Abstract
Background
It is important to understand the cellular responses emanating from environmental perturbations to redesign the networks for practical applications. In particular, the carbon (C) metabolism, nitrogen (N) assimilation, and energy generation are by far important, where those are interconnected and integrated to maintain cellular integrity. In our previous study, we investigated the effect of C/N ratio on the metabolic regulation of gdhA, glnL, glt B,D mutants as well as wild type Escherichia coli (Kumar and Shimizu, MCF, 1-17, 9:8,2010), where it was shown that the transcript levels of cyoA and cydB which encode the terminal oxidases, fnr and fur which encode global regulators were significantly up-regulated under N-limited condition as compared to C-limited condition. In the present study, therefore, the effects of such single-gene knockout on the metabolic regulation were investigated to clarify the roles of those genes in the aerobic continuous culture at the dilution rate of 0.2 h-1.
Results
The specific glucose consumption rates and the specific CO2 production rates of cyoA, cydB, fnr, and fur mutants were all increased as compared to the wild type under both C-limited and N-limited conditions. The former phenomenon was consistent with the up-regulations of the transcript levels of ptsG and ptsH, which are consistent with down-regulations of crp and mlc genes. Moreover, the increase in the specific glucose consumption rate was also caused by up-regulations of the transcript levels of pfkA, pykF and possibly zwf, where those are consistent with the down regulations of cra, crp and mlc genes. Moreover, the transcript levels of rpoN together with glnK, glnB, glnE were up-regulated, and thus the transcript levels of glnA,L,G, and gltB,D as well as nac were up-regulated, while gdhA was down-regulated. This implies the interconnection between cAMP-Crp and PII-Ntr systems. Moreover, cyoA, cydB, fnr and fur gene deletions up-regulated the transcript levels of respiration (nuoA, ndh, cyoA, cydB, and atpA) and the oxidative stress related genes such as soxR, S and sodA, where this was further enhanced under N-limitation. In the cases of cyoA and cydB mutants, arcA, fnr, fur, cydB (for cyoA mutant), and cyoA (for cydB mutant) genes were up-regulated, which may be due to incomplete oxidation of quinol. It was also shown that fur gene transcript level was up-regulated in accordance with the activation of respiratory chain genes. It was shown that the deletion of fur gene activated the enterobactin pathway.
Conclusion
The present result demonstrated how the fermentation characteristics could be explained by the transcript levels of metabolic pathway genes as well as global regulators in relation to the knockout of such single genes as cyoA, cydB, fnr, and fur, and clarified the complex gene network regulation in relation to glycolysis, TCA cycle, respiration, and N-regulated pathways. The present result is quite important in understanding the metabolic regulation for metabolic engineering. Moreover, the present result may be useful in improving the specific glucose consumption rate and activation of the TCA cycle by modulating the respiratory chain genes and the related global regulators. The result obtained under N-limited condition may be useful for the heterologous protein production under N-limitation.
Background
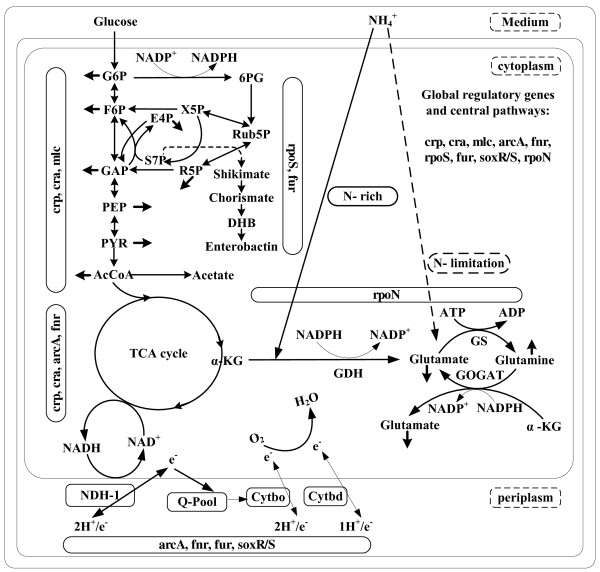
It is important to understand the cellular responses emanating from the environmental perturbations to redesign the networks for practical applications as well as for theoretical studies [1-3]. Microorganisms such as Escherichia coli live in environments which are subject to rapid changes in the availability of carbon (C) and nitrogen (N) sources [4,5]. The carbon metabolism, nitrogen assimilation, and energy generation are integrated to maintain the cellular integrity (Figure 1). The limitation of such nutrients stimulates hunger state responses in bacteria, which turns the emphasis on scavenging substrates and induction for stimulating nutrient acquisition [6-8]. Microbes adapt to the low nutrient conditions by maintaining high metabolic fluxes that may reduce the energetic efficiency of overall metabolism [9,10]. In the case of N-assimilation, energy independent glutamate dehydrogenase (GDH) pathway is used when sufficient amount of nitrogen is present, while energy dependent glutamine synthetase-glutamate synthase (GS-GOGAT) pathway is used under N-limitation (Figure 1) [2,11,12]. The bulk of energy is generated by the respiratory chain in E. coli under aerobic condition, and its efficiency depends upon the cumulative activity of various elements [13-15]. The aerobic respiratory chain consists of multiple elements such as NADH dehydrogenases, catalyzing the generation of proton motive force during NADH oxidation, and quinone pool containing terminal oxidases, transferring electrons to oxygen (Figure 1). It is also crucial for the maintenance of redox balance [16,17].
Figure 1.
Schematic diagram of integrated carbon, nitrogen, energy metabolism and its global regulation in E. coli.
In our previous study, metabolic regulations of E. coli and its single-gene knockout mutants such as gdhA, glnL, gltB and gltD mutants were investigated at various C/N ratios in the aerobic continuous culture [18]. From this study, it was found that transcript levels of the terminal oxidases encoded by such genes as cyoA and cydB, and the global regulatory genes such as fnr and fur were up-regulated under N-limitation especially at the C/N ratio of 16.48. The cyoA and cydB genes code for cytochrome bo oxidase, functional under aerobic condition (H+/e, 2), and cytochrome bd, active under microaerobic condition (H+/e, 1), respectively [19]. The fnr gene product, Fnr protein, is a primary transcriptional regulator that mediates the transition from aerobic to anaerobic growth [20-22]. The fur gene codes for the transcriptional activator Fur (Ferric uptake regulator), which controls its own synthesis as well as the transcription of genes involved in the iron homeostasis [23,24]. It also participates in the regulation of other cellular functions such as oxidative stress, glycolysis, TCA cycle, respiration, 2, 3-dihydroxybenzoate biosynthesis etc. [25-28]. From the practical application point of view, the role of aerobic respiratory chain and global regulators such as Fur has been highlighted in the biofuel production using E. coli [29-31]. The attention has also been paid to understand the role of Fur in iron uptake mechanisms with a view to find novel antimicrobial targets [32,33]. In the present study, therefore, the effects of cyoA and cydB genes knockout as well as fnr and fur genes knockout on the metabolism of E. coli were investigated based on fermentation characteristics and transcriptional mRNA levels in the aerobic continuous culture under both C- and N- limitations.
