Abstract
Background: Samples used for genotyping and transcription studies are obtained and conserved in very specific conditions. The possibility to use autopsy tissue samples, which contain nucleic acids of very poor quality, would open new possibilities for genetic studies. Methods: We have used liver tissue samples from autopsy cases to (i) determine its quality; (ii) study gene expression of 13 genes involved in different cell processes, before and after cDNA pre-amplification (quantitative reverse transcriptase polymerase chain reaction); and (iii) analyze the presence of 2 common polymorphisms of relevance for illness (ACE I/D genotype by PCR amplification, and TNF-α promoter gene polymorphism, by DNA sequencing). Results: Samples were grouped according to different buffered formalin fixation times (group 1, <15 days; group 2, 60-90 days; group 3, 150-180 days; group 4, 240-270 days). Nucleic acids showed a time-dependent degradation. The expression of 13 genes could be studied in all cases from groups 1 and 2, only 7 from group 3 and none from group 4. cDNA preamplification allowed the study of all genes in all samples. DNA genotyping for ACE and TNF-α promoter region was possible in all cases. Conclusions: We conclude that nucleic acids extracted from autopsy specimens after prolonged periods of time in formalin were of sufficient quality to study gene expression and genotyping using currently available methodology and cDNA pre-amplification.
Keywords: Autopsy, formalin, gene expression, polymorphism, pre-amplification, critical illness
Introduction
The identification of certain single nucleotide polymorphisms or gene expression patterns associated to higher risk of developing ARDS or septic shock or to the development of certain outcomes has immediate clinical implications for prognostic and therapeutic predictions [1-6]. The best source of intact RNA and DNA are fresh or snap frozen tissue or blood samples. Recently, formalin fixed and paraffin embedded (FFPE) tissue specimens, usually from biopsy tissue samples, have been used for genome-wide microarray expression analysis or other assays involving nucleic acids [7,13]. These samples have usually been in formalin and embedded in paraffin for short (from hours to a few days) periods of time. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) has been introduced as a sensitive, accurate, and highly reproducible method to study gene expression, allowing the study of tissue containing degraded RNA, such as FFPE specimens that have been in formalin for short periods of time [14,15].
The availability of tissue samples from autopsy specimens that have been in formalin for prolonged periods of time for molecular biology studies would greatly broaden the options for better understanding critical illness [1].
Materials and methods
Tissue specimens
FFPE liver tissue preparations from 12 autopsy cases from patients dying in the Intensive Care Unit of our Institution were used in the present study. Cases were selected to cover a wide range of fixation times, i.e., <15 days (group 1), 60-90 days (group 2), 150-180 days (group 3) and 240-270 days (group 4). Tissues were fixed postmortem in buffered formalin 10 % for variable lengths of time, and included in paraffin.
RNA and DNA isolation
For total RNA isolation from FFPE samples, paraffin was removed and the RecoverAll Total Nucleic Acid Isolation protocol (Ambion) was performed according to the manufacturer's protocol. RNA and DNA quantification were determined with the NanoDrop ND-1000 Spectropho-tometer (NanoDrop Technologies Inc, Wilmington, DE). The OD260/280 ratio was used to evaluate the purity of the nucleic acid samples. To assess RNA quality, we used the RNA integrity number (RIN) value incorporated into the Agilent 2100 bioanalyzer software (Agilent Technologies, Palo Alto, CA) with the RNA 6000 Lab-Chip kit and standardized RNA ladder (Agilent Technologies) [16]. The length of extracted DNA was compared by electrophoresis of sample aliquots in an ethydium bromide stained aga-rose gel (1%-2% in Tris acetate EDTA buffer, pH 8). After electrophoresis, the gel was photographed under ultraviolet light.
Reverse transcription
Applied Biosystems high-Capacity cDNA reverse transcriptional Kit was used following manufacturer's protocol for reverse transcription with 1 μgtotal RNA (Applied Biosystems).
Real time polymerase chain reaction
Real time polymerase chain reaction step amplification was carried out on the Applied Biosystems 7500 Fast Real Time PCR System. Genes involved in inflammation, apoptosis, cell cycle and tissue remodeling were studied. Thirteen TaqMan Gene Expression Assays with an ampli-con size less than 100 pb [PGK1 (Hs99999906_m1), MMP2 (Hs00234422_m1), MMP9 (Hs00234579_m1), TIMP2 (Hs002342 78_m1), MDM2 (Hs00234753_m1, CDK1A (Hs00355782_m1, CDK1B (Hs00135277_m1, P53 (Hs00153349_m1), BCL2 (Hs00608023_ m1), BAX (Hs00180269_m1), ECA (Hs001741 79_m1), eNOS (Hs00167166_m1) and RAT1 (Hs00258938_m1)] were utilized in this study (Applied Biosystems). qRT-PCR reactions were run in triplicate. Cycle threshold values were determined using the SDS software of the 7500 Fast System (Version 2.0.1).
