Abstract
The relationship between circadian clocks and metabolism is intimate and complex and a number of recent studies have begun to reveal previously unknown effects of food and its temporal availability on the clock and the rhythmic transcriptome of peripheral tissues. Nocturnin, a circadian deadenylase, is expressed rhythmically in a wide variety of tissues, but we report here that Nocturnin expression is arrhythmic in epididymal white adipose tissue (eWAT) of mice housed in 12∶12 LD with ad libitum access to food. However, Nocturnin expression becomes rhythmic in eWAT of mice placed on restricted feeding. We show here that Nocturnin's rhythmic expression pattern is not dependent upon feeding, nor is it acutely induced by feeding in the liver or eWAT of ad libitum fed mice. However, Nocturnin is acutely induced by the absence of the expected meal in eWAT of restricted fed mice. A rise in cAMP levels also induces Nocturnin expression, suggesting that Nocturnin's induction in eWAT by fasting is likely mediated through the same pathways that activate lipolysis. Therefore, this suggests that Nocturnin plays a role in linking nutrient sensing by the circadian clock to lipid mobilization in the adipocytes.
Introduction
Circadian clocks orchestrate a variety of physiological and molecular rhythms that allow organisms to anticipate and respond to the daily changes that occur in the environment such as light, temperature, and feeding/fasting cycles. Restricted feeding studies have revealed that food can act as a zeitgeber (an entraining signal or ‘timegiver’), and have demonstrated that peripheral oscillators can be uncoupled from the master pacemaker located in the suprachiasmatic nucleus (SCN) [1], [2], [3]. Besides the entrainment of the circadian clock by food, there is a growing body of evidence that demonstrates that there is cross-talk between circadian clocks and metabolism and that signals from each feedback onto the other [4]. For example, mutations or deficiencies in core clock genes such as Clock or Bmal1 result in obesity, lipodystrophy and/or glucose homeostasis abnormalities [5], [6], [7], [8], [9], and nuclear receptors central to regulating metabolic processes, such as PPARα and PGC-1α, affect the circadian clock [10], [11].
The connection between circadian biology and metabolism is not limited to the core clock machinery, but extends to genes downstream of the clock as well. For example, loss of the rhythmically expressed deadenylase Nocturnin (Ccrn4l) results in metabolic abnormalities [12], [13], [14]. Nocturnin expression peaks in the early night in a variety of tissues such as the liver, the kidney, the spleen, and the retina [13]. Nocturnin deficient mice show no abnormalities in circadian behavior nor in the rhythmic expression of core clock genes in the liver, showing that Nocturnin is not part of the core circadian mechanism, but rather contributes to the rhythmic output of the clock [12]. Nocturnin −/− mice are resistant to diet-induced obesity and hepatic steatosis, and show dampened rhythms or decreased expression levels of genes related to lipid metabolism in the liver, such as Pparγ and Scd1, respectively [12].
It has also been demonstrated that Nocturnin is the only known mammalian deadenylase that responds acutely to extracellular stimuli: It is induced in NIH-3T3 fibroblasts by serum and by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA), and it is also acutely induced by insulin in 3T3-L1 preadipocytes [14], [15]. However, the in vivo significance of this acute responsiveness has not been clear.
In mice that conditionally over-express REV-ERBα in the liver, thus rendering the circadian clock nonfunctional in this tissue, Nocturnin mRNA levels remain rhythmic with a peak expression time occurring in the early night [16]. This finding suggests that Nocturnin's rhythmicity in the liver can be driven by systemic cues. However, it is unclear whether Nocturnin's expression pattern in the liver, and perhaps in other tissues, is due to an acute induction by extracellular stimuli, or whether it is the result of an entrained mechanism independent of the oscillation of the local liver clock.
White adipose tissue plays a pivotal role in energy homeostasis by storing energy in the form of triglycerides after a meal, and undergoing lipolysis during times of fasting. It also functions as an endocrine organ secreting a variety of adipokines that have systemic affects on appetite and glucose homeostasis [17], [18], [19]. Due to Nocturnin's role in metabolism, its inducibility by extracellular stimuli in vitro, and its capability to be driven by systemic cues in the liver, we set out to examine Nocturnin's expression pattern in white adipose tissue and to investigate whether Nocturnin can be acutely induced by feeding or fasting cues in the liver and the white adipose tissue, two tissues central to metabolic homeostasis. We report that Nocturnin's expression pattern is not rhythmic in white adipose tissue in ad libitum feeding conditions, but is rhythmic in white adipose tissue of mice subjected to temporal, not caloric, restriction of food availability. Also, we show that Nocturnin is acutely induced by fasting in the white adipose tissue of restricted-fed mice.