Results
Fermentation characteristics of the mutants
Table 1 shows the fermentation characteristics of cyoA, cyd, fnr and fur mutants for N- rich (C/N ratio 1.68), and N- limited (C/N ratio 8.42) conditions, respectively. It shows that the biomass concentrations of the mutants were reduced as compared to that of the wild type strain. In the case of cyoA mutant, the specific acetate production rate increased, while it decreased for the case of cydB mutant. Table 1 shows that the specific CO2 production rate was increased for both mutants as compared to that of the wild type under both N- rich and N- limited conditions. The specific glucose consumption rates of cyoA mutant were higher than those of the wild type for both N-rich and N-limited conditions. In the case of fnr mutant, biomass concentration reduced, and the specific acetate production rate and the specific glucose consumption rate increased under both N- rich and N- limited conditions as compared to those of the wild type (Table 1). The specific CO2 production rates were increased for the mutant as compared to those of the wild type under both N-rich and N-limited conditions. Similar to the case of fnr mutant, the biomass concentration reduced, while the specific acetate production rate increased for fur mutant under both N- rich and N- limited conditions (Table 1). One of the distinctive features of fur mutant was the pink color of fermentation broth under N-rich condition while it turned to violet under N-limitation [34].
Table 1.
Fermentation characteristics of the wild type E. coli, and its ΔcyoA, ΔcydB, Δfnr, and Δfur mutants under aerobic continuous at dilution rate 0.2h-1: C/N ratios (a) 1.68, (b) 8.42. Biomass (g/l) indicates total cells in the fermentor, while cell yield (g/g) reflects biomass formed on the consumed glucose.
| Wild | ΔcyoA | ΔcydB | Δfnr | Δfur | |
|---|---|---|---|---|---|
| (A) C- limitation (C/N ratio = 1.68) | |||||
| Biomass (g/l) | 3.33 ± 0.17 | 2.86 ± 0.14 | 3.08 ± 0.15 | 2.38 ± 0.12 | 2.63 ± 0.13 |
| Glucose (g/l) | ND* | ND* | ND* | ND* | ND* |
| Acetate (g/l) | 1.73 ± 0.09 | 1.64 ± 0.08 | 1.25 ± 0.06 | 2.06 ± 0.10 | 1.81 ± 0.09 |
| Cell yield (g/g) | 0.330 ± 0.02 | 0.286 ± 0.01 | 0.308 ± 0.02 | 0.238 ± 0.01 | 0.263 ± 0.01 |
| Specific glucose consumption rate (mmol/gdcw.h) | 3.33 ± 0.17 | 3.88 ± 0.19 | 3.60 ± 0.18 | 4.66 ± 0.24 | 4.21 ± 0.21 |
| Specific acetate production rate (mmol/gdcw.h) | 1.73 ± 0.09 | 1.91 ± 0.10 | 1.35 ± 0.07 | 2.88 ± 0.14 | 2.29 ± 0.11 |
| Specific CO2 production rate (mmol/gdcw.h) | 6.74 ± 0.34 | 7.45 ± 0.37 | 7.99 ± 0.40 | 9.00 ± 0.45 | 8.05 ± 0.40 |
| (B) N- limitation (C/N ratio = 8.42) | |||||
| Biomass (g/l) | 1.67 ± 0.08 | 1.56 ± 0.08 | 1.62 ± 0.08 | 1.48 ± 0.07 | 1.40 ± 0.07 |
| Glucose (g/l) | 3.03 ± 0.15 | 3.01 ± 0.15 | 3.22 ± 0.16 | 3.59 ± 0.18 | 3.68 ± 0.18 |
| Acetate (g/l) | 1.41 ± 0.07 | 1.58 ± 0.08 | 1.13 ± 0.06 | 1.32 ± 0.07 | 1.62 ± 0.08 |
| Cell yield (g/g) | 0.240 ± 0.01 | 0.223 ± 0.01 | 0.239 ± 0.01 | 0.231 ± 0.01 | 0.221 ± 0.01 |
| Specific glucose consumption rate (mmol/gdcw.h) | 4.65 ± 0.23 | 4.98 ± 0.25 | 4.67 ± 0.13 | 4.80 ± 0.24 | 5.02 ± 0.25 |
| Specific acetate production rate (mmol/gdcw.h) | 2.82 ± 0.14 | 3.38 ± 0.17 | 2.33 ± 0.12 | 2.98 ± 0.15 | 3.86 ± 0.19 |
| Specific CO2 production rate (mmol/gdcw.h) | 8.25 ± 0.41 | 8.88 ± 0.44 | 8.47 ± 0.42 | 9.23 ± 0.46 | 8.39 ± 0.42 |
ND* = Not detectable
Gene transcript levels of cyoA mutant
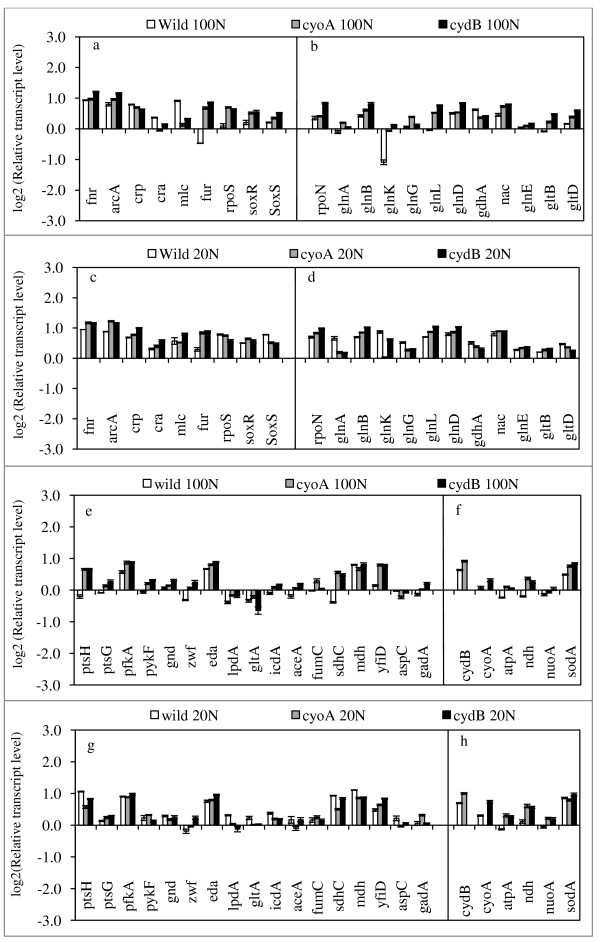
The relative transcript levels for cyoA and cydB mutants are given as compared to those of the wild type in Figure 2, where it indicates that the transcript levels of ptsG, ptsH, pfkA, pykF as well as zwf and eda were all up-regulated (p < 0.01, p < 0.01, p < 0.01, p < 0.01; p < 0.01, p < 0.01, respectively) in accordance with the increase in the specific glucose consumption rate for cyoA mutant as compared to the wild type (Figure 2e). This is consistent with the down-regulations of the transcript levels of cra (p < 0.05), crp (p < 0.01), and mlc (p < 0.01) genes (Figure 2a) (see Additional file 1). Figure 2e also shows that the transcript levels of lpdA, gltA, icdA, aceA, fumC, and sdhC were increased (p < 0.01, p < 0.01, p < 0.01, p < 0.05, p < 0.01, and p < 0.01, respectively) for cyoA mutant as compared to the wild type under N- rich condition. The increased TCA cycle activity caused higher specific CO2 production rate for the mutant as compared to the wild type, and affected respiration. Figure 2f indicates that cyoA gene knockout caused the transcript level of cydB gene to be up-regulated, and those of other respiratory chain genes such as nuoA, ndh, and atpA genes were also up-regulated (p < 0.01, p < 0.01, and p < 0.01, respectively). This is consistent with the up-regulations of soxR, S and sodA (p < 0.01, p < 0.01, and p < 0.01). Moreover, the increase in yfiD may be due to increase of arcA (p < 0.01). Note that aspC was down regulated (p < 0.01) and gadA was up-regulated (p < 0.01), which will be discussed in the discussion section. Figure 2b indicates that the transcript level of rpoN increased, which caused the transcript levels of glnB and glnK to be up-regulated (p < 0.01 and p < 0.01), and those of glnA, L, G and gltB, D genes were up-regulated (p < 0.01, p < 0.01, p < 0.01, and p < 0.01, p < 0.01, respectively). Moreover, the transcript level of nac gene increased (p < 0.01) and gdhA gene decreased (p < 0.01). Those imply that GDH pathway was inativated, while GS pathway was activated for cyoA mutant (as well as cydB mutant) even under N-rich condition (see Additional file 2). This phenomenon was also enhanced under N-limited condition (Figure 2d). Note that cyoA knockout caused rpoS and fur transcript levels to be up-regulated (p < 0.01 and p < 0.01) while fnr transcript level changed little (Figure 2a).