Real time polymerase chain reaction with Pre-Amp
The preamplification was performed using TaqMan PreAmp Master Mix Kit protocol (Applied Biosystems) according to the manufacturers’ instructions.
ACE genotyping
ACE I/D genotypes were determined by the PCR amplification method, as previously described [16]. This method yields amplification products of 84 bp for the D allele and 65 bp for the I al-lele. Products were visualized on 2% agarose gels.
TNF-alpha promoter gene sequencing
Genomic DNA was initially amplified by PCR using specific primers for TNF-alpha promoter gene. PCR amplification reactions were performed in 20 μl total volume containing 100 ng of genomic DNA, 10 pM of forward primer 5'-CTCAAGCCTGCCACCAAG-3', 10 pM of reverse primer 5'- TG'CCAACAACTGCCTTTATATG-3’ (Roche), 100 μM of dNTP mix, 1U EcoTaq (Ecogen) and 2μl of reaction buffer with 1.5 mM MgCl2. Amplification was carried out in a Verity thermocycler (Applied Biosystems) with cycle parameters of 5 min at 94°C followed by 35 cycles at 94 °C for 30 s, 65 °C for 30 s, 72 °C for 30 s, and a final extension at 72°C for 10 min. The PCR products (2 μl) were directly used as template for sequencing PCR, using BigDyeTM terminator v3.1 sequencing kit (Applied Biosystems) on an ABI3100 Avant Genetic Analyzer according to the manufacturer's instructions. The sequencing data were analysed and reviewed using Applied Biosystems DNA Sequencing Analysis Software v5.1.
Statistical analysis
Ct number before and after pre-amplification was compared by the Students’ t test. Data are presented as mean±SEM. We used SPSS.17 statistical package.
Results
Nucleic acid quality and degree of fragmentation
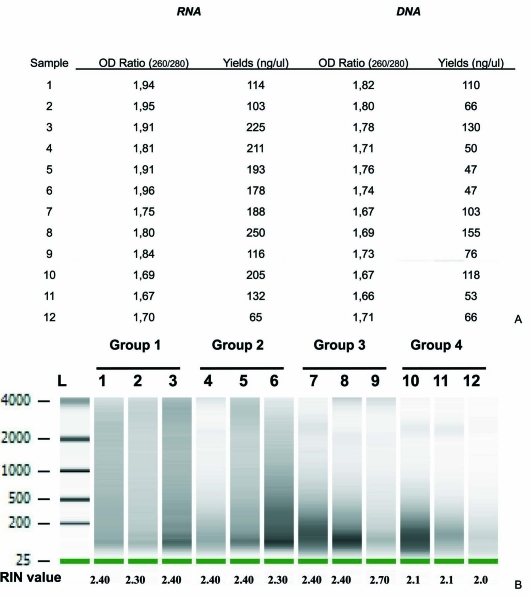
Yield of nucleic acids extracted from human FFPE liver tissue samples (time in buffered formalin from 2 weeks to 8 months) was satisfactory (Figure 1A). RNA and DNA were highly degraded (Figure 1A, 2B). RNA fragments size ranged from 200 to 4000 pb, being smaller for samples with longer fixation times. Very weak signals were detected in the electropherogram of cases 11 and 12, with very long fixation times. DNA obtained from FFPE tissues appeared to be degraded on ethydium-bromide-stained agarose gel electrophoresis and showed short fragments, especially when they were stored in buffered formalin for more than 5 months (Figure 2B).
Figure 1.
Recovery and integrity of isolated nucleic acid from FFPE tissues. A. Extraction yield and quality of nucleic acids from FFPE liver tissue samples. B. RIN and size distribution of RNA, according to formalin fixation time. Group 1, less than 15 days; group 2, 60-90 days; group 3, 150-180 days; group 4, 240-270 days. Lane L contains molecular weight marker RNA
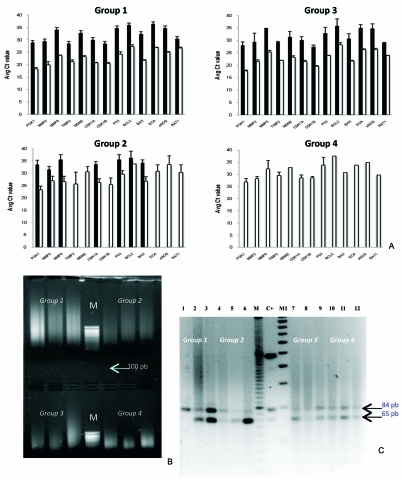
Figure 2.
