Independent predictors of poor survival included presence of vascular invasion (p = 0.0009), late stage disease (p = 0.03), and methylation of the hypermethylated in cancer 1 (HIC1) gene (p = 0.0002). Aberrant methylation of ESR1 and HIC1 signified independent markers of poorer outcome. In this multi-ethnic, primary LSCC cohort, race remained a predictor of late stage disease supporting disparate diagnosis outcomes for African American patients with LSCC.
Keywords: Laryngeal cancer, Hypermethylation, ESR1, HIC1
Introduction
Laryngeal cancer represents the largest subgroup of head and neck cancers (Clayman et al. 2000). Roughly 12,250 new cases of laryngeal cancer are diagnosed each year in the USA (Horner et al. 2009). Given the fundamental role the larynx plays in human speech and communication, determining the optimal management of laryngeal cancers is critical. Treatment options comprise radiotherapy, surgery, chemotherapy or a combination of modalities. Despite refinement of multimodal therapies over the last 20 years, 5-year survival rates of 40% have remained static since the mid-1980s (Parkin et al. 2001).
Although the importance of genetic alterations in cancer has long been recognized, the appreciation of epigenetic changes is more recent and growing. The term “epigenetics” defines all meiotically and mitotically heritable changes in gene expression that are not coded in the DNA sequence itself (Egger et al. 2004). Establishment and maintenance of epigenetic control (gene silencing) has several aspects, which include promoter region hypermethylation, methyl-binding proteins, DNA methyltransferases (DNMTs), histone deacetylases (HDAC), and chromatin state.
DNMTs are enzymes that catalyze the methyl-transfer reaction. Global cytosine methylation patterns in mammals appear to be established by an interplay of three DNMTs: DNMT1, DNMT3A, and DNMT3B. The role of DNMTs is evolving from mere enzymes that copy methylation patterns after replication to being regarded as components of larger complexes actively involved in transcriptional control and chromatin structure modulation (Robertson 2001).
In addition to DNMTs, HDACs play a role in repressing DNA transcription. The HDACs deacetylate core histone tails resulting in tighter packaging of the DNA, making it difficult for transcription factors to access their binding sites (Robertson and Wolffe 2000). Following DNA methylation, methyl-CpG-binding proteins are recruited along with HDACs. This link between DNA methylation and histone deacetylation has been demonstrated by treating cells with a combination of the DNMT inhibitor 5-aza-2′-deoxycytidine (5-azaCdR) and the HDAC inhibitor trichostatin A (TSA). Treatment with 5-azaCdR alone resulted in low-levels of re-expression and minimal demethylation of hypermethylated genes, but a combination of 5-azaCdR and TSA resulted in robust activation of the same genes (Cameron et al. 1999), revealing that DNA methylation and histone deacetylation work together to silence transcription but also that DNA methylation was dominant over histone acetylation status.
Hypermethylation is a well-described DNA modification that has been implicated in normal mammalian development (Costello and Plass 2001; Li et al. 1992), imprinting (Li et al. 1993), and X chromosome inactivation (Pfeifer et al. 1990). CpG islands, which are stretches of DNA with a GC content greater than 55% (Takai and Jones 2002) located in promoter regions of genes, are mainly unmethylated in normal tissues. Methylation of these CpG islands causes stable heritable transcriptional silencing (Egger et al. 2004). This anomalous hypermethylation has been noted in a variety of tumor-suppressor genes, whose inactivation has led many cells down the tumorigenesis continuum (Jones and Laird 1999; Baylin et al. 1998; Chan et al. 2000). Aberrant methylation of CpG islands is a hallmark of human cancers and is found early during carcinogenesis (Egger et al. 2004). Numerous tumor-suppressor genes have been implicated as targets for methylation in other cancers (Cairns 2004; Kim et al. 2004; Roman-Gomez et al. 2004). Promoter hypermethylation of genes in HNSCC have been reported for p16, p14, DAPK, RASSF1A, RARß2, MGMT, a DNA repair gene that functions to remove mutagenic (O6-guanine) adducts from DNA, and E-cadherin, a Ca2+-dependent cell adhesion molecule that functions in cell–cell adhesion, cell polarity, and morphogenesis (Esteller et al. 2001; Sanchez-Cespedes et al. 2000; Worsham et al. 2006; Zou et al. 2001; Pegg 1990; Hirohashi 1998).
Molecular and genetic prognosticators have been shown to play a role in the prevention, diagnosis, radiotherapy outcomes, and appropriateness of adjuvant chemotherapy for a wide spectrum of cancers (Lazarus et al. 1996), including laryngeal squamous cell carcinoma (LSCC). Diagnosis, prognosis, and treatment of these malignancies are expected to be greatly enhanced by the identification of tumor markers specific for LSCC.
Prognostic marker systems based on single parameters have generally proven inadequate. This study incorporated a multi-parametric platform comprising molecular (DNA hypermethylation), clinical, histopathological, demographical, and epidemiological risk variables including smoking and alcohol to model diagnosis (early versus late stage) and prognosis (survival) outcomes in LSCC.
