Abstract
Background
Because many picoplanktonic eukaryotic species cannot currently be maintained in culture, direct sequencing of PCR-amplified 18S ribosomal gene DNA fragments from filtered sea-water has been successfully used to investigate the astounding diversity of these organisms. The recognition of many novel planktonic organisms is thus based solely on their 18S rDNA sequence. However, a species delimited by its 18S rDNA sequence might contain many cryptic species, which are highly differentiated in their protein coding sequences.
Principal Findings
Here, we investigate the issue of species identification from one gene to the whole genome sequence. Using 52 whole genome DNA sequences, we estimated the global genetic divergence in protein coding genes between organisms from different lineages and compared this to their ribosomal gene sequence divergences. We show that this relationship between proteome divergence and 18S divergence is lineage dependant. Unicellular lineages have especially low 18S divergences relative to their protein sequence divergences, suggesting that 18S ribosomal genes are too conservative to assess planktonic eukaryotic diversity. We provide an explanation for this lineage dependency, which suggests that most species with large effective population sizes will show far less divergence in 18S than protein coding sequences.
Conclusions
There is therefore a trade-off between using genes that are easy to amplify in all species, but which by their nature are highly conserved and underestimate the true number of species, and using genes that give a better description of the number of species, but which are more difficult to amplify. We have shown that this trade-off differs between unicellular and multicellular organisms as a likely consequence of differences in effective population sizes. We anticipate that biodiversity of microbial eukaryotic species is underestimated and that numerous “cryptic species” will become discernable with the future acquisition of genomic and metagenomic sequences.
Introduction
Our understanding of the evolution of eukaryotes was revolutionized when it became possible to compare sequenced marker genes, notably the ribosomal genes, among many organisms [1]. In practice, ribosomal genes are often the only markers available for estimating the diversity of unicellular eukaryotes, especially in the Chromalveolates, Excavata and Rhizaria group which have few sequenced representatives. They are also the only markers used in the analysis of environmental or metagenomic DNA sequence datasets [2], [3]. It is thus becoming crucially important to know how well these signatures represent the extent of diversity in the exploding body of data that will become available over the next ten years as revolutionary sequencing technology are used in panoceanic metagenomic campaigns [4], [5]. Marine metagenomics studies rely on a pragmatic species concept; sequences are declared as being from separate species or genera based upon an arbitrary level of sequence divergence at a marker locus, typically the 18S rDNA ribosomal gene [6]. In this study, we analysed how genome divergence, estimated from amino-acid changes in protein coding genes, compares with 18S ribosomal divergence, the universal marker for planktonic eukaryotes biodiversity.
Methods
Whole genome predicted proteins data was downloaded from GenBank, JGI, Genolevure, Ensembl [7], PLAZA [8] and organisms' dedicated databases (Table 1). Complete 18S rDNA sequences were downloaded from GenBank or extracted from the whole genome sequence by screening the complete genome with complete 18S rDNA sequence from a closely related species. For the primate data, 18S rDNA sequenced were reassembled from the GenBank Trace archive (Table 1).
Table 1. Genome data and 18S rDNA data used for analysis.