A. Effect of pre-amplification on TaqMan gene expression pattern using qRTPCR in FFPE tissues. The pre-amplification (open bars) generated lower Ct levels in comparison with no pre-amplification (solid bars) in all groups. B. Electrophoretic pattern of DNA on 1% agarose ethydium bromide-stained gels photographed under ultraviolet light. C. Ethydium-bromide-stained gel showing samples of the ACE genotypes on 2% agarose gels. The longer fragment (84 bp) corresponds to the deletion (D) allele; the shorter fragment (65 bp), to the insertion (I) allele. Groups according to fixation times (as in Figure 1). M: marker (1kb DNA ladder), M1: marker (1000 pb DNA ladder) and C+ (positive control).
Evaluation of TaqMan PreAmp
The expression of 13 genes was studied in 4 different groups (3 cases per group) defined by different buffered formalin fixation times. In groups 1 and 2 all 13 genes could be studied. However, in group 3 only 7 of the 13 genes could be studied, and none in group 4 (Figure 2A). Pre-amplification of cDNA consistently achieved decreased Ct values (p<0.0001) compared with samples without pre-amplification for groups 1, 2 and 3 (Figure 2A), and allowed mRNA identification in all cases from groups 3 and 4 in whom no expression was detected before preamplification.
Analysis of ACE I/D genotyping and TNF-ALPHA-<x promoter sequencing
ACE genotyping could be determined in all samples (Figure 2C). One sample (lane 1) showed the DD genotype, another one presented the II genotype (lane 5), all others showing the ID genotype.
We further analyzed the polymorphism of the complete 540 bp promoter region upstream of the start codon of TNF-ALPHA-α gene by DNA sequencing. We were able to sequence successfully the promoter region of the TNF-ALPHA-α gene in all specimens (data not shown). Only one polymorphism was detected at position -308G/A in one case from group 1.
Discussion
It is not clear whether human tissue samples undergoing formalin fixation for prolonged periods of time, such as those from autopsy cases, can be used for molecular biology studies. Few studies have analyzed the ability to extract and study nucleic acids from samples after prolonged fixation and storage times. Santos et al. [17] reported the DNA extraction yield of human fetal umbilical cord tissue that had been in formalin for 19 years before paraffin embedding. DNA quality was poor (OD260/280=1.5) and only fragments less than 400 pb could be amplified. The present study is the first one reporting RNA and DNA extraction yields from samples under prolonged formalin fixation times, and the ability to study gene expression and DNA polymorphisms in these samples.
RNA was highly fragmented in a time-dependent manner (Figure 1B), confirming previous studies [1-5]. Despite the high degree of degradation, the study of all 13 genes was possible if fixation time was less than 90 days. However, cDNA pre-amplification was necessary to study gene expression in samples with more prolonged fixation times (Figure 2A). Pre-amplification of cDNA before qRT-PCR has been shown to be reliable and useful to study gene expression under conditions of limited copies of mRNA molecules in, for instance, breast cancer biopsy tissue samples [18] or cultured cells [19]. We here report for the first time that cDNA pre-amplification greatly improves the results of qRT-PCR in autopsy tissue samples, and pre-amplification is necessary to study gene expression in those samples with fixation times longer than 60 days. NA also showed a time dependent degradation (Figure 2B). However, DNA quality was fair (OD260/280 from 1.7 to 2.0 in most cases) (Figure 1A). To test the quality of DNA extracted from human liver tissue under conditions of prolonged formalin fixation time for genotyping analysis, we analyzed two commonly studied polymorphisms in these DNA fragments. ACE polymorphism could be determined in all 12 cases, even in those with the longest formalin fixation times (Figure 2C). In addition, gene sequencing for the study of TNF-ALPHA-α promoter gene polymorphism was possible in all cases (data not shown).
In summary, we report for the first time that nucleic acids extracted from human liver tissue samples under prolonged formalin fixation times (i.e., from autopsy specimens) can be used for molecular biology studies using the described methodology. Gene expression is possible even in samples with the longest formalin fixation times (180-240 days) using cDNA pre-amplification. The quality of DNA fragments is sufficient to study polymorphisms involving deletion/insertion or single nucleotide mutations.
