Abstract
Saxitoxin and its analogs cause the paralytic shellfish-poisoning syndrome, adversely affecting human health and coastal shellfish industries worldwide. Here we report the isolation, sequencing, annotation, and predicted pathway of the saxitoxin biosynthetic gene cluster in the cyanobacterium Lyngbya wollei. The gene cluster spans 36 kb and encodes enzymes for the biosynthesis and export of the toxins. The Lyngbya wollei saxitoxin gene cluster differs from previously identified saxitoxin clusters as it contains genes that are unique to this cluster, whereby the carbamoyltransferase is truncated and replaced by an acyltransferase, explaining the unique toxin profile presented by Lyngbya wollei. These findings will enable the creation of toxin probes, for water monitoring purposes, as well as proof-of-concept for the combinatorial biosynthesis of these natural occurring alkaloids for the production of novel, biologically active compounds.
Introduction
Saxitoxin and its derivatives, collectively termed paralytic shellfish toxins (PST), are a group of low molecular weight, highly potent neurotoxic alkaloids which inhibit nerve conduction and muscle contraction by selectively binding and blocking of sodium channels. PSTs are the causative agents of the syndrome termed paralytic shellfish poisoning (PSP) [1], [2], [3]. Saxitoxin (STX), a tricyclic perhydropurine alkaloid, is considered the parent compound of the PSTs and may be substituted at five positions, leading to more than 30 naturally occurring analogs [4], [5], [6], [7], [8], [9]. PSP is a life-threatening affliction with a worldwide distribution, mainly caused by the consumption of shellfish contaminated by PSTs. PSP outbreaks have prompted serious public health concerns as well as significant economic losses due to the closure of fisheries, effects on tourism and costly toxin monitoring programs [10]. The human vectors of PSP are not the primary producers of PSTs, but have been shown to bio-accumulate them via filter feeding on toxic marine dinoflagellates mainly belonging to the genera Alexandrium, Gymnodinium and Pyrodinium [11], [12], [13]. Interestingly, PST biosynthesis has also been identified in several genera of freshwater cyanobacteria, namely Anabaena, Cylindrospermopsis, Aphanizomenon, Planktothrix, Raphidiopsis brookii and Lyngbya wollei [14], [15], [16], [17], [18], [19]. Furthermore, early investigations into PST biosynthesis mechanisms in cyanobacteria and dinoflagellates have indicated that they are synthesised via the same biosynthetic route [20]. Cyanobacteria present a formidable challenge for water management industries, due to toxic and non-toxic strains often being closely related and frequently indistinguishable using traditional taxonomic approaches. The combination of unpredictable formation of dense toxic blooms in water reservoirs and inability to identify toxic strains requires the use of time and resource consuming analytical methods such as HPLC and LC-MS to identify toxins in the water column. Genetic probes specific to toxin biosynthesis genes can afford an early warning system to help manage and control these toxic cyanobacterial blooms, also referred to as Harmful Algal Blooms (HABs).
We have recently identified and characterized putative PST biosynthesis gene clusters in the cyanobacteria Cylindrospermopsis raciborskii T3, Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5 [21], [22]. We have been able to show that the gene clusters are similar in gene content, though several putative tailoring enzymes are variable, leading to indirect evidence of the role of the tailoring gene sxtX. The Anabaena and Aphanizomenon PST gene clusters are apparently more similar in gene content and cluster organization to each other than to the initially characterized PST gene cluster in C. raciborskii, indicating the gene cluster is most probably of ancient origin, and shares a common descent among these closely related species, while the sporadic distribution of toxicity in the genera is explained through the repeated losses of the biosynthetic cluster. Following the lines of that work, we set out to characterize and identify the PST gene cluster in L. wollei, which has been shown to produce novel PST analogs, while lacking more commonly found carbamoylated PST derivatives. In addition, the production of various PST congeners appears to be dependent on the producing species and its geographical origin [7], [9], [23], [24]. This variability in toxin production has a presumable origin in the gene content of the PST gene cluster, its elucidation would thereby provide further insight into the PST biosynthetic machinery and the evolution of this intriguing gene cluster.
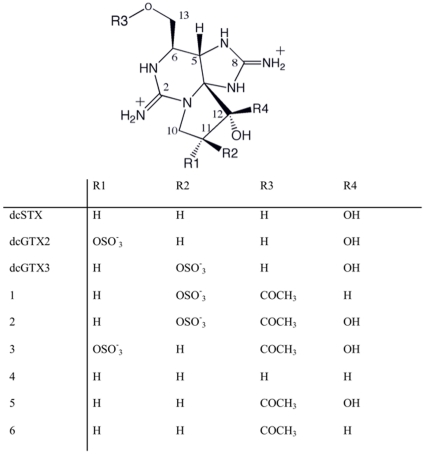
Lyngbya wollei a cyanobacterium belonging to the cyanobacterial order Oscillatoriales, has been reported from south-eastern United States lakes and water reservoirs for the past 100 years and has become increasingly common in the last three decades [25]. L. wollei has been shown to produce various PSTs, including decarbamoyl saxitoxins (dcSTX) and decarbamoyl gonyautoxins (dcGTX2, dcGTX3), as well as six novel analogs denoted 1-6 (Figure 1), which are characterized by the presence of acetate at C-13 and a carbinol at C-12 [7]. The toxin profile of L. wollei does not resemble the profile of any other characterized PST-producing cyanobacteria, such as Anabaena circinalis, Aphanizomenon sp. and Cylindrospermopsin raciborskii. The major PSTs found in A. circinalis are STX, the GTXs, and C1 and C2 [15], [26], while the major PST toxins found in Aph. flos-aquae (Aph. sp. NH-5) are neoSTX and STX [14].
Figure 1. Structure of the saxitoxin analogs identified in Lyngbya wollei (adapted from [7]).