Materials and methods
Cohort
The retrospective study cohort of 79 primary LSCC was examined for a comprehensive set of 41 variables to include eight histopathology factors (Sethi et al. 2009): tumor grade (well, moderate, and poorly differentiated), lymphocytic response (continuous rim/patchy infiltrate/absent), desmoplastic response (prominent and diffuse/patchy and irregular/focal/absent), pattern of invasion (host/tumor interface with pushing cohesive borders (mode 1)/solid cords (mode 2)/thin irregular cords(mode 3)/single cells(mode 4)), vascular invasion (identified/absent), perineural invasion (identified/absent), mitotic index (<5 mitosis per ten high power fields (HPF) and >5 mitosis per ten HPF), and necrosis (extensive/minimal/absent); demographics (four variables: race (as self-reported), gender, age, and marital status); clinical factors (three variables: comorbidity, pneumonia, and family history of cancer); smoking and alcohol; and promoter methylation status of 24 tumor-suppressor genes.
Patient tissue material for this study was obtained according to the Henry Ford Health System institutional review board protocols.
DNA extraction
Whole 5-µm tissue sections or microdissected LSCC lesions and adjacent normal when present were processed for DNA extraction as previously described (Stephen et al. 2007).
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) assay
Archival tissue DNA was interrogated for methylation status using the multi-gene methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) assay. MS-MLPA (Worsham et al. 2006; Chen et al. 2007), a modification of the conventional MLPA assay (Schouten et al. 2002), allows for the simultaneous detection of changes in methylation status as well as copy number changes of approximately 41 different DNA sequences in a single reaction requiring only 20 ng of human DNA.
Briefly, the MS-MLPA panel in the presence of HhaI detects aberrant promoter hypermethylation by taking advantage of an HhaI site in the gene probe of interest. The control gene probes, without an HhaI site, serve as undigested controls. A normal control DNA sample will generate 41 individual peaks for all probes in the absence of HhaI and 15 separate peaks in the presence of HhaI (Fig. 1). Normal controls for methylation assays are run using DNA from paraffin-embedded squamous epithelium from individuals with no evidence of cancer.
Fig. 1.
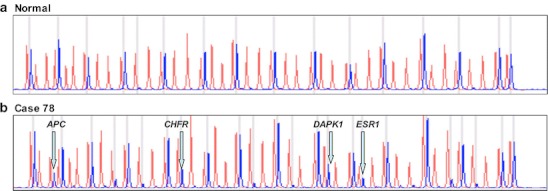
a A normal control DNA sample generates 41 individual peaks for all probes in the absence of HhaI (red) and 15 separate peaks in the presence of HhaI (blue). b Aberrant methylation identified in tumor sample as the appearance of a signal peak that is otherwise absent in normal DNA samples, seen here for APC, CHFR, DAPK1, and ESR1
Gene probe panels
The 41 gene probe panel (ME001B, www.mlpa.com) used in this cohort interrogates 38 unique genes (24 tumor-suppressor genes) implicated in squamous head and neck cancer (HNSCC) for methylation status in two separate reactions (one in the absence of the methyl-sensitive enzyme HhaI and one in the presence of the HhaI enzyme). There are two probes each for MLH1, RASSF1, and BRCA2, and a normal control DNA sample will generate 41 individual peaks in the absence of HhaI and 15 individual peaks in the presence of HhaI (Fig. 1).
Data analysis
Logistic regression and Cox regression models were used to determine risk factors for diagnosis (early vs. late stage) and for prognosis (survival), respectively. Univariate analysis was followed by multivariable modeling. Variables with p < 0.10 in the univariate analysis were tested as independent predictors in the multivariable modeling process. The final multivariate logistic regression (diagnosis) and Cox (survival) models included variables with p < 0.05. In order to address the issue of multiple comparisons, the multivariable stage model was selected by following the ten observations/events for each variable guideline (Harrell et al. 1984). Kaplan–Meier curves were generated to illustrate survival outcomes for independent risk factors.
Results
Of the 79 primary LSCC, 45 were Caucasian American (CA), 32 (41%) were African American (AA), and 2 were other race; 38 were with early stage, 40 late stage, and 1 unknown stage. There were 59 males and 20 females. Other cohort characteristics including age, smoking status, and alcohol use are presented in Table 1.
Table 1.