Figure 2.
Comparison of the transcriptional mRNA levels between the wild type E. coli, cyoA and cydB mutant genes at C/N ratio 1.68 and 8.42.
Under N- limitation, the transcript level of arcA was up-regulated (p < 0.01), and this caused lpdA and TCA cycle genes such as gltA, icdA, sdhC, and mdh to be down-regulated (p < 0.01, p < 0.01, p < 0.01 and p < 0.01, respectively). The up-regulation of arcA also caused yfiD to be up-regulated (p < 0.01) for cyoA mutant as compared to wild type. Moreover, the respiratory chain genes such as cydB, nuoA, ndh, atpA were all up-regulated (p < 0.01, p < 0.01, p < 0.01, p < 0.01, respectively).
Gene transcript levels of cydB mutant
Figure 2 also shows the transcript levels of cydB mutant as compared to those of the wild type and cyoA mutant. The transcript levels of nitrogen regulated genes were similar for both cyoA and cydB mutants. While cydB gene knockout caused the transcript level of cyoA to be up-regulated, the overall changing patterns of the transcript levels were quite similar to the case of cyoA mutant (Figure 2).
Gene transcript levels of fnr mutant
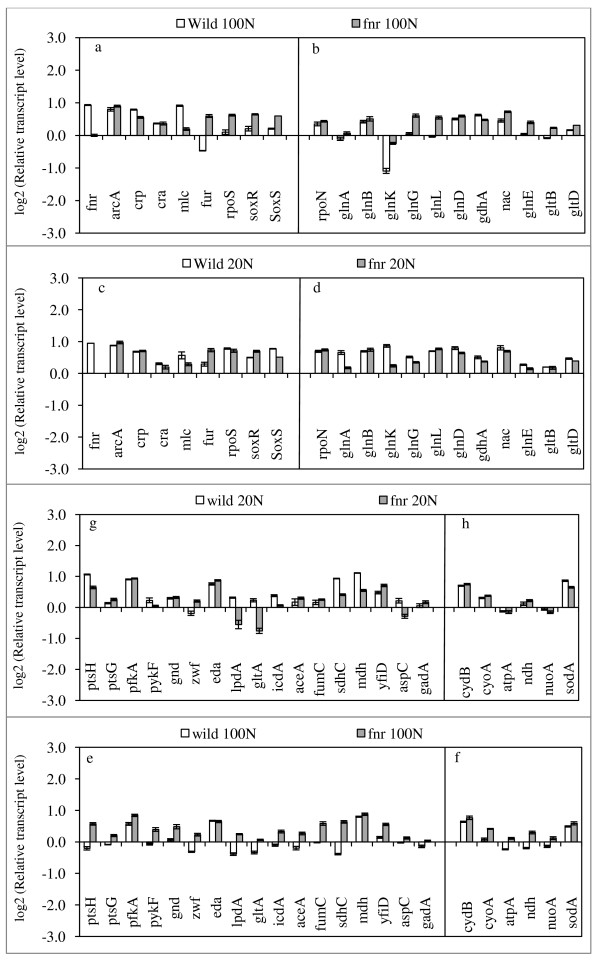
Figure 3 shows the effect of fnr gene knockout on the transcript levels, where it indicates that the transcript levels of the glucose uptake pathway and glycolysis genes such as ptsG, ptsH, pfkA as well as the oxidative PP pathway genes such as zwf and gnd were up-regulated for fnr mutant (p < 0.01, p < 0.01, p < 0.05; p < 0.01, and p < 0.05, respectively) in accordance with the increase of the specific glucose consumption rate as compared to the wild type under N- rich condition (Figure 3e). Some of those are consistent with the down regulation of the transcript levels of crp and mlc (p < 0.01 and p < 0.01) (Figure 3a). Although the transcript level of arcA changed little, the transcript levels of lpdA (p < 0.01) and TCA cycle genes such as gltA, icdA, fumC, sdhC, mdh as well as glyoxylate pathway gene aceA were up-regulated (p < 0.01, p < 0.01, p < 0.01, p < 0.01, p < 0.01; p < 0.01, respectively) for the mutant as compared to the wild type under N- rich condition (Figure 3e), which is consistent with the increased specific CO2 production rate for the mutant as compared to the wild type (Table 1). This is also consistent with up-regulations of soxR, S genes (Figure 3a). Figure 3b indicates that the transcript levels of rpoN, glnB, glnK, and glnE were up-regulated (p < 0.01, p < 0.01, p < 0.01, and p < 0.01 respectively), and those of glnA, L, G, and gltB, D were also up-regulated. The transcript level of nac gene was up-regulated (p < 0.01), while that of gdhA gene was down-regulated (p < 0.01) (Figure 3b). Those indicate that GDH pathway was inactivated, while GS pathway was activated even under N-rich condition (see Additional file 2). Note that the transcript levels of rpoS and fur genes were up-regulated for the mutant (Figure 3a).
Figure 3.
Comparison of the transcriptional mRNA levels between the wild type E. coli and fnr mutant at C/N ratio 1.68 and 8.42.
Under N- limitation, the transcript levels of ptsH and pykF were decreased (p < 0.01 and p < 0.01) (Figure 3g), and those of lpdA, gltA, sdhC, and mdh were down-regulated for the mutant as compared to the wild type (p < 0.01, p < 0.01, p < 0.01 and p < 0.01, respectively). Moreover, the transcript levels of glnA, glnG, and glnK were decreased as compared to the wild type under N-limitation.