Here we report the identification and sequencing of a homologous PST gene cluster in L. wollei. We present the bioinformatically inferred functions for most of the open reading frames (ORFs) in this gene cluster. The identification of saxitoxin biosynthesis gene sequences in various producer organisms enables the creation of probes for the monitoring of toxic blooms.
Results and Discussion
Identification of the Lyngbya PST gene cluster
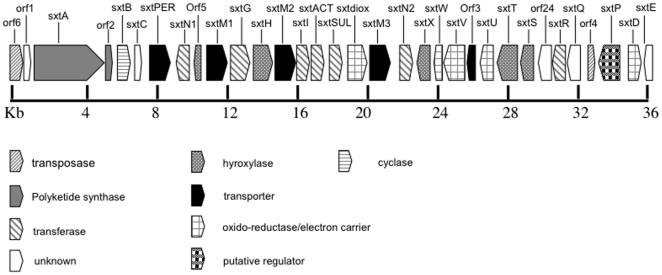
Using the degenerate PCR described, a single amplicon of 400 bp was amplified, cloned and sequenced from L. wollei genomic DNA. These gene fragments represented good candidates for saxitoxin biosynthesis genes due to their homology to the saxitoxin genes (sxtT and sxtH) previously identified in C. raciborskii T3 [21]. Numerous rounds of gene-walking outwards from these known regions revealed a SXT gene cluster in L. wollei spanning approximately 36 kb (Figure 2) and encoding thirty-one genes predicted to be involved in the biosynthesis and export of the these neurotoxins (Table 1). Homology analysis of the regions flanking the PST biosynthetic cluster confirmed that the sequences obtained are most related to Lyngbya species.
Figure 2. Organization of the PST biosynthesis cluster in Lyngbya wollei, scale indicates gene size in base pairs, direction of arrowed boxes indicates direction of transcription.
Table 1. The saxitoxin biosynthesis genes in Lyngbya wollei and their homology-based functions identified using BlastX (percentage identity is reported).
| Gene | Size (bp) | Closest BLAST match (%) | Putative Function | |
| Orf6 | 1009 | ABZ02176.1 Planktothrix agardhii NIVA-CYA 126/8 | 66 | Transposase/frame shift/inactive |
| Orf 1 | 240 | No significant similarity | --- | Unknown |
| sxtA | 3732 | ACG63801.1 SxtA Aphanizomenon sp. NH-5 | 90 | Loading of ACP, Methylation, ACP, Claisen condensation |
| Orf 2 | 112 | ABX60161.1 CyrB (AoaB) C. raciborskii AWT205 * | 61 | Truncated/inactive |
| sxtB | 969 | ACG63800.1 SxtB Aphanizomenon sp. NH-5 | 91 | Cyclization |
| sxtC | 285 | ABI75092.1 SxtC Cylindrospermopsis raciborskii T3 | 78 | Unknown |
| sxtPER | 1218 | ABI75130.1 SxtPER Anabaena circinalis AWQC131C | 89 | Export of PST's |
| sxtN1 | 837 | ABI75104.1 SxtN Cylindrospermopsis raciborskii T3 | 93 | Sulfotransfer |
| Orf 5 | 75 | ACG63814.1 SxtH Aphanizomenon sp. NH-5 | 92 | Truncated/inactive |
| sxtM 1 | 1440 | ACG63815.1 SxtM Aphanizomenon sp. NH-5 | 81 | Export of PSTs |
| sxtG | 1134 | ABI75136.1 SxtG Anabaena circinalis AWQC131C | 94 | Amidinotransfer |
| sxtH | 1029 | ABI75098.1 SxtH Cylindrospermopsis raciborskii T3 | 81 | C-12 hydroxylation |
| sxtM 2 | 1458 | ACG63815.1 SxtM Aphanizomenon sp. NH-5 | 80 | Export of PSTs |
| sxtI | 1071 | ACC69003.1 SxtI Cylindrospermopsis raciborskii T3 | 80 | Truncated/inactive |
| sxtACT | 1197 | ZP_05376006.1 H. denitrificans ATCC51888 | 33 | C13 acylation |
| sxtSUL | 909 | CAJ70870.1 Candidatus Kuenenia stuttgartiensis | 35 | Sulfotransfer |
| sxtdiox | 1005 | ACG63810.1 SxtT Aphanizomenon sp. NH-5 | 83 | C-12 reduction |
| sxtM 3 | 1512 | ACG58379.1 SxtM Anabaena circinalis AWQC131C | 88 | Export of PSTs |
| sxtN 2 | 837 | ABI75104.1 SxtN Cylindrospermopsis raciborskii T3 | 94 | Sulfotransfer/inactive |
| sxtX | 774 | ACF94656.1 SxtX Cylindrospermopsis raciborskii T3 | 98 | N-1 hydroxylation |
| sxtW | 330 | ABI75106.1 SxtW Cylindrospermopsis raciborskii T3 | 100 | Electron carrier |
| sxtV | 1680 | ABI75107.1 SxtV Cylindrospermopsis raciborskii T3 | 95 | Dioxygenase reductase |
| Orf 3 | 358 | ABI75130.1 SxtPER Anabaena circinalis AWQC131C | 83 | Truncated/inactive |
| sxtU | 750 | ABI75108.1 SxtU Cylindrospermopsis raciborskii T3 | 92 | Reduction of C-1 |
| sxtT | 1005 | ACG63810.1 SxtT Aphanizomenon sp. NH-5 | 90 | C-12 hydroxylation |
| sxtS | 801 | ABI75110.1 SxtS Cylindrospermopsis raciborskii T3 | 89 | Ring formation |
| Orf 24 | 747 | ABI75131.1 Orf24 Anabaena circinalis AWQC131C | 82 | Unknown |
| sxtR | 777 | ABI75112.1 SxtR Cylindrospermopsis raciborskii T3 | 94 | Unknown |
| stxQ | 777 | ACG63806.1 SxtQ Aphanizomenon sp. NH-5 | 93 | Unknown |
| Orf 4 | 279 | ACC85294.1 transposase Nostoc punctiforme PCC73102 | 67 | Transposition/inactive |
| sxtP | 1482 | ABI75126.1 SxtP Anabaena circinalis AWQC131C | 86 | Regulator/pilli formation |
| sxtD | 759 | ABI75125.1 SxtD Anabaena circinalis AWQC131C | 85 | Desaturation |
| sxtE | 363 | ABI75124.1 SxtE Anabaena circinalis AWQC131C | 90 | Unknown |
*Indicates a PsiBLAST search was used.