Results
Nocturnin expression is not dependent upon or induced by feeding
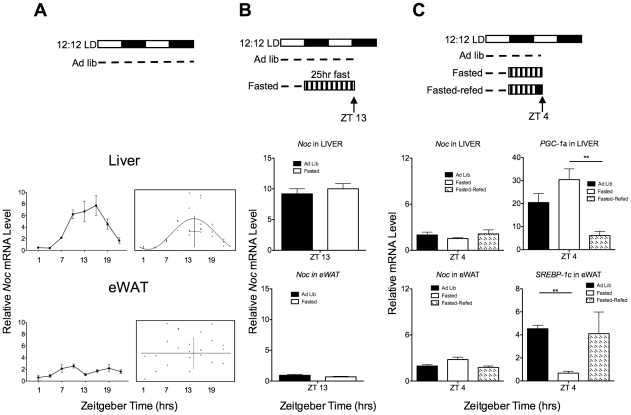
The fact that Nocturnin was acutely induced by stimuli such as TPA and insulin in vitro raised the possibility that Nocturnin may be acutely induced in response to feeding [14], [15]. Nocturnin expression peaks in the early night in the livers of ad libitum fed mice housed in 12∶12 light:dark (LD) conditions (Figure 1A; [12], [13]). However, quantitative real-time PCR analysis of the epididymal white adipose tissue (eWAT) from ad libitum fed mice housed in 12∶12 light:dark revealed that Nocturnin is expressed at low levels, and is statistically arrhythmic (Figure 1A, bottom panel). Nocturnin expression is arrhythmic despite rhythmic core clock gene expression in eWAT [20], [21]. The observation that Nocturnin's expression pattern can be driven by systemic cues in the liver with a peak expression in the early night [16] led us to hypothesize that this expression pattern could be the result of an acute induction of Nocturnin caused by the large bout of feeding which begins at the onset of darkness [22]. To test this, mice were habituated to a 12∶12 LD cycle and then fasted for 25 hours, beginning at ZT12 (ZT refers to “zeitgeber time” in hours, where ZT0 is light onset and ZT12 is light offset) and ending at ZT13 the next day (Figure 1B, top panel). Contrary to our hypothesis, we found that Nocturnin expression both in the liver and in the white adipose tissue was not dependent upon the presence of food (Figure 1B, bottom panel).
Figure 1. Nocturnin expression is not dependent upon or induced by feeding.
(A) Nocturnin expression is not dependent upon feeding. The top panel is a schematic depicting the experimental setup. Open bars represent lights on, and the closed bars represent lights off. The dotted line depicts food availability. Graphs (left) represent the mean relative mRNA level (±SEM) for each time point in liver (top) and eWAT (bottom), n = 3–6. CircWave analysis (right panels of A) shows the corresponding statistical analyses for rhythmicity. In the CircWave graphs, each dot represents the normalized relative mRNA level from the eWAT of an individual mouse. The curve represents the best-fit fourier curve, with the center of gravity (phase) represented by the vertical bar intersecting with the horizontal bar which represents the mean of the entire data set (liver; p≤0.002). Data set for which a fourier curve could not be fitted (eWAT) is indicated by ‡. (B) Nocturnin expression is not induced by fasting. Schematic of fasting experiment (top panel) as described in (A). The bar with vertical stripes represents the fasting period. Graphs represent the mean (±SEM) for each group, n = 4. (C) Nocturnin expression is not induced by feeding after a fast. Schematic representation of experiment is as described in (A) and (B). The solid black bar in the “refed” condition represents the time period in which food was made available after the fast. Gene expression analysis of the liver (middle panel) and eWAT (bottom panel) collected at ZT4 shows that Nocturnin is not induced by feeding after a fast, though the expected changes in expression patterns of Srebp-1c and Pgc-1α were observed (see text). Data represent the mean (±SEM), n = 3 for each group.
Though Nocturnin peak expression in the early night was not affected by the absence of food, it remained to be determined if Nocturnin could be acutely induced by feeding if the feeding occurred at the nadir of its regular expression pattern. To test this, a group of mice were fasted overnight, beginning at ZT12, and were re-fed at ZT2 (Figure 1C, top panel). Gene expression analysis of the liver and of the white adipose tissue, taken two hours after feeding began, revealed that Nocturnin expression levels remained unchanged from the ad libitum and fasted controls, and were not induced by feeding (Figure 1C). As previously reported, Srebp-1c expression was down-regulated in eWAT and Pgc-1α expression was up-regulated in liver in response to the fast (Figure 1C; [23]). Therefore, these experiments demonstrate that Nocturnin expression is not dependent upon, nor induced, by feeding.