Gene transcript levels of fur mutant
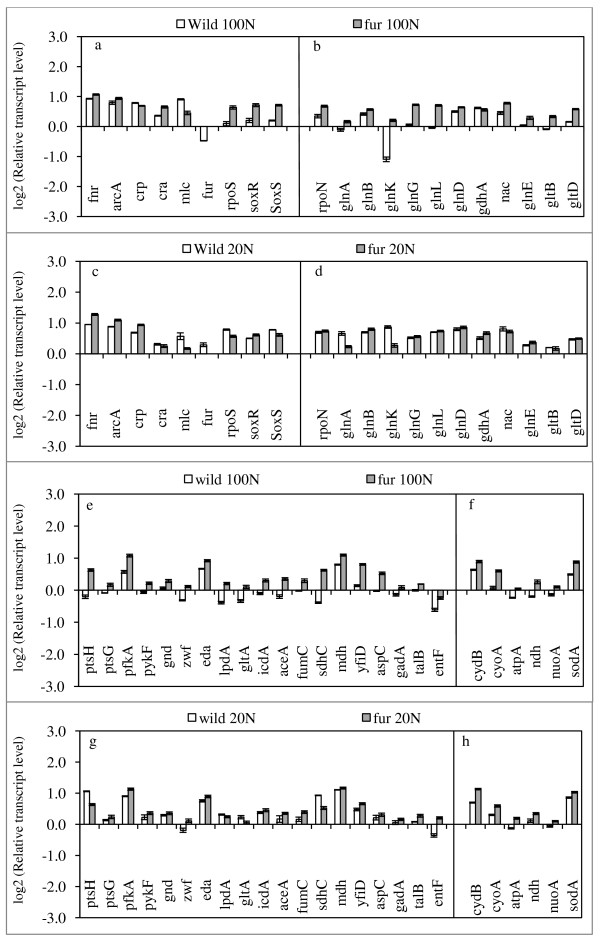
Figure 4 shows the effect of fur gene knockout on the RNA transcript levels, where it indicates that the transcript levels of PTS genes such as ptsH and ptsG, and the glycolysis genes such as pfkA and pykF as well as the oxidative pentose phosphate (PP) pathway genes such as zwf, gnd, and the ED pathway gene eda were all up-regulated as compared to wild type under N- rich condition (p < 0.01, p < 0.01; p < 0.01, p < 0.01; p < 0.01, p < 0.01, and p < 0.01, respectively) (Figure 4e). This is consistent with the fermentation data where the specific glucose consumption rate was higher for the mutant as compared to the wild type (Table 1). The activation of TCA cycle genes is consistent with the activation of the respiration where the transcript levels of cyoA, cydB, nuoA, ndh, atpA as well as sodA genes were all up-regulated (p < 0.01, p < 0.01, p < 0.01, p < 0.01, p < 0.01, and p < 0.01, respectively) (Figure 4f). Moreover, the transcript level of rpoN increased (p < 0.01), and the transcript levels of glnK, glnB, glnE were up-regulated (p < 0.05, p < 0.01, p < 0.01), and those of glnA, L, G, and gltB, D genes as well as nac gene were up-regulated (p < 0.01, p < 0.01, p < 0.01, and p < 0.01, p < 0.01, respectively), while gdhA gene transcript expression was decreased (p < 0.01)(Figure 4b). Those imply that GDH pathway was inactivated, while GS pathway was activated even under N-rich condition as also seen by enzyme activities (see Additional file 2).
Figure 4.
Comparison of the transcriptional mRNA levels between the wild type E. coli and fur mutant at C/N ratio 1.68 and 8.42.
Under N- limitation, the transcript levels of lpdA, gltA, fumC, sdhC, and aceA were down-regulated (p < 0.05, p < 0.01, p < 0.01, p < 0.01 and p < 0.01, respectively), which may be partly due to the up-regulation of arcA (p < 0.01) (Figure 4c). The up-regulations of arcA, possibly caused the increase of the transcript level of cydB (p < 0.01) (Figure 4h). Moreover, the respiratory chain genes such as nuoA, ndh and atpA were all up-regulated for the mutant as compared to the wild type (p < 0.01, p < 0.01 and p < 0.01, respectively) (Figure 4h). The transcript levels of talB and entF genes in fur mutant were up-regulated (p < 0.01, p < 0.01) under both N- rich and N- limited conditions as compared to the wild type (Figure 4e, g).
Discussion
Figure 2 indicates that cyoA gene knockout caused the transcript level of cydB gene to be up-regulated, while cydB gene knockout caused cyoA transcript level to be up-regulated under both N- rich and N- limiting conditions. The aerobic respiratory chain of E. coli can function with either of the two different membrane-bound NADH dehydrogenase I (nuo operon) and NADH dehydrogenase II (ndh), where those generate quinones, and serve as important electron carriers for respiratory chain, where cytochrome bo3 (cyoABCD) and cytochrome bd (cydAB) utilize them for proton motive force. Malfunction of these would diminish the ability to convert NADH to NAD+ and transfer electron to oxygen by converting quinone to quinol and generate proton motive force. Figure 2 also indicates that the transcript levels of arcA were up-regulated for both cyoA and cydB mutants. ArcA is activated through phosphorylation by a membrane protein ArcB, where this phosphorylation occurs after quinone inhibition of ArcB autophosphorylation is released [35]. Quinones are electron carriers where isoprenoid chain anchors them to the membrane, and those are thought to regulate the ArcB-ArcA two component system in response to redox state, which results in a release of quinone inhibition on ArcB, and subsequent autophosphorylation of ArcB and activation of arcA. Moreover, the increase in redox ratio NADH/NAD+ , caused by the decreased NADH oxidation due to cyoA or cydB knockout and by the activation of TCA cycle implied by the specific CO2 production rate (Table 1), may have also activated the transcript levels of arcA gene [17,36]. The activation of arcA (or phosphorylated ArcA) may have caused nuoA, ndh, and fumC transcript levels to be up-regulated for cyoA and cydB mutants as compared to the wild type. In the case where removal of all of three cytochlome oxidases such as cyo, cyd, and cbd would result in anaerobic growth characteristics even under oxic conditions and may produce small amount of lactate for NADH reoxidation [37]. However, in the present study, cydB was active in cyoA mutant and cyoA was active in cydB mutant, and thus the quinol oxidase could have been worked, though NADH/NAD+ ratio tended to increase, which may have caused arcA to be up-regulated. Moreover, NADH can be converted to NADPH by PntAB [38], and thus the cell tries to minimize the inbalance of NADH/NAD+ ratio.
Figure 2c shows the up-regulation of fnr gene, where it has been known that the transcriptional regulator Fnr of E. coli functions as an O2 sensor, and the protein is in the active form and is predominately found as a homo-dimer with one [4Fe-4S] cluster per monomer under anoxic conditions. In the presence of oxygen, [4Fe-4S] Fnr is converted to [2Fe-2S] cluster and finally to apoFnr, which is no longer active in gene regulation [39,40]. Nevertheless, fnr gene transcript level increased for cyoA and cydB mutants, and fnr mutant gives some effect under N-limitation even under aerobic condition. This indicates that Fnr does not play its conventional role, but may have some role under aerobiosis.
It was previously shown that the specific glucose consumption rate as well as the specific CO2 production rate was up-regulated under N- limitation as compared to N-rich condition for the wild type [18]. The same phenomenon was observed in cyoA and cydB mutants even under N- rich condition. Part of the reason may be explained as follows: In the cultivation of cyoA and cydB mutants, cAMP-Crp was down-regulated, where this caused mlc gene to be down-regulated, which caused ptsG and ptsH genes to be up-regulated as seen in Figure 2e, where ptsG is also under control of ArcA. Note that the increase in glucose concentration causes cAMP to be decreased due to the increase in unphosphorylated E IIAGlc, which does not activate Cya. The expression of crp may be indirectly related with this. In the upstream of the mlc gene, there is a binding region of Crp, and mlc expression changed in accordance with crp gene expression [41]. Moreover, the increase in the glucose concentration caused cra to be down-regulated (Figure 2a), and caused pfkA, pykF, and zwf genes to be up-regulated (Figure 2e). Those together caused the specific glucose consumption rate to be up-regulated. Under N- limited condition, the specific growth rate and the transcript levels of glucose uptake, glycolysis, and the oxidative PP pathway genes were further up-regulated as compared to those of the wild type. As for N- regulation genes, nac was up-regulated, while gdhA gene was down-regulated for cyoA and cydB mutants as compared to the wild type under N- rich condition. Moreover, glnB, K, and D transcript levels were up-regulated, and glnA, L, G and gltB, D transcript levels were also up-regulated, which is consistent with the up-regulation of rpoN. This implies the interplay between cAMP-Crp and PII-Ntr systems between C metabolism and N assimilation, where it was shown that Crp can be recruited by sigma 54 holoenzyme (Eσ54) to a site upstream of glnAp2 [42].