Characterization of the PST gene cluster
L. wollei possesses a toxin profile that is markedly different to that of other known PST producers. The production of these novel saxitoxin analogs, that are either decarbamoylated, acetylated, or hydroxylated (Figure 1) is hypothesized to be the result of the comparative differences between the genetic background of L. wollei and other cyanobacterial producers of PSTs. The putative saxitoxin gene cluster in L. wollei is highly similar in gene content, however, it includes additional enzymes not present, in the so far characterized PST biosynthetic clusters [19], [21], [22]. Specifically, an extra dioxygenase (sxtdiox, present in R. brookii), two additional sulfotransferases (sxtN2 and sxtSUL (present in R. brookii), two additional exporters (sxtM1 and sxtM2), and a novel acyl transferase (sxtACT) define the L. wollei pathway. Furthermore, the L. wollei PST gene cluster encodes a truncated carbamoyltransferase (sxtI) and does not contain the gene sxtL, coding for a GDSL-lipase-like enzyme identified in the other recently characterized saxitoxin gene clusters [19], [21], [22]).
Bioinformatic analysis of the L. wollei saxitoxin gene cluster further revealed a putative recombination event, which presumably results in the synthesis of the novel C-13 acetate saxitoxin analogs, and accounts for the lack of any analogs containing a carbamoyl group at this position (C-13). The gene responsible for the formation of the carbamoyl group originally identified in C. raciborskii T3 [21], is the 1839 bp long O-carbamoyltransferase denoted sxtI. The sxtI gene homolog from L. wollei was only 1071 bp in length. Recently, Kellmann et al. [27] have shown that the L. wollei sxtI first 912 base-pairs had 92.9% identity to sxtI from C. raciborskii T3, base-pairs 913 to 1059 and 1177 to 1189 (C. raciborksii T3 numbering) were deleted, and the 3′-end was truncated after base-pair 1227. As a result of these deletions the partial L. wollei sxtI translated enzyme does not contain the putative catalytic site GPRALGGRS [27]. It is therefore, most probably inactive. The apparent truncation of sxtI might be the result of a partial gene deletion event or alternatively by the insertion of a gene, denoted sxtACT (Figure 2). sxtACT codes for an enzyme similar to an O-acyltrasferase belonging to the CoA-dependent acyltransferase superfamily. A member of this family, Deacetylvindoline 4-O-acetyltransferase (EC:2.3.1.107) catalyzes the last step in vindoline biosynthesis [28]. It is also possible that sxtACT could be an ancient sxt gene that has been lost in the other cyanobacterial lineages. Irrespective of whether sxtI and sxtACT had been recently acquired or lost from the PST biosynthesis clusters, genetic rearrangement has resulted in the absence of carbamoylated saxitoxins and the presence of the novel C-13 acetylated saxitoxin analogs in L. wollei. Furthermore, the three novel genes identified in the L. wollei saxitoxin gene cluster, namely sxtACT, sxtSUL and sxtdiox, appear to have been inserted as one locus, thereby conferring the ability to produce analogs 1-6 in L. wollei.
sxtL, a gene which is not present in the L. wollei PST gene cluster codes for an enzyme similar to a GDSL lipase, which has been proposed to catalyze the hydrolytic cleavage of the carbamoyl group from STX analogs in C. raciborskii T3. The absence of an active carbamoyltransferase in L. wollei would explain the lack of carbamoylated saxitoxin derivatives produced by L. wollei. In this modified genetic background, that is, without an active sxtI homolog, and therefore no carbamoylated saxitoxins, the lack of a decarbamoylating enzyme, SxtL, seems evolutionally logical.
L. wollei synthesizes a new group of derivatives that contain only one carbinol residue at C-12 (denoted 1,4 and 6 Figure 1), whereas all other previously identified saxitoxins have a hydrated ketone function at that position, a so called geminal hydroxyl [7]. Only one epimer, the b-H form, of these 12-dihydro derivatives has been identified in L. wollei, and this is therefore a specific enzymatic step in the biosynthesis of these compounds [7]. The L. wollei gene cluster encodes three distinct dioxygenases, denoted sxtH, sxtT and sxtdiox, which are most similar to phenylpropionate dioxygenases. These enzymes contain two domains; an N-terminal Rieske domain with an [2Fe-2S] cluster and a C-terminal catalytic domain with a mononuclear Fe(II) binding site [29]. sxtT and sxtH, originally identified in C. raciborskii T3, are putatively involved in the hydroxylation of the terminal diol in saxitoxin biosynthesis [21]. However, sxtdiox is unique to the L. wollei and R. brookii D9 PST gene clusters. In a recent detailed evolutionary analysis of the sxt genes, Murray et al. [30] found that the sxtH/T genes form a monophyletic sxt clade, which is under positive selection and has undergone significant intragenomic or intraspecific recombination. sxtH has likely duplicated in L. wollei, or the ancestral strain, giving rise to sxtT, and duplicated again to give rise to sxtdiox in R. brookii D9 and L. wollei [30].