| Species | Database | URL | Release | Gene | 18S rDNA sequence |
| DIPTERA | |||||
| Aedes aegypti | VectorBase | http://aaegypti.vectorbase.org/ | AaegL1.1 | 16789 | from genome assembly |
| Culex pipiens | VectorBase | http://cpipiens.vectorbase.org/ | CpipJ1.2 | 18883 | from genome assembly |
| Drosophila ananassae | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 15070 | from genome assembly |
| Drosophila melanogaster | flybase | ftp://ftp.flybase.net/genomes/ | r5.9 | 21064 | M21017.1 |
| Drosophila erecta | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 15048 | from genome assembly |
| Drosophila yakuba | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 16082 | from genome assembly |
| Drosophila grimshawi | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 14986 | from genome assembly |
| Drosophila willistoni | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 15513 | from genome assembly |
| Drosophila persimilis | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 16878 | from genome assembly |
| Drosophila pseudoobscura | flybase | ftp://ftp.flybase.net/genomes/ | r2.3 | 16071 | AY03717 |
| Drosophila sechellia | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 16471 | from genome assembly |
| Drosophila simulans | flybase | ftp://ftp.flybase.net/genomes/ | r1.3 | 15415 | AY037174.1 |
| VERTEBRATA | |||||
| Homo sapiens | Ensembl | http://archive.ensembl.org/ | v54 | 47509 | M10098 |
| Pan troglodytes | Ensembl | http://archive.ensembl.org/ | v54 | 34142 | rebuilt from Trace |
| Mus musculus | Ensembl | http://archive.ensembl.org/ | v38 | 31986 | X00686.1 |
| Rattus norvegicus | Ensembl | http://archive.ensembl.org/ | v54 | 32948 | X01117 |
| Macaca Mulatta | Ensembl | http://archive.ensembl.org/ | v54 | 36384 | rebuilt from Trace |
| Pongo pygmaeus | Ensembl | http://archive.ensembl.org/ | v54 | 23533 | rebuilt from Trace |
| Bos Taurus | Ensembl | http://archive.ensembl.org/ | v54 | 26977 | DQ222453.1 |
| Equus caballus | Ensembl | http://archive.ensembl.org/ | v54 | 22641 | AJ311673.1 |
| Gallus gallus | Ensembl | http://archive.ensembl.org/ | v47 | 22195 | AF173612 |
| Xenopus tropicalis | Ensembl | http://archive.ensembl.org/ | v54 | 27710 | from genome assembly |
| STREPTOPHYTA | |||||
| Oryza sativa | Rice | http://rice.plantbiology.msu.edu/ | v6 | 67393 | from genome assembly |
| Sorghum bicolor | JGI | http://genome.jgi-psf.org/Sorbi1/Sorbi1.download.ftp.html | Sbi1_4 | 34496 | from genome assembly |
| Populus trichocharpa | JGI | http://genome.jgi-psf.org/ | v1.1 | 45555 | from genome assembly |
| Medicago truncatula | Medicago | http://www.medicago.org/ | 44830 | AF093506.1 | |
| Arabidopsis thaliana | TAIR | http://www.arabidopsis.org/index.jsp | 27855 | X16077.1 | |
| Arabidopsis lyrata | JGI | http://www.jgi.doe.gov/genome-projects/ | 32670 | from genome assembly | |
| Carica papaya | Carica | asgpb.mhpcc.hawaii.edu/papaya/ | 24782 | from genome assembly | |
| Vitis vinifera | Genoscope | http://www.genoscope.cns.fr/ | 30434 | from genome assembly | |
| CHLOROPHYTA | |||||
| Micromonas pusilla CCMP1545 | JGI | http://www.jgi.doe.gov/genome-projects/ | V2 | 10242 | from genome assembly |
| Micromonas pusilla RCC299 | JGI | http://www.jgi.doe.gov/genome-projects/ | V3 | 10109 | from genome assembly |
| Ostreococcus lucimarinus | JGI | http://www.jgi.doe.gov/genome-projects/ | v2 | 7651 | from genome assembly |
| Ostreococcus RCC809 | JGI | http://www.jgi.doe.gov/genome-projects/ | v1 | 7773 | from genome assembly |
| Bathycoccus prasinos | Genoscope | http://bioinformatics.psb.ugent.be/ | V1 | 8747 | from genome assembly |
| Ostreococcus tauri | Bogas | http://bioinformatics.psb.ugent.be/ | v2 | 7725 | from genome assembly |
| SACCHAROMYCETACEAE | |||||
| Saccharomyces cerevisiae | SGD | http://www.yeastgenome.org/ | 5914 | Z75578 | |
| Saccharomyces paradoxus | MIT | http://www.broad.mit.edu/annotation/ | 4774 | X97806 | |
| Saccharomyces mikatae | Broad | http://fungal.genome.duke.edu/ | 5884 | AB040998 | |
| Saccharomyces kudriavzevi | WUSTL | http://fungal.genome.duke.edu/ | 6371 | AACI02000378.1 | |
| Saccharomyces bayanus | MIT | http://www.broad.mit.edu/annotation/ | 4492 | X97777 | |
| Saccharomyces castellii | WUSTL | http://fungal.genome.duke.edu/ | 5864 | AACF01000230.1 | |
| Lachancea waltii | Genolevure | http://fungal.genome.duke.edu/ | 5350 | AADM01000401.1 | |
| Lachancea thermotolerans | Genolevure | http://fungal.genome.duke.edu/ | 5092 | X89526.1 | |
Twenty six phylogenetic independent comparisons were inferred from couple of species with less than 5% 18S rDNA divergences (all species pairs, number of genes and phylogenies within each lineage are available in Figure S1).