Cohort characteristics
| Variable | Response | Early stage, N = 38 | Late stage, N = 40 | Unknown stage, N = 1 |
|---|---|---|---|---|
| Race | African American | 11 | 20 | 1 |
| Caucasian American | 27 | 18 | – | |
| Other | – | 2 | – | |
| Gender | Male | 29 | 29 | 1 |
| Female | 9 | 11 | – | |
| Age | Less than 50 years | 18 | 16 | 1 |
| 51–65 years | 14 | 17 | – | |
| Over 65 years | 6 | 7 | – | |
| Smoking | Current smoker | 21 | 26 | 1 |
| Past smoker | 16 | 11 | – | |
| Never smoker | 1 | 3 | – | |
| Alcohol usea | No | 1 | 1 | – |
| Yes | 31 | 35 | 1 |
aMissing for ten LSCC
Of the 24 tumor-suppressor genes, 17 were aberrantly methylated in at least one case to include TIMP3, APC, CDKN2A, MLH1, RARB, CDKN2B, hypermethylated in cancer 1 (HIC1), CHFR, BRCA2, RASSF1, DAPK1, estrogen receptor 1 (ESR1), TP73, IGSF4, CDH13, GSTP1, and CDKN1B. The most frequently methylated genes were GSTP1 (34/79), CDH13 (27/79), TP73 (18/79), RARB (17/79), APC (13/79), CHFR (12/79), DAPK1 (11/79), CDKN2A, and ESR1 (10/79).
Four variables, ESR1, APC, tumor necrosis, and race, with univariate effects for late stage (p < 0.10) were included in the first multivariable model. After modeling, aberrant methylation of ESR1 (p = 0.014, OR = 16.35; 95% CI, 1.75, 152.5), race (p = 0.035, OR = 3.17; 95% CI, 1.08, 9.26), and extensive tumor necrosis (p = 0.018, OR = 5.15; 95% CI, 1.33, 20.01) remained in the final model as independent predictors of late stage LSCC (Table 2). The area under the curve, a measure of the model's predictive ability, was 0.78.
Table 2.
Multivariable stage model
| Variable | OR | 95% confidence limits | p value | |
|---|---|---|---|---|
| ESR1: methylation vs no methylation | 16.35 | 1.75 | 152.5 | 0.014 |
| Tumor necrosis: extensive vs none | 5.15 | 1.33 | 20.01 | 0.018 |
| Race: African American vs Caucasian American | 3.17 | 1.08 | 9.26 | 0.035 |
The median survival for patients in this cohort was 4.40 years (range, 0.04 to 16.21). Five variables, HIC1, DAPK1, vascular invasion, comorbidity, and stage, with individual effects (p < 0.10) for poor survival were included in the initial multivariable model. The final multivariate survival (Otterson et al. 1995) model indicated vascular invasion (p = 0.0009, HR = 4.51; 95% CI, 1.86, 10.93), late stage disease (p = 0.029, HR = 2.16; 95% CI, 1.08, 4.32), and methylation of the HIC1 gene (p = 0.0002, HR = 9.52; 95% CI, 2.92, 31.01) as independent predictors of poor survival (Table 3). Kaplan–Meier curves, generated for each risk variable retained in the final multivariable model, are illustrated in Figs. 2, 3, and 4. LSCC patients with vascular invasion (n = 8, adjusted p < 0.01) had a significantly shorter survival time as compared to patients without vascular invasion (n = 69, Fig. 2). LSCC patients with late stage disease (n = 40, stages 3 and 4, adjusted p = 0.029) had poorer survival as compared to those with early stage disease (n = 38, stages 0, 1, and 2, Fig. 3). LSCC patients without HIC1 methylation (n = 74) had a median survival of 4.40 (range, 0.04 to 16.21) as compared to a median survival of 1.02 years (range, 0.044 to 2.88) for patients with HIC1 methylation (n = 5, adjusted p < 0.01, Fig. 4).
Table 3.
Multivariable survival model
| Variable | Hazard ratio | 95% confidence limits | p value | |
|---|---|---|---|---|
| Stage: late vs early | 2.16 | 1.08 | 4.32 | 0.029 |
| HIC1: methylation vs no methylation | 9.52 | 2.92 | 31.01 | 0.0002 |
| Vascular invasion: identified vs not identified | 4.51 | 1.86 | 10.93 | 0.0009 |
Fig. 2.
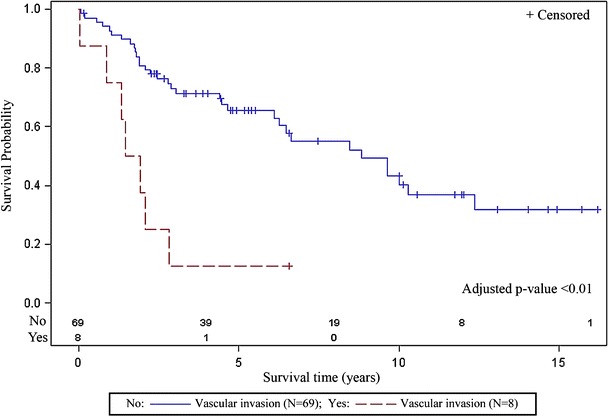
Patients with vascular invasion (n = 8) had a significantly shorter survival time as compared to patients without vascular invasion (n = 69)
Fig. 3.