Analysis of the dioxygenase protein alignment revealed that three positions at the C-terminus of the enzyme, conserved in the saxitoxin dioxygenases, are modified in sxtdiox (Figure 3). As the mechanism of catalysis is unknown in this enzyme family it is not possible to determine the effect of these alterations [31]. However, we propose that that sxtdiox carries out the hydroxylation of C-12 in the saxitoxin derivatives, 1, 4, 6, thereby forming the novel 12-dihydro-derivatives. It will be of biochemical significance to confirm this novel activity by heterologous expression and characterization of this enzyme.
Figure 3. Amino acid alignment of the dioxygenases encoded by the various saxitoxin clusters.
The conserved regions that are modified in sxtdiox are boxed.
L. wollei also produces O-sulfated saxitoxin analogs (dcGTX2/3, 1,2,3) and the L. wollei gene cluster encodes two sulfotransferases (sxtN1 and sxtN2) homologous to sxtN identified in C. raciborskii T3 [21], which presumably transfers a sulfate group to O-22 forming dcGTX2/3. sxtN1 and sxtN2 are highly similar, with a nucleotide sequence identity of 97%. However, sxtN2 contains a stop codon in its ORF, and therefore is most probably inactive and possibly represents a gene duplication event. The L. wollei saxitoxin gene cluster also contains another gene bioinformatically identified as a PAPS dependent sulfotransferase, namely sxtSUL, which is unique to the L. wollei and R. brookii D9 PST gene clusters. We postulate that due to its uniqueness in this gene cluster and its homology to other characterized sulfotransferases, that sxtSUL might sulfate the novel saxitoxin analogs produced by L. wollei (5,6) forming the saxitoxin analogs 1,2,3, though its involvement in the formation of the GTX toxins cannot be excluded.
Surprisingly, the L. wollei PST gene cluster contains the PST tailoring enzyme sxtX, a gene similar to cephalosporin hydroxylase, and presumably responsible for N-1 hydroxylation of STX, thereby converting STX to neoSTX [21], [22]. This finding is not in agreement with the analysis of Onodera et al. [7], which did not reveal any N-hydroxylated PST derivatives in the L. wollei isolate analyzed. This discrepancy might be attributed to low levels of N-hydroxylated derivatives in the analyzed sample, bellow the limit of detection of methods used, or alternatively might be the result of novel STX derivatives which were not elucidated with the methods used. On the other hand, the L. wollei biomass used in our study, although from the same source (Guntersville reservoir), might be of a dissimilar strain with a different genetic background to the sample analyzed by Onodera et al. [7], and therefore might have a distinct toxin profile. A further explanation would include a different catalytic activity for sxtX than was previously proposed, clarifying its actual activity will require the heterologous expression of sxtX to determine its catalytic activity and natural substrates. In order to reconcile these apparent discrepancies a culturable strain of L. wollei needs to be analyzed for the presence of PST biosynthesis genes as well as PST derivatives produced, though this was not possible at the time of this study as the available freeze-dried L. wollei biomass was not suitable for reliable PST composition analysis.
Recent studies into the kinetics of PST accumulation in cyanobacterial cells and growth media, suggest an active transport mechanism [32], [33]. The recently identified saxitoxin gene clusters [21], [22] also contain genes coding for Multidrug And Toxic compound Extrusion (MATE) proteins of the NorM family. The L. wollei saxitoxin gene cluster contains three genes belonging to this family, sxtM1, sxtM2 and sxtM3. Furthermore, the L. wollei PST cluster also contains a gene, denoted sxtPER, which is most similar to permeases of the drug and metabolite transporter (DMT) family. This gene is also present in A. circinalis 131C and Aph. sp. NH-5 [22] but absent in the gene cluster of C. raciborskii T3 [21]. The observed multitude of transporters present in the L. wollei PST gene cluster might be attributed to the variety and uniqueness of PST analogs produced by this species, or conversely to an adaptation of its transport mechanism to pressures in its environmental niche, requiring increased extracellular transport of PSTs.
Curiously, orf3, which is located between sxtV and sxtU also shows high similarity to sxtPER, though it is truncated and presumed inactive, and might be a result of a further gene duplication/recombination event. In addition, there are further indications of previous gene duplication and deletion events in the L. wollei PST biosynthesis gene cluster. Orf 5, which is located 5 prime to sxtM1, and contains a short truncated sequence which is highly similar to sxtH, a hydroxylase putatively involved in the hydroxylation of C-12 of saxitoxin. This fragment might be the result of the gene duplication event that formed the L.wollei unique transporters SxtM1 and sxtM2, as the intact copy of sxtH is also 5 prime and adjacent to sxtM2.
Evolution and distribution of PST biosynthesis in Lyngbya
Lateral gene transfer (LGT) and gene deletions are probable explanations for the sporadic distribution of saxitoxin producers among the different cyanobacterial species and across kingdoms [22], [27], [30]. The variations observed in the structure and gene content of the saxitoxin gene cluster in L. wollei may be a direct result of its mobility via the recombination events that are inherent to transposition. This is further supported by the presence of multiple copies of transposases identified in many secondary metabolite gene clusters, including those of other cyanobacterial toxins such as cylindrospermopsin, microcystin, nodularin and saxitoxin [34], [35], [36], [37,]. Interestingly, a small gene fragment denoted orf 2, which is located between sxtA and sxtB shows high similarity to the gene cyrB of C. raciborskii AWT205, which is involved in the biosynthesis of the cyanotoxin cylindrospermopsin [35]. This finding could indicate that part of the cylindrospermopsin biosynthesis gene cluster was present in this cyanobacterium and was lost through gene deletion and/or recombination event, while also possibly acquiring the PST biosynthesis genes. Curiously, L. wollei seems to present a geographical segregation in toxin production, as isolates from North America (such as the sample analyzed in this study) have so far only been shown to produce PSTs, while Australian isolates have only been shown to produce cylindrospermopsin [7], [38], [39]. A similar geographical distribution of toxicity is seen in the genera Cylindrospermopsis, that produce PSTs in South America and cylindrospermopsin in Europe and Australia [17], [40], [41], [42], and Anabaena, which only produce PSTs in Australia [15], [23], [24]. These results further support the view that LGT and gene deletions contribute to the evolution and distribution of cyanobacterial toxin biosynthesis genes. In agreement with these findings, recent phylogenetic analysis of cyanobacterial PST genes [30], [37] has indicated that the PST genes are of ancient origin, with a complex history involving horizontal gene transfer from different sources, whereby the ability to produce PST was subsequently lost multiple times in non-PST producing cyanobacteria.