All orthologous gene pairs between species were inferred by reciprocal best hit (e-value 10−3). We retrieved the common set of orthologous genes within each lineage by extracting the orthologous genes present in all pairwise species comparisons. We thus obtained 2151 common gene pairs in Chlorophyta, 5051 in Diptera, 2925 in Saccharomyceta, 4160 in Streptophyta and 5949 in Vertebrata. Protein sequences were aligned with the Needleman Wunsch algorithm [9] and processed with custom C codes to compute amino-acid identities over the concatenated alignments. Substitution rates dAA were estimated via maximum likelihood with the PAML package (Jones [10] substitution matrix) [11].
We manually inspected multiple sequence alignments to identify common sites of the 18S rDNA : large insertions occurring in some sequences were excluded from the alignment to get consistent divergence estimate across pairwise comparions. All 18S rDNA pairs were aligned with the Needleman Wunsch algorithm to estimate pairwise differences, The nucleotide substitution rates of the 18S rDNA were estimates with the PAML package (HKY85 substitution model).
Statistical analyses were performed with the R software.
Results
The rate of 18S rDNA and protein evolution
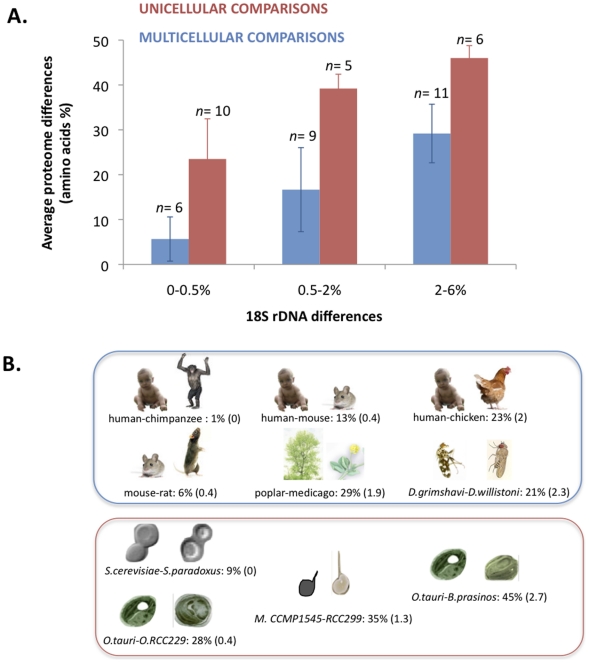
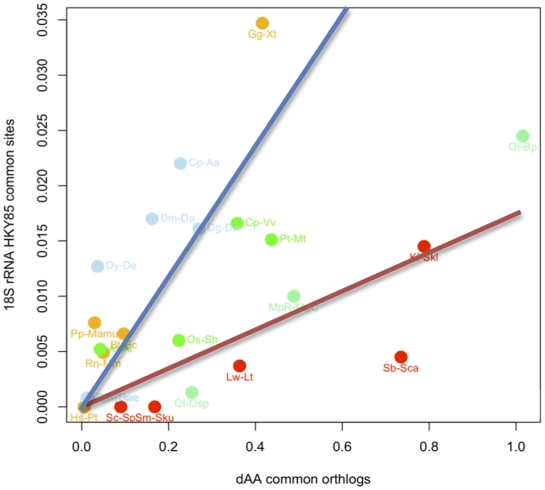
Recent genome and metagenomic projects have highlighted the surprising discrepancy between 18S rDNA divergence and whole genome divergence in some phytoplanktonic species [12], [13], [14], [15], that are keystone players in the global carbon cycling [16]. Here we investigated the generality of this observation among both unicellular and muticellular eukaryotes. We compared the 18S rDNA and the proteome divergence across all available eukaryotic genomes in 2 unicellular (Baker's yeast and green alga) and 3 multicellular lineages (Vertebrates, Diptera and Land plants). We found that for a given level of rDNA divergence, unicellular eukaryotes had substantially greater proteome divergence than multicellular eukaryotes (Figure 1A). This can be more formally tested using an analysis of covariance of proteome versus rDNA divergence, forcing the regression lines through the origin and testing for equality of slopes : the test is highly significantly different (p<0.0001) (Figure 1A). Identical 18S rDNA sequences between two unicellular species may correspond to proteome divergences of the same order as those observed between Xenopus and Chicken or the Poplar tree and the grass Medicago (Figure 1B). Amino-acid divergences between orthologous genes are only one of the many hallmarks of evolutionary divergence after speciation. A genomic species definition for protists based on proteome divergence is stringent, because genomic rearrangements, the acquisition of new genes via duplication or even a few mutations within a subset of genes may be sufficient to delineate two species [17], [18]. To reduce possible effects of amino-acid content, base composition and non-independency of observations, we computed the substitution rates on a common set of orthologs within each lineage across all independent pairwise comparisons. Consistent with the raw number of difference estimates, the evolution rate of the 18S rDNA relative to the proteome is much lower in unicellular species (analysis of covariance unicellulars versus multicellulars p = 0.048) (Figure 2).