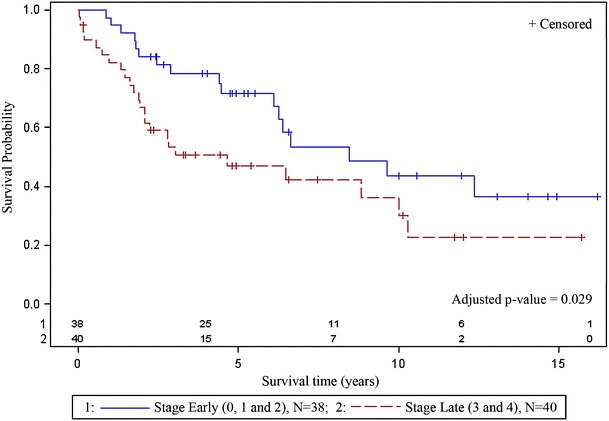
Patients with late stage disease (n = 40, stage 3 and 4) had poorer survival as compared to those with early stage disease (n = 38, stage 0, 1, and 2)
Fig. 4.
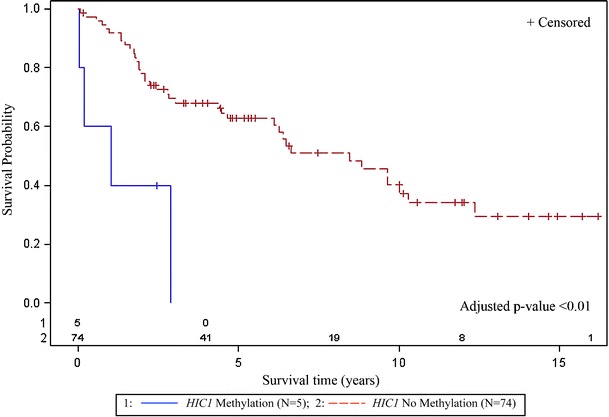
Methylation of HIC1 was an independent predictor of poorer survival. LSCC patients without HIC1 methylation (n = 74) had a median survival of 4.40 (range, 0.04 to 16.21) as compared to a median survival of 1.02 years (range, 0.044 to 2.88) for patients (n = 5) with HIC1 methylation
Discussion
Epigenetic mechanisms involve DNA and histone modifications resulting in the heritable silencing of genes without a change in their coding sequence. The major form of epigenetic information in mammalian cells is DNA methylation, or the covalent addition of a methyl group to the fifth position of cytosine within CpG dinucleotide predominantly located in the promoter region, which normally remains unmethylated in normal cells (Jones and Laird 1999; Baylin et al. 1998). The consequence of CpG island hypermethylation, especially for those islands associated with tumor-suppressor gene promoters, is the loss of tumor-suppressor gene function, which contributes to tumorigenesis (Worsham et al. 2003). Gene silencing, as a consequence of promotor hypermethylation, can be partially relieved by demethylation of the promoter region (Jones and Laird 1999; Baylin and Herman 2000). Recent work has revealed that DNA methylation is an important player in many processes, including DNA repair, genome instability, and regulation of chromatin structure (Jones and Laird 1999; Jones and Baylin 2002).
Promoter methylation-mediated silencing is a hallmark of many established tumor-suppressor genes. Aberrant methylation of promoter CpG islands, as an alternative to gene mutation or deletion in tumorigenesis, is now recognized as an important mechanism for gene inactivation (Baylin et al. 1998). Previous studies from our group and others have demonstrated aberrant DNA methylation patterns in HNSCC, underscoring a role for epigenetics in tumor pathogenesis.
Genes found to be methylated in LSCC and HNSCC include CDKN2A, CDKN2B, DAPK1, IGSF2, MLH1, and RB1. Inactivation of the CDKN2B/p15, CDKN2A/p14, and CDKN2A/p16 genes is a frequent event in human oral squamous cell carcinomas (Worsham et al. 2006; Shintani et al. 2001). The presence of aberrant methylation of p15 and p16 in precancerous oral tissues (Shintani et al. 2001) implicates methylation of p15 and p16 as early events in the pathogenesis of oral lesions.
Aberrant promoter methylation of DAPK1 has been shown to frequently occur in human head and neck cancers (Worsham et al. 2006; Sanchez-Cespedes et al. 2000), non-small-cell lung carcinomas (Esteller et al. 1999), gastric and colorectal carcinomas (Lee et al. 2002; Satoh et al. 2002), and uterine cervical carcinomas (Dong et al. 2001). In HNSCC, DAPK1 promoter hypermethylation has been associated with metastasis to lymph nodes as well as advanced disease stage (Sanchez-Cespedes et al. 2000). Promoter hypermethylation of IGSF2, a novel immunoglobulin-like intercellular adhesion molecule first characterized as a tumor suppressor of non-small cell lung cancer and termed TSLC1 (Kuramochi et al. 2001), has been reported in nasopharyngeal carcinomas (Hui et al. 2003). In esophageal squamous cell carcinomas, loss of expression correlated with promoter methylation status and TSLC1 expression was restored by demethylating agents in cell lines (Ito et al. 2003).