Biosynthesis of PST congeners
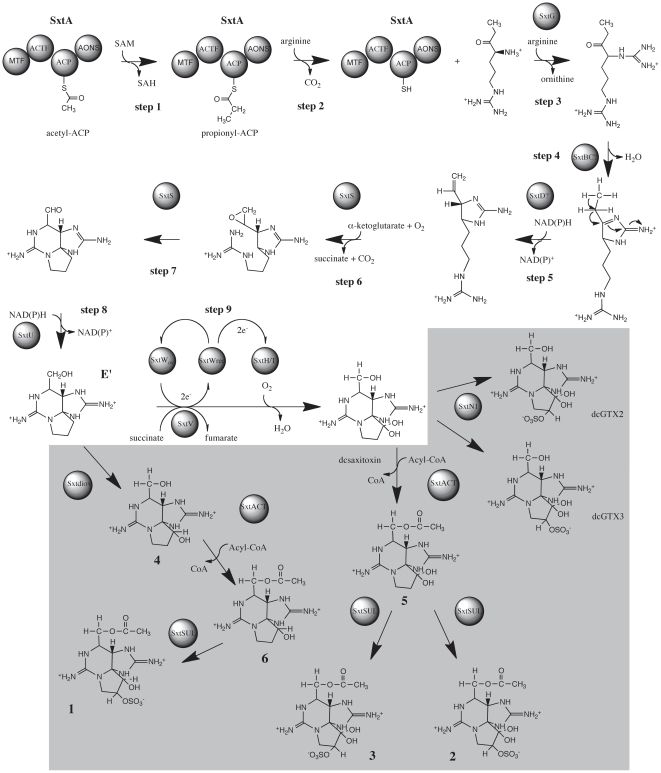
Based on the in silico analysis of the L. wollei PST gene cluster, the recently elucidated putative saxitoxin biosynthetic pathway [21], [22] has been correspondingly modified (Figure 4). In L. wollei, we propose that the biosynthetic steps leading up to the formation of intermediate E' are identical to the pathway described for C. raciborskii, A. circinalis and Aph. sp. [21], [22] (steps 1- 8 Figure 4). Thereafter, sxtH and sxtT, each coding a terminal oxygenase subunit of bacterial phenyl-propionate and related ring-hydroxylating dioxygenases, catalyze the consecutive hydroxylation of C-12, forming dcSTX (Figure 4 step 9). Unique to L.wollei are the proposed novel tailoring reactions leading to the formation of Lyngbya-specific saxitoxin isoforms (Figure 1,4). dcGTX2 and dcGTX3 are putatively formed by a sulfur transfer onto O-11 of dcSTX catalyzed by sxtN1 (sulfotransferase). sxtACT, an acyl-CoA dependant acyltransferase moves an acyl group onto the hydroxyl at C-13 of dcSTX resulting in analog 5. The sulfotransfer onto analog 5, catalyzed by SxtSUL, synthesizes analogs 2 and 3. The sxtdiox catalyzed, single hydroxylation of the biosynthetic intermediate produced by step 8 (Figure 4) forms analog 4. An acyltransfer onto the analog 4 C-13 hydroxyl, via the action of sxtACT results in the formation of analog 6, which can then be sulfated by SxtSUL to form analog 1.
Figure 4. Proposed biosynthetic pathway for the PSTs produced by Lyngbya wollei.
The gray shaded area highlights the steps that are unique to L. wollei.
As noted by Onodera et al. [7], the lack of an 11-a-O-sulfate epimer of saxitoxin analog 1 (Figure 1) in L. wollei indicated that the reduction of C-12 occurs prior to the introduction of 11-O-sulfate in a stereo specific manner from the b-side of the molecule [7]. This predicted biosynthesis pathway should be confirmed by future heterologous expression of the enzymes and molecular intermediate analysis by mass-spectrometry.
Analysis of saxitoxin gene clusters in the numerous and divergent producing organisms provides insights into the biosynthetic machinery and the variations that give rise to different toxin profiles. This in turn enables the construction of hypotheses regarding the role of the genes present, which will direct validation by recombinant expression in heterologous hosts. A case in point are the genetic rearrangements identified in the L. wollei PST biosynthesis gene cluster, which include a putative truncation and inactivation of the carbamoyltransferase gene (sxtI), coupled with the presence of an acyltransferase gene (sxtACT), a novel sulfotransferase (sxtSUL) and a dioxygenase (sxtdiox) adjacent to each other. This observed genetic variance presumably resulted in a gene cluster that does not confer the ability to produce carbamoylated saxitoxin derivatives. The new family of saxitoxin analogs made possible by this genetic rearrangement, contain an acetate group at the same position (C-13), as well as a carbinol at C-12, and have shown reduced neurotoxicity [7]. The deletion identified in the L. wollei sxtI gene further confirms the putative role assigned to sxtI as a carbamoyltransferase, based on bioinformatic analysis of the gene [27] and in vitro biosynthesis studies [43] that show carbamoylphosphate, the natural substrate for carbamoyltransferases, to be a precursor for saxitoxin biosynthesis. This example of natural combinatorial biosynthesis indicates the ability to produce unnatural compounds from alkaloid biosynthetic pathways, enabling the production of novel biologically active compounds. The in silico functional assignment of sxt genes in L. wollei, has enabled the construction of a putative PST biosynthetic pathway, thereby elucidating the production of the novel saxitoxin analogs by this organism (Figure 1). The novel PST tailoring genes identified in L. wollei, add to the catalytic collective available for the growing field of combinatorial biosynthesis.