When a particular nutrient becomes limiting, E. coli increases the production of proteins that forage for the limiting nutrient. Among the scavenging regulons, Crp, which allows the use of alternative carbon sources, and the two component NtrB/NtrC system, which controls scavenging for nitrogen are by far important in practice. Crp surveys carbon status with cAMP, while Ntr system surveys nitrogen status by glutamine. In the case of carbon catabolism in relation to glucose, the affinity to glucose may change depending on the glucose concentration in the fermentor. Under the culture condition of higher glucose concentration, the affinity of glucose is constant, where glucose forms complex with E IICBGlc and phosphorylated by E IIAGlc-P. Under low glucose concentration such as shown in Table 1, where glucose concentration could not be detected, the affinity to glucose may change depending on the activities of Lam (in outer membrane) and MglBAC (in inner membrane) under such hunger state [43]. The transcript levels of crp as shown in Figure 2, 3, &4 may help estimating the relative glucose concentration. In the case of nitrogen regulation, the affinity of NH4 + is also different depending on the NH4+ concentration. Namely, under N-limited condition (NH4+ < 5 μM), AmtB is used with high affinity, while Amt is blocked by binding GlnK when NH4+ > 50 μM [44], and its transport is made mainly by diffusion.
In the case of fnr mutant under N-rich condition, the similar mechanism may exists. Namely, the cell concentration decreased and the glucose concentration increased, which caused crp transcript level to be down-regulated and thus mlc gene was also down-regulated as state above (Figure 3a). This caused ptsG and ptsH to be up-regulated (Figure 3e). This, together with up-regulations of pfkA and pykF, caused the specific glucose consumption rate to be increased as compared to the wild type. The fnr gene deletion caused cyoA and gltB,D to be up-regulated (Figure 3b) (see Additional file 1), which were also caused by the down-regulation of crp transcript level (see Additional file 1). Loss of ability of an fnr or narXL mutant to reduce nitrate will result in more reduction of the ubiquinone pool, and hence repression of components of the ArcA regulon such as cyoA, and thus Fnr should indirectly activate rather than repress cyoA. This effect is compounded by the fact that arcA transcription is directly activated by Fnr, which in turn is essential for oxidation of ubiquinol by the cytoplasmic or periplasmic nitrate reductase, NarG or Nap [45]. The down-regulation of crp also caused glnA,L, aceA, and gltA to be up-regulated (see Additional file 1). Those are consistent with the increased specific CO2 production rate. The reason why gdhA was repressed while glnA,L,G, gltBD were up-regulated may be due to increased rpoN, though not significant (Figure 3b).
In the case of fur gene knockout, cyoA, sdhC, and sodA genes transcript levels were up-regulated, where those genes are repressed by Fur (see Additional file 1). The increase in rpoN transcript level is consistent with the up-regulations of nac, glnA,L,G, gltB,D together with glnB,K,E and down-regulation of gdhA.
Under N-limitation, the up-regulation of arcA may have caused down-regulation of lpdA and gltA transcript levels (see Additional file 1). Those are also repressed in the media containing excess glucose known as glucose catabolite repression [36,25]. Under N- limitation, the increased glycolytic flux results in the increase of pyruvate, a substrate of PDH encoded by complex pdhR-aceEF-lpdA operon, that has been reported to induce yfiD gene [46]. The gene products of yfiD and gadA have been reported to confer oxidative and acid resistance, respectively [47,48]. Meanwhile, aspC transcript level decreased by the deletion of terminal oxidases under both N-rich and N- limited conditions as compared to the wild type, while it was up-regulated for fur mutant under both conditions. The aspC gene encodes aspartate aminotransferase (AspC) that synthesize aspartate and also provides alternate route to glutamate synthesis [49].
Moreover, consider why cydB gene transcript level was up-regulated for cyoA, fnr, fur mutants, and also for the wild type under N- limitation, where it has been reported to scavenge O2 under micro-aerobic condition [50]. Each cytochrome redox reaction is catalyzed by a low-spin heme which oxidizes quinon and feeds electrons to a highspin heme center that reduces O2 [51,52]. The cydAB operon is transcribed from a complex element containing total five promoters. These promoters P1, P2, P3, and P4 are regulated by O2, arcA and fnr gene products. The P5 promoter provides significant fraction of total cydAB transcription in aerobic environment [50,53].
The catabolic oxidation of electron carriers, which are mostly reduced flavoproteins, by molecular oxygen results in the production of reactive oxygen species that could damage DNA, lipid membranes, and proteins [54,55]. The cytotoxic effects of the reactive oxygen species are mostly mediated by iron, and the regulation of iron uptake has been reported to minimize oxidative stress induced by iron [56]. The aerobic respiratory chain activity appears to be changed little in fnr mutant as compared to the wild type under N- limitation. As opposed to the case of fnr mutant, the deletion of fur gene highly activates aerobic respiratory chain under N- limitation. It could be due to the de-repression of the iron uptake system which is repressed by Fur [57]. Although both fnr and fur genes appear to contribute to the oxidative stress regulation, the mechanisms may be different. It may be considered that fnr increases the synthesis of iron containing proteins, while fur regulates the iron uptake mechanism. The cellular regulations in E. coli are adapted to avoid toxicity from endogenous oxidants generated as a result of oxidative stress, where present study indicates that fnr and fur genes play important roles in these regulations. The effects of fnr and fur genes on oxidative stress were implied by the up-regulations of the transcript levels of soxR, soxS and sodA genes.
The metal levels are often sensed by metal-sensing regulatory RNA, which encodes metal-sensing proteins involved in the transport and storage of intra-cellular metals [58,59]. In the native environment, E. coli continuously faces iron deficiency which is one of the essential trace metals functioning as cofactor in many of the cellular constituents such as flavoproteins, and therefore, it is evolutionarily furnished with the mechanism to regulate iron uptake and storage system [54,57]. However, excessive iron may cause oxygen toxicity by catalyzing the formation of reactive free radicals through such reactions as Fenton/Haber-Weiss [55]. It has been known that aerobic respiration generates superoxide ions (O2-), with NDH II as the main generator of endogenous superoxide and NDH I and SDH as small contributors [60]. The malfunction of these enzymes would also decrease the O2-. Combination with inability to convert NADH to NAD+, a decrease in endogeneous O2- would cause reductive stress, which may activate Fur [61]. Fur requires binding to Fe2+ to become active. It was shown previously that O2- deactivates Fur after its conversion to H2O2 by superoxide dismutase, through Fenton reaction [62] (H2O2 + Fe2+ → HO• + OH- + Fe3+). Therefore, a decrease in endogeneous O2- generation would increase the availability of Fe2+, through a decrease in H2O2 level, and in effect activate Fur relative to control [30]. Namely, Fur senses the reductive stress and represses genes in which Fe-S clusters are safe from damage by reactive oxygen species. It is essential for the cell to use iron economically, which is achieved to some extent by a combination of regulations such as siderophores synthesis, iron transport genes regulation etc. [56]. For these reasons, iron transport and siderophores (e.g. enterobactin) pathway related genes such as talB, entF are repressed by Fur protein [33,63,64]. In the present study, the transcript level of fur gene was up-regulated in particular under N- limitation, which may not be due to the iron deficiency by considering sufficient iron concentration in the medium. In the absence of fur gene, E. coli needs an alternate mechanism of iron homeostasis, and activated enterobactin pathway might have contributed to this regulation.