MLH1 belongs to the group of genes controlling mismatch repair (Arzimanoglou et al. 2002), and its frequent methylation in dysplastic lesions of HNSCC samples indicates MLH1 methylation as an early event in HNSCC tumorigenesis (Ghosh et al. 2010). RB1 plays an important role in cell cycle control (RB pathway) (Sherr 1996; Yokoyama et al. 1996). RB1 has been found to be methylated in oral squamous cell cancers, where it is highly correlated with tobacco use and/or alcohol consumption (Malekzadeh et al. 2009).
In this LSCC cohort, aberrant methylation of ESR1 and HIC1 was an independent predictor of late stage diagnosis and poorer survival outcomes, respectively. ESR1, at 6q25.1, is important for hormone binding, DNA binding, and activation of transcription (Ponglikitmongkol et al. 1988). ESR1 has metastasis-suppressor properties, suggesting a tumor-suppressor role for ESR1 (Issa et al. 1994). Methylation of CpG sites in the ESR1 promoter, with concordant loss or downregulation of ESR1 expression, is the primary mechanism in prostate cancer (Li et al. 2000). ESR1 exhibits age-dependent methylation in colon mucosa (Issa et al. 1994), the cardiovascular system (Post et al. 1999), ulcerative colitis (Issa et al. 2001), and prostate cancer, suggesting that ESR1 may be involved in age-dependent increase in cancer incidence.
Epigenetic silencing of HIC1 has been shown to significantly influence tumorigenesis (Rathi et al. 2003). The underlying mechanism is via HIC1's regulation of p53-dependent apoptotic DNA-damage responses through the HIC1-SIRT1-p53 circular loop (Chen et al. 2004). HIC1 encodes a transcriptional repressor with five Kruppel-like C2H2 zinc finger motifs and an N-terminal BTB/POZ domain (Wales et al. 1995). SIRT1 is an NAD+-dependent deacetylase, which is important for chromatin silencing, gene regulation, metabolism, and longevity (Haigis and Guarente 2006) and is a direct target of HIC1 via the POZ domain (Chen et al. 2005). Under normal physiological conditions, actively expressed HIC1 represses SIRT1 transcription, thereby allowing acetylation of p53 to enhance its function to control growth arrest and apoptosis in response to stress, such as DNA damage (Chen et al. 2004). During the course of aging, the HIC1 promoter undergoes hypermethylation, and this could set up transcriptional silencing of HIC1 and release its repressive effects on SIRT1 resulting in deacetylation of core histone and non-histone (p53) proteins. Chronic p53 deacetylation would attenuate its ability to transactivate or repress the expression of its downstream target genes for growth arrest and apoptosis, allowing cells to bypass these events and survive DNA damage (Chen et al. 2004). Deregulation of HIC1-SIRT1-p53 is a potential prognostic biomarker in lung cancer (Tseng et al. 2009). In pediatric tumor cell lines with aberrantly methylated HIC1, re-expression of HIC1 mRNA was induced by treatment with demethylating agent 5-aza 2′ deoxycytidine (Moscow et al. 1988).
In this cohort, survival of the five LSCC patients with HIC1 methylation of less than 3 years was remarkably poorer when compared to those without HIC1 methylation (Fig. 4). LSCC patients without HIC1 methylation (n = 74) had a median survival of 4.40 (range, 0.04 to 16.21) as compared to a median survival of 1.02 years (range, 0.044 to 2.88) for patients with HIC1 methylation. As an independent predictor of poor survival in this LSCC cohort, an aberrantly methylated HIC1 gene suggests a potential demethylating therapeutic target.
The most frequently methylated genes in this study cohort were GSTP1 (34/79), CDH13 (27/79), TP73 (18/79), RARB (17/79), APC (13/79), and CHFR (12/79) and underscore their involvement in the pathogenesis of LSCC.
Glutathione S-transferase pi (GSTP1) encodes for the glutathione S-transferase pi enzyme which plays an important role in detoxification. Promoter hypermethylation pattern of the p16, MGMT, GSTP1, and DAPK genes have been used as molecular markers for cancer cell detection in the paired serum DNA, and almost half of the HNSCC patients with methylated tumors were found to display these epigenetic changes in the paired serum (Sanchez-Cespedes et al. 2000).
Aberrant methylation of CDH13 gene was reported in colorectal, breast, lung cancers, and as a primary event in HNSCC cell lines (Worsham et al. 2006). In its tumor suppressor role of maintaining cell adhesion integrity, methylation-mediated silencing of CDH13 would allow tumor cells to spread, facilitating metastasis and poorer survival.
TP73 codes a product which has significant structural homology to the TP53 gene product in the domains involving transactivation, DNA binding, and oligomerization (Dong et al. 2002). In HNSCC, hypermethylation of TP73 occurred as a primary as well as a disease progression event (Worsham et al. 2006).
Promoter hypermethylation of APC has been reported in 25% of oral cancers (Uesugi et al. 2005). Aberrant promoter methylation of APC and RARB in early and late stage HNSCC suggests these occur as earlier epigenetic events when compared to methylation of CHFR (Chen et al. 2007). In this LSCC cohort, CHFR methylation occurred in eight early and four late stage tumors and, unlike HIC1, did not emerge as in independent predictor of late stage disease.