Figure 1. 18S rDNA versus proteome divergence in unicellular and multicellular lineages.
A. Average proteome (amino-acid) and 18S rDNA differences (%) for 21 unicellular and 26 multicellular pairwise comparisons. The first class of 18S rDNA sequence differences limit, 0.5%, is the smallest threshold used to delineate Operational Taxonomic Units (OTU) in planktonic eukaryotes [26]. B. Selected examples of pairwise comparisons in each 18S rDNA divergence class: percent of amino-acid divergence (percent of 18S rDNA differences).
Figure 2. 18s rDNA evolution rates versus Amino-acid evolution rates for all common orthologous genes within lineages for independent pairs of species.
Yellow: Vertebrates, Green: Streptophytes, Light blue: Diptera, Light green: Chlorophyta, Red: Saccharomyceta.
Discussion
A population genetic explanation
What could be the cause of this decoupling between 18S rDNA and proteome divergence in unicellar versus multicellular species? There are two general explanations; first, the proportion of mutations that are strongly deleterious is higher in 18S rDNA, when compared to protein sequences, in unicells compared to multicells. One could argue that the 18S rDNA may be under much more stronger selection in unicells, where fitness may depend more directly from transcription efficiency than in multicellular species. Second, the rate of adaptive evolution could be higher in protein sequences in unicells compared to multicells. It is difficult to differentiate between these possibilities. However, unicells and multicells are likely to differ in their effective population sizes and this suggests a simple explanation; that the proportion of effectively neutral mutations changes more in response to differences in the effective population size in the 18S rDNA than in the proteome. This can be formalised as follows. Let us assume that all mutations are deleterious (or effectively neutral) and that the distribution of fitness effects is a gamma distribution. Under a gamma distribution it can be shown that the rate of evolution, R, is a function of the mutation rate, μ, divergence time, t, and the Distribution of Fitness effects of new mutations, fully described by the shape parameters, ß, and the effective population size, Ne [19], [20], [21].
We can thus express the relative ratio between the rate of evolution of the 18S rDNA, Rr, and the rate of evolution of the proteome, Rp, in one lineage as a function of three parameters, where Ne is the average effective population size within a lineage:
This ratio can be estimated from our observations (Figure 2) by taking the linear regression coefficient for each lineage (slope = 0.017 for unicellulars and slope = 0.059 for multicellular organims).
If we assume that unicells have an effective population size, Ne, that is 1000 to 1,000,000 times larger than in multicells, then ßr−ßp would be between −0.2 and −0.1 to explain the differences in the regression slopes. So quite modest differences in the distribution of fitness effects, and effective population sizes can lead to substantial differences in the relative rates at which the 18S rDNA and protein coding sequences evolve. Recent estimates of ß p for nuclear genes in Humans and Drosophila are 0.2 and 0.35 respectively [22] [23]and we thus expect ß r to take values smaller than 0.25.