The gene sequence data presented in this study will enable future investigations into the regulation of PST biosynthesis gene expression in this organism using techniques such as promoter analysis and real-time PCR whereby providing insights into the physiological roles of the PSTs and furthering our ability to predict and prevent the formation of harmful algal blooms. The availability of more saxitoxin biosynthesis gene sequences will enable better monitoring of algal blooms for water authorities, including PCR-based early warning systems such as qPCR and toxin gene specific PCRs. This work may also facilitate the identification of the genes involved in saxitoxin biosynthesis in dinoflagellates, which are the cause of human mortality and great economic damage to the shellfish and tourism industries.
Materials and Methods
Cyanobacterial sampling
Lyngbya wollei (Farlow ex Gomont) was originally isolated from Guntersville Reservoir (Guntersville, AL) and its taxonomy and toxicity subsequently determined [16]. A field sample of a Lyngbya wollei unialgal bloom from Guntersville Reservoir was freeze-dried, stored at −20°C and used for the subsequent DNA isolation.
DNA extraction
Total genomic DNA was extracted from freeze-dried cyanobacterial cells using the Mo Bio PowerPlant DNA isolation kit (Carlsbad) in accordance with the manufacturers instructions. Genomic DNA was stored at −20°C.
16S rRNA gene analysis
Genomic DNA isolated from L. wollei was amplified using the 16S rRNA gene primers 27F and 809R as previously described [44], and deposited in GenBank under accession number EU603708.
Saxitoxin gene amplification
We have recently identified the gene clusters responsible for the biosynthesis of saxitoxin in the cyanobacteria C. raciborskii T3, A. circinalis AWQC131C, and Aph. sp. NH-5 [21], [22]. Based on this sequence information, amino acid alignments of two highly similar enzymes in the saxitoxin gene clusters, namely sxtT and sxtH, which are putatively involved in the formation of the terminal diol at C-12 in saxitoxin [21], were created. Degenerate PCR primers targeting sxtT and sxtH used in this study (DioxF 5′ CCNGARTGGGAYGAYCCNAAYTA 3′ DioxR 5′ ATRTCYTGYTCDATNGTYTCRTC 3′) were designed in silico from sequence alignments produced using ClustalX [45]. Degenerate PCR was performed in 20 µL reaction volumes containing 1× Taq polymerase buffer, 2.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphates, 25 pmol each of the forward and reverse primers, 50 ng of genomic DNA and 0.2 U of Taq polymerase (Fischer Biotech). Thermal cycling was performed in a GeneAmp PCR System 2400 Thermal cycler (Perkin Elmer Corporation). Cycling began with a denaturing step at 94°C for 4 min followed by 35 cycles of denaturation at 94°C for 10 s, primer annealing at 50°C for 30 s and a DNA strand extension at 72°C for 45 s. Amplification was completed by a final extension step at 72°C for 5 min.
Amplified DNA was analyzed by agarose gel electrophoresis in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 7.8), and visualized by UV transillumination after staining with ethidium bromide (0.5 µg · mL). Where multiple amplicons were detected during the gel electrophoresis, single amplicons were excised from the gels and purified using the Promega Wizard® SV Gel and PCR Clean-Up, prior to sequencing.
Gene cloning
Clone libraries were created using the pGemT-easy cloning kit (Promega) in accordance with the manufacturers instructions.
DNA sequencing. Automated DNA sequencing was performed using the PRISM Big Dye cycle sequencing system and a model 373 sequencer (Applied Biosystems).
Gene walking
Characterization of unknown regions of DNA flanking the putative saxitoxin biosynthesis genes, sxtT and sxtH, in L. wollei was performed using an adaptor-mediated PCR as previously described [21], [22], [34]. PCRs were performed in 20 µL reaction volumes containing 1× Taq polymerase buffer, 2.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphates, 10 pmol each of the forward and reverse primers, between 10 and 100 ng of genomic DNA and 0.5 U of a mixture of 10:1 Taq polymerase/PFU (Fischer Biotech). Thermal cycling was performed in a GeneAmp PCR System 2400 Thermal cycler (Perkin Elmer Corporation). Cycling began with a denaturing step at 94°C for 2 min followed by 30 cycles of denaturation at 94°C for 10 sec, primer annealing between 55°C and 65°C for 20 sec and a DNA strand extension at 72°C for 3 min. Amplification was completed by a final extension step at 72°C for 7 min.
Bioinformatic analysis
Sequence data were analyzed using ABI Prism-Autoassembler software, while identity/similarity values to other translated sequences were determined using BLAST in conjunction with the National Center for Biotechnology Information (NIH, Bethesda, MD). Fugue blast (http://www-cryst.bioc.cam.ac.uk/fugue/) was used to identify distant homologs via sequence-structure comparisons. The gene clusters were assembled using the software package Phred, Phrap, and Consed (http://www.phrap.org/phredphrapconsed.html), open reading frames were manually identified. Alignments were performed using ClustalX [45].