A current shortcoming in the more rigorous analysis of racial disparities in HNSCC is a dearth of study cohorts with adequate representation of minority patients. In this LSCC cohort, with 41% AA patient representation, AA were more likely to have advanced stage disease than their CA counterparts, and this is consistent with previous HNSCC studies from our group (Sethi et al. 2009).
There is substantial evidence that lack of adequate health insurance coverage is associated with less access to care and poorer outcomes for cancer patients (Ward et al. 2008), supporting insurance and cost-related barriers to high-quality prevention, early detection, and treatment as important measures to assess cancer disparities. Patients in this study cohort were primary LSCC within a primary health care setting. Of the 79 LSCC, insurance status was available in 69 (missing in 10/79), and only 1/69 lacked insurance, presenting a limitation of this variable in data analyses outcomes for this cohort.
Tumor behavior is dependent on a complex interrelationship between the tumor and patient (Sethi et al. 2009), and several studies have suggested expansion of the current TNM staging system to include host factors to augment the clinical utility and progress in cancer staging. In the present study, we evaluated the association of a broad spectrum of tumor histopathology characteristics at primary diagnosis in a diverse primary care LSCC cohort. Extensive tumor necrosis (p = 0.018) was an independent predictor of late stage disease and concurs with a highly significant association of necrosis and higher node-positive disease in HNSCC (Kuhnt et al. 2005).
Vascular invasion has been significantly correlated with cancers of the floor of the mouth (Suzuki et al. 2007), but there is a lack of information with respect to vascular invasion and LSCC. In this cohort, presence of vascular invasion (p = 0.0009) was a predictor of poor survival.
Smoking and alcohol abuse are well-established risk factors for LSCC (Hashibe et al. 2007). In this study cohort, the majority of patients were either current or past smokers (75/79) and alcohol users (67/69), reiterating the role of these risk factors in the pathogenesis of LSCC.
Epigenetic events of promoter hypermethylation are emerging as promising molecular targets for cancer detection and represent an important tumor-specific marker in tumorigenesis. Aberrant methylation of ESR1 and HIC1 were independent predictors of late stage LSCC and poorer survival, respectively. A limitation of this study remains the relatively small number of patient samples and its retrospective analysis. Validation of these findings in larger LSCC cohorts would further support these genes as important DNA methylation markers with a role in treatment given the reversible nature of promoter methylation-associated gene silencing. Race remained a predictor of late stage disease supporting disparate diagnosis outcomes for African American patients with LSCC.
Acknowledgements
Drs. Stephen and Worsham had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
This study was supported by R01 NIH DE 15990 (Dr. Worsham).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Arzimanoglou II, Hansen LL, Chong D, Li Z, Psaroudi MC, Dimitrakakis C, Jacovina AT, Shevchuk M, Reid L, Hajjar KA, Vassilaros S, Michalas S, Gilbert F, Chervenak FA, Barber HR. Frequent LOH at hMLH1, a highly variable SNP in hMSH3, and negligible coding instability in ovarian cancer. Anticancer Res. 2002;22(2A):969–975. [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16(4):168–174. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. doi: 10.1016/S0065-230X(08)60702-2. [DOI] [PubMed] [Google Scholar]
- Cairns P. Detection of promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Ann N Y Acad Sci. 2004;1022:40–43. doi: 10.1196/annals.1318.007. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Chan MF, Liang G, Jones PA. Relationship between transcription and DNA methylation. Curr Top Microbiol Immunol. 2000;249:75–86. doi: 10.1007/978-3-642-59696-4_5. [DOI] [PubMed] [Google Scholar]
- Chen W, Cooper TK, Zahnow CA, Overholtzer M, Zhao Z, Ladanyi M, Karp JE, Gokgoz N, Wunder JS, Andrulis IL, Levine AJ, Mankowski JL, Baylin SB. Epigenetic and genetic loss of hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 2004;6(4):387–398. doi: 10.1016/j.ccr.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor hic1 directly regulates sirt1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Chen K, Sawhney R, Khan M, Benninger MS, Hou Z, Sethi S, Stephen JK, Worsham MJ. Methylation of multiple genes as diagnostic and therapeutic markers in primary head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2007;133(11):1131–1138. doi: 10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- Clayman GL, Lippman SM, Laramore GF, et al. Neoplasms of the head and neck. Cancer medicine. 5. New York: BC Becker Inc; 2000. [Google Scholar]
- Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38(5):285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7(7):1982–1986. [PubMed] [Google Scholar]