Large effective population sizes of unicellular eukaryotes may thus provide an explanation for the surprising low divergence of 18S rDNA relative to the genome divergence. More generally, this conclusion applies to any barcoding gene sufficiently constrained to provide a large phylogenetic spread over the eukaryotic tree of life, suggesting that biodiversity studies have to make a trade-off between phylogenetic spread and phylogenetic depth for a given barcoding gene. Given the present diversity estimates of eukaryotic unicells from conserved barcoding genes like the 18S rDNA [24], [25], we thus anticipate that future eukaryotic planktonic metagenomic and genomic analysis will lead to an increase in the number of species.
Supporting Information
Phylogenetic relationships and number of genes used for independent comparison.
(TIFF)
Acknowledgments
We would like to thank Linda Medlin for insightful comments, the Genomics of phytoplankton team, Romain Blanc-Mathieu, Camille Clerissi, Evelyne Derelle, Yves Desdevises, Rozenn Thomas, Eve Toulza and Lucie Subirana for stimulating discussions and Severine Jancek for help with a previous analysis. We would also like to acknowledge Timo Gourbiere for providing pictures in Fig 1B.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded by the European Community's 7th Framework program FP7 under grant agreement n°254619 and the AAP-FRB2009-PICOPOP (http://www.fondationbiodiversite.fr/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Garcia P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 3.Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 4.TARA. Available: http://oceans.taraexpeditions.org/. Accessed 2010 Jan 26.
- 5.Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, et al. The Sorcerer II Global Ocean Sampling Expedition: Metagenomic Characterization of Viruses within Aquatic Microbial Samples. Plos One. 2008;3 doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Oceanogr. 2004;49:784–798. [Google Scholar]
- 7.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, et al. Ensembl's 10th year. Nucleic Acids Research. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, et al. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 10.Jones DT, Taylor WR, Thornton JM. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Computer Applications in the Biosciences. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 12.Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci U S A. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 14.Cuvelier ML, Allen AE, Monier A, McCrow JP, Messie M, et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14679–14684. doi: 10.1073/pnas.1001665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jancek S, Gourbiere S, Moreau H, Piganeau G. Clues about the Genetic Basis of Adaptation Emerge from Comparing the Proteomes of Two Ostreococcus Ecotypes (Chlorophyta, Prasinophyceae). Molecular Biology and Evolution. 2008;25:2293–2300. doi: 10.1093/molbev/msn168. [DOI] [PubMed] [Google Scholar]
- 16.Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnology And Oceanography. 2004;49:168–179. [Google Scholar]
- 17.Coyne J, Orr H. 2004. 545 Speciation, Sinauer Associates.
- 18.Gourbiere S, Mallet J. Are Species Real? The Shape of the Species Boundary with Exponential Failure, Reinforcement, and the “Missing Snowball”. Evolution. 2010;64:1–24. doi: 10.1111/j.1558-5646.2009.00844.x. [DOI] [PubMed] [Google Scholar]
- 19.Crow J, Kimura M. In: An Introduction to Population Genetics Theory. Crow J, Kimura M, editors. Harper and Row; 1970. [Google Scholar]
- 20.Welch JJ, Eyre-Walker A, Waxman D. Divergence and polymorphism under the nearly neutral theory of molecular evolution. J Mol Evol. 2008;67:418–426. doi: 10.1007/s00239-008-9146-9. [DOI] [PubMed] [Google Scholar]
- 21.Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 22.Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. Plos Genetics. 2008;4 doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piganeau G, Desdevises Y, Derelle E, Moreau H. Picoeukaryotic sequences in the Sargasso sea metagenome. Genome Biol. 2008;9:R5. doi: 10.1186/gb-2008-9-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Not F, del Campo J, Balague V, de Vargas C, Massana R. New insights into the diversity of marine picoeukaryotes. PLoS One. 2009;4:e7143. doi: 10.1371/journal.pone.0007143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viprey M, Guillou L, Ferreol M, Vaulot D. Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ Microbiol. 2008;10:1804–1822. doi: 10.1111/j.1462-2920.2008.01602.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationships and number of genes used for independent comparison.
(TIFF)