Nucleotide sequence accession number
The L. wollei PST gene cluster sequences were submitted to GenBank and are available under the accession number EU603711.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Australian Research Council. BAN is a fellow of the Australian Research Council. The funding body had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kao CY, Levinson SR, editors. New York: The New York Academy of Science; 1986. Tetrodotoxin, Saxitoxin, and the molecular biology of the sodium channel. [PubMed] [Google Scholar]
- 2.Wang J, Salata JJ, Bennett PB. Saxitoxin is a gating modifier of HERG K+ channels. The Journal of General Physiology. 2003;121:583–598. doi: 10.1085/jgp.200308812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su Z, Sheets M, Ishida H, Li F, Barry WH. Saxitoxin blocks L-type ICa. Journal of Pharmacology & Experimental Therapeutics. 2004;308 doi: 10.1124/jpet.103.056564. [DOI] [PubMed] [Google Scholar]
- 4.Arakawa O, Noguchi T, Shida Y, Onoue Y. Occurrence of carbamoyl-N-hydroxy derivatives of saxitoxin and neosaxitoxin in a xanthid crab Zosimus aeneus. Toxicon. 1994;32:175–183. doi: 10.1016/0041-0101(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa O, Nishio S, Noguchi T, Shida Y, Onoue Y. A new saxitoxin analogue from a xanthid crab Atergatis floridus. Toxicon. 1995;33:1577–1584. doi: 10.1016/0041-0101(95)00106-9. [DOI] [PubMed] [Google Scholar]
- 6.Oshima Y. Postcolumn derivatization liquid chromatographic method for paralytic shellfish toxins. Journal of AOAC International. 1995;78:528–532. [Google Scholar]
- 7.Onodera H, Satake M, Oshima Y, Yasumoto T, Carmichael Wayne W. New saxitoxin analogues from the freshwater filamentous cyanobacterium Lyngbya wollei. Natural Toxins. 1997;5:146–151. doi: 10.1002/1522-7189(1997)5:4<146::AID-NT4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 8.Zaman L, Arakawa O, Shimosu A, Shida Y, Onoue Y. Occurrence of a methyl derivative of saxitoxin in Bangladeshi freshwater puffers. Toxicon. 1998;36:627–630. doi: 10.1016/s0041-0101(97)00086-x. [DOI] [PubMed] [Google Scholar]
- 9.Negri A, Stirling D, Quilliam M, Blackburn S, Bolch C, et al. Three novel hydroxybenzoate saxitoxin analogues isolated from the dinoflagellate Gymnodinium catenatum. Chemical Research in Toxicology. 2003;16:1029–1033. doi: 10.1021/tx034037j. [DOI] [PubMed] [Google Scholar]
- 10.Hallegraeff GM. Harmful algal blooms: A global overview. In: Hallegraeff GM, Anderson DM, Cembella AD, editors. Manual on Harmful Marine Microalgae. Paris: UNESCO; 1995. pp. 1–22. [Google Scholar]
- 11.Shimizu Y. Chemistry and distribution of deleterious dinoflagellate toxins. In: Faulkner D, Fenical W, editors. New York: Plenum; 1977. pp. 261–269. [Google Scholar]
- 12.Harada T, Oshima Y, Yasumoto T. Structure of two paralytic shellfish toxins, gonyautoxins V and VI, isolated from a tropical dinoflagellate Pyrodinium bahamense var. compressa. Agricultural & Biological Chemistry. 1982;46:1861–1864. [Google Scholar]
- 13.Oshima Y, Hasegawa M, Yasumoto T, Hallegaeff G, Blackburn S. Dinoflagellate Gimnodium catenatum as the source of paralytic shellfishtoxins in Tasmanian shellfish. Toxicon. 1987;25:1105–1111. doi: 10.1016/0041-0101(87)90267-4. [DOI] [PubMed] [Google Scholar]
- 14.Mahmood NA, Carmichael WW. Paralytic shellfish poisons produced by the freshwater cyanobacterium Aphanizomenon flos-aquae NH-5. Toxicon. 1986;24:175–186. doi: 10.1016/0041-0101(86)90120-0. [DOI] [PubMed] [Google Scholar]
- 15.Humpage AR, Rositano J, Bretag AH, Brown R, Baker P, et al. Paralytic shellfish poisons from australian cyanobacterial blooms. Australian Journal of Marine & Freshwater Research. 1994;45:761–771. [Google Scholar]
- 16.Carmichael WW, Evans WR, Yin QQ, Bell P, Moczydlowski E. Evidence for paralytic shellfish poisons in the freshwater cyanobacterium Lyngbya wollei (Farlow ex Gomont) comb. nov. Applied & Environmental Microbiology. 1997;63:3104–3110. doi: 10.1128/aem.63.8.3104-3110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagos N, Onodera H, Zagatto PA, Andrinolo D, Azevedo S, et al. The first evidence of paralytic shellfish toxins in the freshwater cyanobacterium Cylindrospermopsis raciborskii, isolated from Brazil. Toxicon. 1999;37:1359–1373. doi: 10.1016/s0041-0101(99)00080-x. [DOI] [PubMed] [Google Scholar]
- 18.Pomati F, Sacchi S, Rossetti C, Giovannardi S, Onodera H, et al. The freshwater cyanobacterium Planktothrix sp. FP1: Molecular identification and detection of paralytic shellfish poisoning toxins. Journal of Phycology. 2000;36:553–562. doi: 10.1046/j.1529-8817.2000.99181.x. [DOI] [PubMed] [Google Scholar]
- 19.Stucken K, John U, Cembella A, Murillo AA, Soto-Liebe K, et al. The smallest known genomes of multicellular and toxic cyanobacteria: Comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS ONE. 2010;5(5) doi: 10.1371/journal.pone.0009235. doi: 10.1371/journal.pone.0009235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu Y. Microalgal metabolites. Chemical Reviews. 1993;93:1685–1698. [Google Scholar]