There are functional interactions between the carbon and ion utilization regulators Crp and Fur, and it was shown that the TCA cycle was repressed by the loss of both transcription factors [25]. In the present study, it was shown that TCA cycle was activated for cyoA, cydB, fnr, and fur mutants as compared to wild type under both C-limited and N-limited conditions. The sdh operon (sdhCDAB) encoding for subunits of iron-dependent SDH showed strong glucose repression, and strong apparent activation by Crp by both wild type and fur mutant [25]. The present result indicates that fur was significantly up-regulated in cyoA, cydB, and fnr mutants as compared to that of wild type, and sdhC was down-regulated (Figure 2, 3), while it was up-regulated in fur mutant (Figure 4).
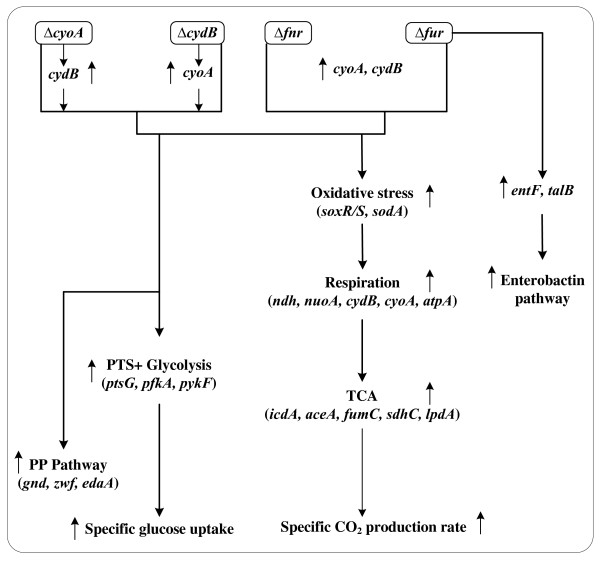
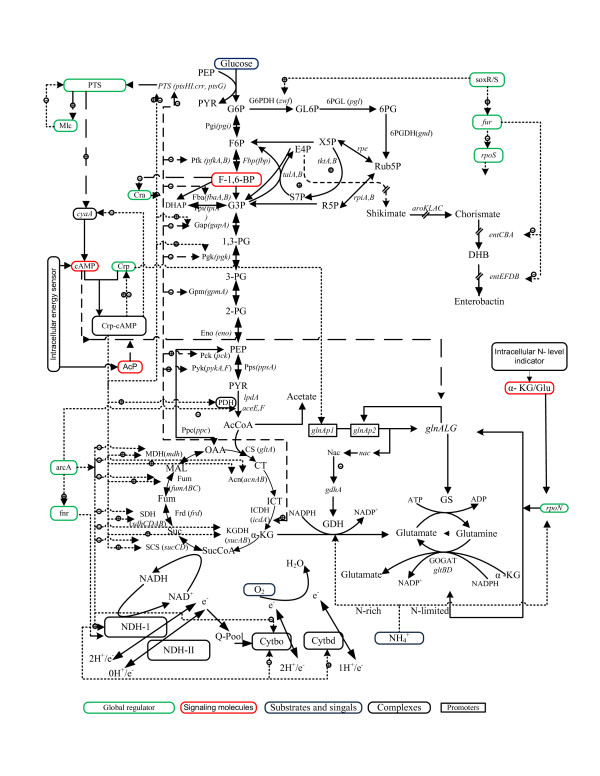
The overall fermentation characteristics were summarized in relation to glycolysis, TCA cycle, and respiratory chain genes in Figure 5. This can be interpreted in more detail by considering the effect of global regulators as shown in Figure 6.
Figure 5.
Overall illustration for the effects of cyoA, cydB, fnr, and fur gene knockout on the metabolism based on the fermentation characteristics.
Figure 6.
Overall illustration of the gene regulation in terms of global regulators in central carbon metabolism of E. coli.
Conclusion
The present result demonstrated how the fermentation characteristics could be explained by the transcript levels of metabolic pathway genes as well as global regulators in relation to a single gene knockout such as cyoA, cydB, fnr, and fur mutants, and clarified the complex gene network regulation in relation to glycolysis, TCA cycle, respiration, and N-regulated pathway. The present result is quite important in understanding the metabolic regulation in relation to metabolic engineering. Moreover, the present result may be useful in improving the specific glucose consumption rate and activation of the TCA cycle by modulating the respiratory chain genes and the related global regulators. The result obtained under N-limited condition may be useful for the heterologous protein production under N-limitation.
Materials and methods
Strains, media composition, and cultivation conditions
The strains used were Escherichia coli BW25113 (F- λ- rph-1 ΔaraBADAH33 lacIq ΔlacZWJ16 rrnBT14 ΔrhaBADLD78 hsdR514), and its single gene knockout mutants lacking such genes as cyoA (JW0422), cydB (JW0723), fnr (JW1318) and fur (JW0669). These single gene knockout mutants were obtained from Keio collection [65]. All the strains were first precultured in the Luria-Bertani medium. The second preculture and the main culture were carried out using M9 minimal medium containing 10 g of glucose together with the following components (per liter): 6.81 g Na2HPO4, 2.99 g KH2PO4, 0.58 g NaCl and 5.94 g (NH4)2SO4. The following components were filter sterilized and then added (per liter) with 1 ml of 1 M MgSO4.7H2O, 1 ml of 0.1 mM CaCl2 2H2O, 1 ml of 1 mg/l thiamine HCl and 10 ml of trace element solution containing (per liter): 0.55 g CaCl2.2H2O, 1.67 g FeCl3.6H2O, 0.1 g MnCl2.4H2O, 0.17 g ZnCl2, 0.043 g CuCl2.2H2O, 0.06 g CoCl2.2H2O, and 0.06 g Na2MoO4.2H2O. The concentrations of nitrogen source such as (NH4)2SO4 were 1.188 g/l and 5.94 g/l, where the concentrations of all the other medium components were kept constant. The continuous culture was conducted in a 1-l fermenter (MDL 100, Marubishi Co., Tokyo, Japan) with a working volume of 500 ml. The pH was controlled at 7.0 ± 0.05 using 2 N HCl or 2 N NaOH, and the temperature was set at 37°C. The air flow rate was 1 vvm (air volume/working volume/min), and the agitation speed was 350 rpm to maintain the dissolved oxygen concentration at 35-40% (v/v) of air saturation [66]. The CO2 concentrations were monitored using an off-gas analyzer (BMJ-02 PI, ABLE Co., Japan). The dilution rate was 0.2 h-1 for all the continuous cultures. The samples were collected at the steady state which was confirmed by the constant off-gas and cell density. It generally took 5-6 residence times to achieve the steady state.
Analytical method
Bacterial growth was monitored by measuring the optical density of the culture broth at 600 nm (OD600) using a spectrophotometer (Ubet-30, Jasco, Tokyo, Japan). It was converted to dry cell weight (DCW) based on the OD600-DCW relationship previously obtained [67]. Glucose and acetate concentrations in the medium were measured using commercially available kits (Wako Co., Osaka, Japan for glucose; Roche, Molecular Biochemical, Mannheim, Germany for acetate).