- Dong S, Pang JC, Hu J, Zhou LF, Ng HK. Transcriptional inactivation of tp73 expression in oligodendroglial tumors. Int J Cancer. 2002;98(3):370–375. doi: 10.1002/ijc.10204. [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59(1):67–70. [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- Ghosh A, Ghosh S, Maiti GP, Sabbir MG, Zabarovsky ER, Roy A, Roychoudhury S, Panda CK (2010) Frequent alterations of the candidate genes hmlh1, itga9 and rbsp3 in early dysplastic lesions of head and neck: clinical and prognostic significance. Cancer Sci 101(6):1511–1520 [DOI] [PMC free article] [PubMed]
- Haigis MC, Guarente LP. Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Harrell FE, Jr, Lee KL, Califf RM, Pryor DB, Rosati RA. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3(2):143–152. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- Hashibe M, Boffetta P, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Mates D, Fabianova E, Rudnai P, Brennan P. Contribution of tobacco and alcohol to the high rates of squamous cell carcinoma of the supraglottis and glottis in Central Europe. Am J Epidemiol. 2007;165(7):814–820. doi: 10.1093/aje/kwk066. [DOI] [PubMed] [Google Scholar]
- Hirohashi S. Inactivation of the e-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153(2):333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MJ, Ries LAG, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinchcomb DG, Edwards BK. Seer cancer statistics review, 1975-2006. Bethesda: National Cancer Institute; 2009. [Google Scholar]
- Hui AB, Lo KW, Kwong J, Lam EC, Chan SY, Chow LS, Chan AS, Teo PM, Huang DP. Epigenetic inactivation of tslc1 gene in nasopharyngeal carcinoma. Mol Carcinog. 2003;38(4):170–178. doi: 10.1002/mc.10156. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor cpg island links ageing and neoplasia in human colon. Nat Genet. 1994;7(4):536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related cpg island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- Ito T, Shimada Y, Hashimoto Y, Kaganoi J, Kan T, Watanabe G, Murakami Y, Imamura M. Involvement of tslc1 in progression of esophageal squamous cell carcinoma. Cancer Res. 2003;63(19):6320–6326. [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21(2):163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Kim H, Kwon YM, Kim JS, Lee H, Park JH, Shim YM, Han J, Park J, Kim DH. Tumor-specific methylation in bronchial lavage for the early detection of non-small-cell lung cancer. J Clin Oncol. 2004;22(12):2363–2370. doi: 10.1200/JCO.2004.10.077. [DOI] [PubMed] [Google Scholar]
- Kuhnt T, Mueller AC, Pelz T, Haensgen G, Bloching M, Koesling S, Schubert J, Dunst J. Impact of tumor control and presence of visible necrosis in head and neck cancer patients treated with radiotherapy or radiochemotherapy. J Cancer Res Clin Oncol. 2005;131(11):758–764. doi: 10.1007/s00432-005-0018-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi M, Fukuhara H, Nobukuni T, Kanbe T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura T, Sekiya T, Reeves RH, Murakami Y. Tslc1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27(4):427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- Lazarus CL, Logemann JA, Pauloski BR, Colangelo LA, Kahrilas PJ, Mittal BB, Pierce M. Swallowing disorders in head and neck cancer patients treated with radiotherapy and adjuvant chemotherapy. Laryngoscope. 1996;106(9 Pt 1):1157–1166. doi: 10.1097/00005537-199609000-00021. [DOI] [PubMed] [Google Scholar]
- Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, Chung SC, Sung JJ, To KF. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res. 2002;8(6):1761–1766. [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li LC, Chui R, Nakajima K, Oh BR, Au HC, Dahiya R. Frequent methylation of estrogen receptor in prostate cancer: correlation with tumor progression. Cancer Res. 2000;60:702–706. [PubMed] [Google Scholar]
- Malekzadeh K, Sobti RC, Nikbakht M, Shekari M, Hosseini SA, Tamandani DK, Singh SK. Methylation patterns of rb1 and casp-8 promoters and their impact on their expression in bladder cancer. Cancer Invest. 2009;27(1):70–80. doi: 10.1080/07357900802172085. [DOI] [PubMed] [Google Scholar]
- Moscow JA, Townsend AJ, Goldsmith ME, Whang-Peng J, Vickers PJ, Poisson R, Legault-Poisson S, Myers CE, Cowan KH. Isolation of the human anionic glutathione s-transferase cdna and the relation of its gene expression to estrogen-receptor content in primary breast cancer. Proc Natl Acad Sci U S A. 1988;85(17):6518–6522. doi: 10.1073/pnas.85.17.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterson GA, Khleif SN, Chen W, Coxon AB, Kaye FJ. Cdkn2 gene silencing in lung cancer by DNA hypermethylation and kinetics of p16ink4 protein induction by 5-aza 2′deoxycytidine. Oncogene. 1995;11(6):1211–1216. [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94(2):153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Mammalian o6-alkylguanine-DNA alkyltransferase: Regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50(19):6119–6129. [PubMed] [Google Scholar]