- 21.Kellmann R, Mihali TK, Jeon YJ, Pickford R, Pomati F, et al. Biosynthetic intermediate analysis and functional homology reveal a putative saxitoxin gene cluster in cyanobacteria. Applied & Environmental Microbiology. 2008a;74:4044–4053. doi: 10.1128/AEM.00353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mihali TK, Neilan BA. Identification of the saxitoxin biosynthesis cluster in Anabaena circinalis 131C and Aphanizomenon flos-aquae NH-5. BMC Biochemistry. 2009;10:8. doi: 10.1186/1471-2091-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velzeboer RMA, Baker PD, Rositano J, Heresztyn T, Codd GA, et al. Geographical patterns of occurrence and composition of saxitoxins in the cyanobacterial genus Anabaena (Nostocales, Cyanophyta) in Australia. Phycologia. 2000;39:395–407. [Google Scholar]
- 24.Beltran EC, Neilan BA. Geographical segregation of the neurotoxin-producing cyanobacterium Anabaena circinalis. Applied & Environmental Microbiology. 2000;66:4468–4474. doi: 10.1128/aem.66.10.4468-4474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speziale BJ, Dyck LA. Lyngbya infestations: comparative taxonomy of Lyngbya wollei comb. nov. (Cyanobacteria). Journal of Phycology. 1992;28:613–706. [Google Scholar]
- 26.Baker PD, Humpage AR. Toxicity associated with commonly occurring cyanobacteria in surface waters of the Murray-Darling Basin. Australian Journal of Marine & Freshwater Research. 1994;45:773–786. [Google Scholar]
- 27.Kellmann R, Mihali TK, Neilan BA. Identification of a saxitoxin biosynthesis gene with a history of frequent horizontal gene transfers. Journal of Molecular Evolution. 2008;67:526–538. doi: 10.1007/s00239-008-9169-2. [DOI] [PubMed] [Google Scholar]
- 28.St-Pierre B, Laflamme P, Alarco A-M, De Luca V. The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. The Plant Journal. 1998;14:703–713. doi: 10.1046/j.1365-313x.1998.00174.x. [DOI] [PubMed] [Google Scholar]
- 29.Mason JR, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annual Review of Microbiology. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 30.Murray S, Mihali TK, Neilan BA. Molecular Biology and Evolution; 2010. Extraordinary conservation, gene loss and positive selection in the evolution of an ancient neurotoxin. doi: 10.1093/molbev/msq295. [DOI] [PubMed] [Google Scholar]
- 31.Kauppi B, Lee K, Carredano E, Parales RE, Gibson DT, et al. Structure of an aromatic-ring-hydroxylating dioxygenase - naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 32.Pomati F, Rossetti C, Manarolla G, Burns BP, Neilan BA. Interactions between intracellular Na+ levels and saxitoxin production in Cylindrospermopsis raciborskii T3. Microbiology. 2004;150:455–461. doi: 10.1099/mic.0.26350-0. [DOI] [PubMed] [Google Scholar]
- 33.Castro D, Vera D, Lagos N, Garcia C, Vasquez M. The effect of temperature on growth and production of paralytic shellfish poisoning toxins by the cyanobacterium Cylindrospermopsis raciborskii C10. Toxicon. 2004;44:483–489. doi: 10.1016/j.toxicon.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Moffitt MC, Neilan BA. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Applied & Environmental Microbiology. 2004;70:6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mihali TK, Kellmann R, Muenchhoff J, Barrow KD, Neilan BA. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Applied & Environmental Microbiology. 2008;74:716–722. doi: 10.1128/AEM.01988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tillett D, Dittmann E, Erhard M, von Dohren H, Borner T, et al. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chemistry & Biology. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- 37.Moustafa A, Loram JE, Hackett JD, Anderson DM, Plumley FG, et al. Origin of saxitoxin biosynthetic genes in cyanobacteria. PLoS One. 2009;4:e5758. doi: 10.1371/journal.pone.0005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carmichael WW, Evans WR, Yin QQ, Bell P, Moczydlowski E. Evidence for paralytic shellfish poisons in the freshwater cyanobacterium Lyngbya wollei (Farlow ex Gomont) comb. nov. Applied & Environmental Microbiology. 1997;63:3104–3110. doi: 10.1128/aem.63.8.3104-3110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seiferta M, McGregor G, Eaglesham G, Wickramasinghe W, Shaw G. First evidence for the production of cylindrospermopsin and deoxy-cylindrospermopsin by the freshwater benthic cyanobacterium, Lyngbya wollei (Farlow ex Gomont) Speziale and Dyck. Harmful Algae. 2007;6:73–80. [Google Scholar]
- 40.Messineo V, Melchiorre S, Di Corcia A, Gallo P, Bruno M. Seasonal succession of Cylindrospermopsis raciborskii and Aphanizomenon ovalisporum blooms with cylindrospermopsin occurrence in the volcanic Lake Albano, Central Italy. Environmental Toxicology. 2010;25:18–27. doi: 10.1002/tox.20469. [DOI] [PubMed] [Google Scholar]
- 41.Neilan BA, Saker ML, Fastner J, Törökné A, Burns BP. Phylogeography of the invasive cyanobacterium Cylindrospermopsis raciborskii. Molecular Ecology. 2003;12:133–140. doi: 10.1046/j.1365-294x.2003.01709.x. [DOI] [PubMed] [Google Scholar]
- 42.Hawkins PR, Chandrasena NR, Jones GJ, Humpage AR, Falconer IR. Isolation and toxicity of Cylindrospermopsis raciborskii from an ornamental lake. Toxicon. 1997;35:341–346. doi: 10.1016/s0041-0101(96)00185-7. [DOI] [PubMed] [Google Scholar]
- 43.Kellmann R, Neilan BA. Biochemical characterization of paralytic shellfish toxin biosynthesis in vitro. Journal of Phycology. 2007;43:497–508. [Google Scholar]
- 44.Neilan BA. Identification and phylogenetic analysis of toxigenic cyanobacteria by multiplex randomly amplified polymorphic DNA PCR. Applied & Environmental Microbiology. 1995;61:2286–2291. doi: 10.1128/aem.61.6.2286-2291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]