RNA preparation, design of PCR primers
Total RNA was isolated from E. coli cells by Qiagen RNeasy Mini Kit (QIAGEN K.K., Japan) according to the manufacturer's recommendation. The quantity and purity of the RNA were determined by the optical density measurements at 260 and 280 nm and by 1% formaldehyde agarose gel electrophoresis. The sequences of primers for respective genes used in this study were reported elsewhere [18,68], except such genes as entF and talB. The primer sequences of these additional genes are as follows:
entF TTTATTGCCGATCCTTTTGC (Left)
GGTAACGGCTTGTTCGACAT (Right)
talB CGTTTGTTGGCCGTATTCTT (Left)
AGAATTTCGCCGATGTTACG (Right)
Criteria for the design of the gene-specific primer pairs were followed according to Molecular Cloning: A Laboratory Manual [69]. The primers used in this study were synthesized at Hokkaido System Science Co. (Sapporo, Hokkaido, Japan). In all cases, the primer-supplied company confirmed the purity and absolute specificity of primers.
c DNA synthesis and PCR amplification
RT-PCR reactions were carried out in a TaKaRa PCR Thermal Cycler (TaKaRa TP240, Japan) using Qiagen OneStep RT-PCR Kit (QIAGEN K.K., Japan). The reaction mixture was incubated for 30 min at 50°C for reverse transcription (cDNA synthesis) followed by 15 min incubation at 95°C for initial PCR activation. Then, the process was subjected to 30 cycles of amplification which consisted of a denaturing step (94°C for 1 min), an annealing step (approximately 5°C below melting temperature of primers for 1 min), and an extension step (72°C for 1 min), and then finally the reaction mixture was incubated for 10 min at 72°C for final extension. To check for nucleic acid contamination, one negative control was run in every round of RT-PCR. This control lacks the template RNA in order to detect possible contamination of the reaction components. 5 ml of amplified products were run on a 1% agarose gel. Gels were stained with 1 μg ml-1 of ethidium bromide, photographed using a Digital Image Stocker (DS-30, FAS III, Toyobo, Osaka, Japan) under UV light and analyzed using Gel-Pro Analyzer 3.1 (Toyobo, Osaka, Japan) software. Although the PCR products obtained for all the genes showed the predicted sizes on agarose gel, the identity of amplified fragments of some genes was demonstrated by DNA sequencing. In order to determine the optimal amount of input RNA, the two-fold diluted template RNA was amplified in RT-PCR assays under identical reaction conditions to construct a standard curve for each gene product. When the optimal amount of input RNA was determined for each gene product, RTPCR was carried out under identical reaction conditions to detect differential transcript levels of genes. The gene dnaA, which encodes E. coli DnaA transcription dual regulator and is not subjected to variable expression, i.e. abundant expression at relatively constant rate in most cells, was used as an internal control for the RT-PCR determinations [68]. To calculate the standard deviation, RT-PCR was independently performed three times for each gene under identical reaction condition. To ensure that the observed changes were statistically significant, the Student's t-test was applied.
Enzyme activities
The enzyme activities were measure for GDH, GS, and GOGAT, where assay methods are described elsewhere [18].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RK carried out fermentation experiments, assayed, made statistical analysis, analyzed the results, and drafted the manuscript. KS considered the experimental design, analyzed the results, and prepared manuscript together with RK. All authors read and approved the final manuscript.
Supplementary Material
Effect of global regulators on metabolic pathway related genes. The table lists the known effects of global regulators on metabolic pathway related genes.
Enzyme Activity (Unit*/mg protein). The data shows the activities of the nitrogen assimilatory enzymes (GDH, GS, and GOGAT).
Contributor Information
Rahul Kumar, Email: rahul_iitd@yahoo.co.uk.
Kazuyuki Shimizu, Email: shimi@bio.kyutech.ac.jp.
Acknowledgements
This research was supported by Strategic International Cooperative Program, Japan Science and Technology Agency (JST). The authors are pleased to mention about the fruitful discussions with Prof. Johnjoe McFadden of Surrey University and Prof. Hans Westerhoff of Manchester University.
References
- Fuhrer T, Sauer U. Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J Bacteriol. 2009;191:2112–2121. doi: 10.1128/JB.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Doucette CD, Fowler WU, Feng XJ, Piazza M, Rabitz HA, Wingreen NS, Rabinowitz JD. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol Syst Biol. 2009;5:302. doi: 10.1038/msb.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q, Yang C, Oshima T, Mori H, Shimizu K. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl Environ Microbiol. 2004;70:2354–2366. doi: 10.1128/AEM.70.4.2354-2366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B. Global regulation of gene expression. Proc Natl Acad Sci USA. 2000;97:14044–14045. doi: 10.1073/pnas.97.26.14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer DP, Soupene E, Lee HL, Khodursky AB, Peter BJ, Bender RA, Kustu S. Nitrogen regulatory protein C-controlled genes of Escherichia coli: Scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci USA. 2000;97:14674–14679. doi: 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CN, Mandel MJ, Silhavy TJ. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J Bacteriol. 2005;187:7549–7553. doi: 10.1128/JB.187.22.7549-7553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. Hungry Bacteria-definition and properties of a nutritional state. Environ Microbiol. 2001;3:605–611. doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- Harder W, Dijkhuizen L. Physiological responses to nutrient limitation. Annu Rev Microbiol. 1983;37:1–24. doi: 10.1146/annurev.mi.37.100183.000245. [DOI] [PubMed] [Google Scholar]
- Molenaar D, van Berlo R, de Ridder D, Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol Syst Biol. 2009;5:323. doi: 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mattos MJT, Neijssel OM. Bioenergetic consequences of microbial adaptation to lownutrient environments. J Biotechnol. 1997;59:117–126. doi: 10.1016/S0168-1656(97)00174-0. [DOI] [PubMed] [Google Scholar]
- Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat Chem Biol. 2006;2:529–530. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- Pramanik J, Keasling JD. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng. 1997;56:398–421. doi: 10.1002/(SICI)1097-0290(19971120)56:4<398::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Neijssel OM, De Mattos MJT, Tempest DW. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt FC, editor. Chapter 107. American Society for Microbiology, Washington, DC; 1996. Growth yield and energy distribution. [Google Scholar]
- Varma A, Boesch BW, Palsson BO. Biochemical production capabilities of Escherichia coli. Biotechnol Bioeng. 1993;42:59–73. doi: 10.1002/bit.260420109. [DOI] [PubMed] [Google Scholar]
- Bekker M, De Vries D, Beek AT, Hellingwerf KJ, De Mattos MJT. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd- II oxidase. J Bacteriol. 2009;191:5510–5517. doi: 10.1128/JB.00562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol. 2006;72:3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Shimizu K. Metabolic regulation of Escherichia coli and its gdhA, glnL, gltB, D mutants under different carbon and nitrogen limitations in the continuous culture. Microbial Cell Factories. 2010;9:8. doi: 10.1186/1475-2859-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen A, Finel M, Haltia T, Gennis RB, Wikstrom M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry. 1991;30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon K, Hung SP, Mekjian K, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J Biol Chem. 2003;278:29837–29855. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Lin EC. Adaptation of Escherichia coli to redox environments by gene expression. Mol Microbiol. 1993;9:9–15. doi: 10.1111/j.1365-2958.1993.tb01664.x. [DOI] [PubMed] [Google Scholar]
- Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 2007;35:6762–6777. doi: 10.1093/nar/gkm631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gosset G, Barabote R, Gonzalez CS, Cuevas WA, Saier MH Jr. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J Bacteriol. 2005;187:980–990. doi: 10.1128/JB.187.3.980-990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassinova N, Kozyrev D. A method for direct cloning of fur-regulated genes: identification of seven new fur-regulated loci in Escherichia coli. Microbiology. 2000;146:3171–3182. doi: 10.1099/00221287-146-12-3171. [DOI] [PubMed] [Google Scholar]