Acknowledgments
This study was funded by FIS PI04/08/42, Instituto de Salud Carlos III, Spain. The study sponsor had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; nor in the decision to submit the paper for publication.
References
- 1.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive-gene expression in paraffin-embedded tissue. J Pathol. 2001;195:66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Jeffrey SS, Lonning PE, Hillner BE. Genomicsbased prognosis and therapeutic prediction in breast cancer. J Natl Compr Canc Netw. 2005;3:291–300. doi: 10.6004/jnccn.2005.0016. [DOI] [PubMed] [Google Scholar]
- 3.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–71. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suffredini AF, Chanock SJ. Genetic variation and the assessment of risk in septic patients. Intensive Care Med. 2006;32:1679–1680. doi: 10.1007/s00134-006-0328-x. [DOI] [PubMed] [Google Scholar]
- 5.Clark MF, Baudouin SV. A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med. 2006;32:1679–80. doi: 10.1007/s00134-006-0327-y. [DOI] [PubMed] [Google Scholar]
- 6.Flores C, Pino-Yanes MM, Villar J. A quality assessment of genetic association studies supporting susceptibility and outcome in acute lung injury. Crit Care. 2008;12:R130. doi: 10.1186/cc7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolinay T, Kaminski N, Felgendreher M, Kim HP, Reynolds P, Watkins SC, Karp D, Uhlig S, Choi AM. Gene expression profiling of target genes in ventilator-induced lung injury. Physiol Genomics. 2006;26:68–75. doi: 10.1152/physiolgenomics.00110.2005. [DOI] [PubMed] [Google Scholar]
- 8.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–43. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25:409–18. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Smyth P, Cahill S, Denning K, Flavin R, Aherne S, Pirotta M, Guenther SM, O'Leary JJ, Sheils O. Improved RNA quality and TaqMan Pre-amplification method (PreAmp) to enhance expression analysis from formalin fixed paraffin embedded (FFPE) materials. BMC Biotechnology. 2008;8:10. doi: 10.1186/1472-6750-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goulter AB, Harmer DW, Clark KL. Evaluation of low density array technology for quantitative parallel measurements of multiple genes in human tissue. BMC Genomics. 2006;7:34. doi: 10.1186/1471-2164-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noutsias M, Rohde M, Block A, Klippert K, Lettau O, Blunert K, Hummel M, Kuhl U, Lehmkuhl H, Hetzer R, Rauch U, Poller W, Pauschinger M, Schultheiss HP, Volk HD, Kotsch K. Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies. BMC Molecular Biology. 2008;9:3. doi: 10.1186/1471-2199-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mengual L, Burset M, Marín-Aguilera M, Ribal MJ, Alcaraz A. Multiplex preamplification of specific cDNA targets prior to gene expression analysis by TaqMan arrays. BMC Research Notes. 2008;1:21. doi: 10.1186/1756-0500-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DT, Jordan RC. Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol. 2002;15:979–87. doi: 10.1097/01.MP.0000026054.62220.FC. [DOI] [PubMed] [Google Scholar]
- 15.Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH. Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5' nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn. 2000;2:84–91. doi: 10.1016/S1525-1578(10)60621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The Ran RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans AE, Poirier O, Kee F, Lecerf L, McCrum E, Falconer T, Crane J, O´Rourke DF, Cambien F. Polymorphisms of the angiotensin-convertingenzyme gene in subjects who die from coronary heart disease. Q J Med. 1994;87:211–4. [PubMed] [Google Scholar]
- 18.Santos M, Saito C, Line S. Extraction of genomic DNA from paraffin-embedded tissue sections of human fetuses fixed and stored in formalin for long periods. Pathology-Research and Practice. 2008;204:633–36. doi: 10.1016/j.prp.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Ciotti P, Garuti A, Ballestrero A, Cirmena G, Chiaramondia M, Baccini P, Bellone E, Mandich P. Reliability and reproducibility of a RNA pre-amplification method for low-density array analysis from formalin-fixed paraffin-embedded breast cancer samples. Diagn Mol Pathol. 2009;18:112–8. doi: 10.1097/PDM.0b013e3181831320. [DOI] [PubMed] [Google Scholar]
- 20.Scicchitano MS, Dalmas DA, Bertiaux MA, Anderson SM, Turner LR, Thomas RA, Mirable R, RW Boyce. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J Histochem Cytochem. 2006;54:1229–37. doi: 10.1369/jhc.6A6999.2006. [DOI] [PubMed] [Google Scholar]