- Pfeifer GP, Tanguay RL, Steigerwald SD, Riggs AD. In vivo footprint and methylation analysis by pcr-aided genomic sequencing: comparison of active and inactive x chromosomal DNA at the cpg island and promoter of human pgk-1. Genes Dev. 1990;4(8):1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- Ponglikitmongkol M, Green S, Chambon P. Genomic organization of the human oestrogen receptor gene. EMBO J. 1988;7(11):3385–3388. doi: 10.1002/j.1460-2075.1988.tb03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Milliken EE, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999;43:985–991. doi: 10.1016/S0008-6363(99)00153-4. [DOI] [PubMed] [Google Scholar]
- Rathi A, Virmani AK, Harada K, Timmons CF, Miyajima K, Hay RJ, Mastrangelo D, Maitra A, Tomlinson GE, Gazdar AF. Aberrant methylation of the hic1 promoter is a frequent event in specific pediatric neoplasms. Clin Cancer Res. 2003;9(10 Pt 1):3674–3678. [PubMed] [Google Scholar]
- Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J, Jimenez-Velasco A, Castillejo JA, Agirre X, Barrios M, Navarro G, Molina FJ, Calasanz MJ, Prosper F, Heiniger A, Torres A. Promoter hypermethylation of cancer-related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood. 2004;104(8):2492–2498. doi: 10.1182/blood-2004-03-0954. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Jen J, Herman JG, Sidransky D. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60(4):892–895. [PubMed] [Google Scholar]
- Satoh A, Toyota M, Itoh F, Kikuchi T, Obata T, Sasaki Y, Suzuki H, Yawata A, Kusano M, Fujita M, Hosokawa M, Yanagihara K, Tokino T, Imai K. DNA methylation and histone deacetylation associated with silencing dap kinase gene expression in colorectal and gastric cancers. Br J Cancer. 2002;86(11):1817–1823. doi: 10.1038/sj.bjc.6600319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Lu M, Kapke A, Benninger MS, Worsham MJ. Patient and tumor factors at diagnosis in a multi-ethnic primary head and neck squamous cell carcinoma cohort. J Surg Oncol. 2009;99(2):104–108. doi: 10.1002/jso.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Sciences. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Shintani S, Nakahara Y, Mihara M, Ueyama Y, Matsumura T. Inactivation of the p14(arf), p15(ink4b) and p16(ink4a) genes is a frequent event in human oral squamous cell carcinomas. Oral Oncol. 2001;37(6):498–504. doi: 10.1016/S1368-8375(00)00142-1. [DOI] [PubMed] [Google Scholar]
- Stephen JK, Vaught LE, Chen KM, Shah V, Schweitzer VG, Gardner G, Benninger MS, Worsham MJ. An epigenetically derived monoclonal origin for recurrent respiratory papillomatosis. Arch Otolaryngol Head Neck Surg. 2007;133(7):684–692. doi: 10.1001/archotol.133.7.684. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Suzuki T, Asai M, Ichimura K, Nibu K, Sugasawa M, Kaga K. Clinicopathological factors related to cervical lymph node metastasis in a patient with carcinoma of the oral floor. Acta Otolaryngol Suppl. 2007;559:129–135. doi: 10.1080/03655230701600020. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of cpg islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng R, Lee C, Hsu H, Tzao C, Wang Y. Distinct hic1-sirt1-p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia. 2009;11(8):763–770. doi: 10.1593/neo.09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi H, Uzawa K, Kawasaki K, Shimada K, Moriya T, Tada A, Shiiba M, Tanzawa H. Status of reduced expression and hypermethylation of the apc tumor suppressor gene in human oral squamous cell carcinoma. Int J Mol Med. 2005;15(4):597–602. [PubMed] [Google Scholar]
- Wales MM, Biel MA, Deiry W, Nelkin BD, Issa JP, Cavenee WK, Kuerbitz SJ, Baylin SB. P53 activates expression of hic-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1(6):570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- Ward E, Halpern M, Schrag N, Cokkinides V, DeSantis C, Bandi P, Siegel R, Stewart A, Jemal A. Association of insurance with cancer care utilization and outcomes. CA Cancer J Clin. 2008;58(1):9–31. doi: 10.3322/CA.2007.0011. [DOI] [PubMed] [Google Scholar]
- Worsham MJ, Pals G, Schouten JP, Spaendonk RM, Concus A, Carey TE, Benninger MS. Delineating genetic pathways of disease progression in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2003;129(7):702–708. doi: 10.1001/archotol.129.7.702. [DOI] [PubMed] [Google Scholar]
- Worsham MJ, Chen KM, Meduri V, Nygren AO, Errami A, Schouten JP, Benninger MS. Epigenetic events of disease progression in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2006;132(6):668–677. doi: 10.1001/archotol.132.6.668. [DOI] [PubMed] [Google Scholar]
- Yokoyama J, Shiga K, Sasano H, Suzuki M, Takasaka T. Abnormalities and the implication of retinoblastoma locus and its protein product in head and neck cancers. Anticancer Res. 1996;16:641–644. [PubMed] [Google Scholar]
- Zou CP, Youssef EM, Zou CC, Carey TE, Lotan R. Differential effects of chromosome 3p deletion on the expression of the putative tumor suppressor rar beta and on retinoid resistance in human squamous carcinoma cells. Oncogene. 2001;20(47):6820–6827. doi: 10.1038/sj.onc.1204846. [DOI] [PubMed] [Google Scholar]