- Tseng CP. Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur, and hemA gene products. FEMS Microbiol Lett. 1997;157:67–72. doi: 10.1111/j.1574-6968.1997.tb12754.x. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, Ozenberger BA, McIntosh MA. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J Mol Biol. 1990;212:669–682. doi: 10.1016/0022-2836(90)90229-F. [DOI] [PubMed] [Google Scholar]
- Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- Brynildsen MP, Liao JC. An integrated network approach indentifies the isobutanol response network of Escherichia coli. Mol Syst Biol. 2009;5:34. doi: 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasling JD, Chou H. Metabolic engineering delivers next generation biofuels. Nature Biotechnol. 2008;26:298–299. doi: 10.1038/nbt0308-298. [DOI] [PubMed] [Google Scholar]
- Frederick RE, Mayfield JA, DuBois JL. Iron trafficking as an antimicrobial target. Biometals. 2009;22:583–593. doi: 10.1007/s10534-009-9236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz MF, Lavińa M. Involvement of enterobactin synthesis pathway in production of Microcin H47. Antimicrob Agents Chemother. 2004;48:1235–1241. doi: 10.1128/AAC.48.4.1235-1241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TE. Structural characterization of Escherichia coli periplasmic ferric siderophores binding proteins. Dissertation, University of Calgary, Alberta; 2001. [Google Scholar]
- Georgellis D, Kwon O, Lin ECC. Quinones as the Redox Signal for the Arc Two-Component System of Bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Shalel-Levanon S, San KY, Bennett GN. Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol Bioeng. 2005;89:556–564. doi: 10.1002/bit.20381. [DOI] [PubMed] [Google Scholar]
- Portnoy VA, Herrgard MJ, Palsson BO. Aerobic fermentation of D-glucose by an evolved cytochrome oxidase-deficient Escherichia coli strain. Appl Environ Micorobiol. 2008;74:7561–7569. doi: 10.1128/AEM.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- Unden G, Achebach S, Holighaus G, Tran HG, Wackwitz B, Zeuner Y. Control of Fnr function of Escherichia coli by O2 and reducing conditions. J Mol Microbiol Biotechnol. 2002;4:263–268. [PubMed] [Google Scholar]
- Reinhart F, Achebach S, Koch T, Unden G. Reduced Apo-Fumarate Nitrate Reductase Regulator (ApoFNR) as the major form of FNR in aerobically growing Escherichia coli. J Bacteriol. 2008;190:879–886. doi: 10.1128/JB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Lim S, Seok YJ, Ryu S. Heat shock RNA polymerase (Eσ32) is involved in the transcription of mlc and crucial for induction of the Mlc regulon by glucose in Escherichia coli. J Biol Chem. 2001;276:25871–25875. doi: 10.1074/jbc.M101757200. [DOI] [PubMed] [Google Scholar]
- Mao XJ, Huo YX, Buck M, Kolb A, Wang YP. Interplay between CRP-cAMP and PII-Ntr systems forms novel regulatory network between carbon metabolism and nitrogen assimilation in Escherichia coli. Nucleic Acids Res. 2007;35:1432–1440. doi: 10.1093/nar/gkl1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. Hungry bacteria-definition and properties of a nutritional state. Environ Microbiol. 2001;3:605–611. doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- Javalle A, Severi E, Thorton J, Merrick M. Ammonium sensing in Escherichia coli. J Biol Chem. 2004;279:8530–853. doi: 10.1074/jbc.M312399200. [DOI] [PubMed] [Google Scholar]
- Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, Overton TW. A renassessment of the Fnr regulon and transcriptomic analysis of the effects of nitrate, nitrite, narXL, and narQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem. 2006;281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Ishida Y, Yamada K, Yamamoto K, Ishihama A. PdhR (Pyruvate Dehydrogenase Complex Regulator) Controls the Respiratory Electron Transport System in Escherichia coli. J Bacteriol. 2007;189:5534–5541. doi: 10.1128/JB.00229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AFV, Schultz S, Bomke J, Pils T, Lehmann WD, Knappe J. YfiD of Escherichia coli and Y061 of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem Biophys Res Commun. 2001;285:456–462. doi: 10.1006/bbrc.2001.5186. [DOI] [PubMed] [Google Scholar]
- Moreau PL. The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids. J Bacteriol. 2007;189:2249–61. doi: 10.1128/JB.01306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotheringham IG, Dacey SA, Taylor PP, Smith TJ, Hunter MG, Finlay ME, Primrose SB, Parker DM, Edwards RM. The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K12. J Biochem. 1986;234:593–604. doi: 10.1042/bj2340593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Melville SB, Albrecht JA, Gunsalus RP. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, Cooper CE. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat Chem Biol. 2009;5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- Verkhovsky MI, Morgan JE, Puustein A, Wikström M. Kinetic trapping of oxygen in cell respiration. Nature. 1996;380:268–270. doi: 10.1038/380268a0. [DOI] [PubMed] [Google Scholar]
- Govantes F, Orjalo AV, Gunsalus RP. Interplay between three global regulatory proteins mediates oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon. Mol Microbiol. 2000;38:1061–1073. doi: 10.1046/j.1365-2958.2000.02215.x. [DOI] [PubMed] [Google Scholar]
- Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J Biotechnol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/S1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- Braun V. Regulation of iron uptake minimizes iron-mediated oxidative stress. J Biosci. 1998;23:483–489. doi: 10.1007/BF02936142. [DOI] [Google Scholar]
- McHugh JP, Rodríguez-Quiñones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. Global iron-dependent gene regulation in Escherichia coli. J Biol Chem. 2003;278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- Dann CE III, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winker WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Helmann JD. Measuring metals with RNA. Mol Cell. 2007;27:859–860. doi: 10.1016/j.molcel.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennis RB, Stewart V. In: Respiration in Escherichia coli and Salmonella: Cellular and Molecular Biology. Bock A, Curtiss R III, Kaper JB, Neidhardt FC, Nystrom T, Rudd KE, Squires CL, editor. Washington, DC: ASM press; 1996. [Google Scholar]
- Jovanovic G, Lloyde LJ, Stumpf MP, Mayhew AJ, Buck M. Induction and function of the phase shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem. 2006;281:21147–21161. doi: 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- Blanchard JR, Wholey WY, Conlon EM, Pomposiello PJ. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional network. PLoS One. 2007;2:e1186. doi: 10.1371/journal.pone.0001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semsey S, Andersson AM, Krishna S, Jensen MH, Masse E, Sneppen K. Genetic regulation of fluxes: iron homeostasis of Escherichia coli. Nucleic Acids Res. 2006;34:4960–4967. doi: 10.1093/nar/gkl627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantash FM, Ammerlaan M, Earhart CF. Enterobactin synthase polypeptides of Escherichia coli are present in an osmotic-shock-sensitive cytoplasmic locality. Microbiology. 1997;143:147–156. doi: 10.1099/00221287-143-1-147. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Hassan MR, Shimizu K. Growth phase-dependent changes in the expression of global regulatory genes and associated metabolic pathways in Escherichia coli. Biotechnol Lett. 2008;30:853–860. doi: 10.1007/s10529-007-9621-1. [DOI] [PubMed] [Google Scholar]
- Peng L, Shimizu K. Effect of fadR gene knockout on the metabolism of Escherichia coli based on analyses of protein expressions, enzyme activities and intracellular metabolite concentrations. Enzyme Microbiol Technol. 2006;38:512–520. doi: 10.1016/j.enzmictec.2005.07.019. [DOI] [Google Scholar]
- Kabir MM, Shimizu K. Gene expression patterns for metabolic pathway in pgi knockout Escherichia coli with and without phb genes based on RT-PCR. J Biotechnol. 2003;105:11–31. doi: 10.1016/S0168-1656(03)00170-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of global regulators on metabolic pathway related genes. The table lists the known effects of global regulators on metabolic pathway related genes.
Enzyme Activity (Unit*/mg protein). The data shows the activities of the nitrogen assimilatory enzymes (GDH, GS, and GOGAT).