Restricted feeding induces the rhythmicity of Nocturnin in white adipose tissue
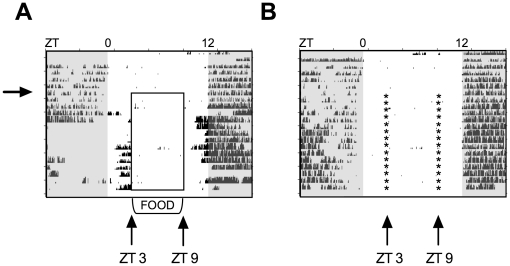
To investigate the effect that restricted feeding would have on the expression patterns of Nocturnin, mice were placed on a feeding paradigm in which food availability was temporally restricted to six hours during the middle of the day, from ZT3 to ZT9 (Figure 2A). Mice in the control group were mock restricted fed by disturbing the cages at ZT3 and ZT9, though the mice had ad libitum access to food during the entire study (Figure 2B). Restricted fed mice displayed food anticipatory wheel running activity within three or four days after the onset of food restriction, whereas the ad libitum fed group did not (Figure 2A,B). This indicates that food entrainment has occurred in the experimental group. Gene expression analysis of ad libitum fed mice revealed that Nocturnin expression, along with other core clock genes, was rhythmic in the liver, as previously reported (Figure 1A and 3A, left panel; [12], [13]).
Figure 2. Wheel-running actograms of restricted fed or ad libitum fed C57BL/6 mice.
(A) Restricted fed mice. Each horizontal line of the actogram represents a twenty-four hour period, and each tick mark on the graph represents one running-wheel revolution. Lights off are indicated by the shaded areas of the actograms. Mice were habituated to a 12∶12 LD cycle for 7 days with ad libitum access to food. At ZT9 on Day 7 (marked by the arrow), food was removed from the cages. At ZT3, a small bowl containing food was placed in the bottom of the cage. At ZT9 the food was removed. This was repeated daily until the conclusion of the experiment on Day 21. Food availability is marked by the open box. This actogram is a representative example from an individual mouse. (B) Ad libitum fed controls. Actogram is as described in (A). Mice were habituated to a 12∶12 LD cycle for 7 days. On day 8, small bowls of food were placed into the cages, and food was moved in and out of the bowl at ZT3 and ZT9, respectively. Controls were maintained on ad libitum feeding throughout the study, but on day 8 small bowls of food were placed into the cages, and food was moved in and out of the cage in the same manner as the restricted fed mice. Times food bowls were moved in and out are represented by *.
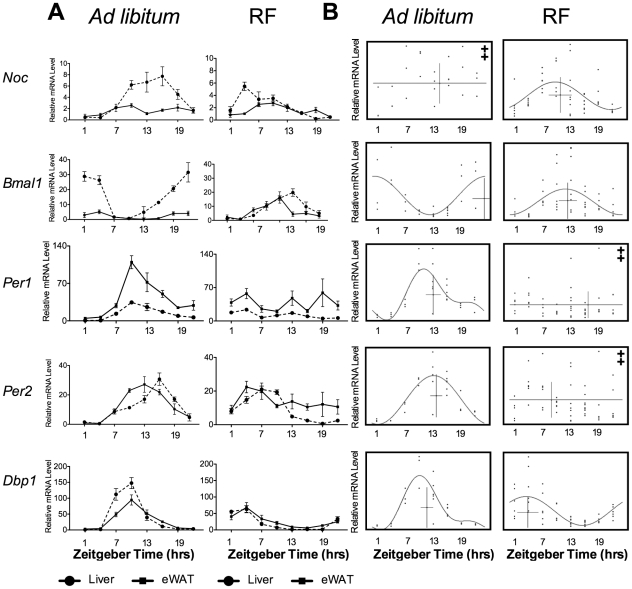
Figure 3. Restricted feeding results in the rhythmic expression of Nocturnin in the epididymal white adipose tissue.
(A) Gene expression analysis conducted on the liver (dashed line) and on the eWAT tissue (solid line) in ad libitum fed mice (left panel) and restricted fed mice (RF, right panel). Data represent the mean (± SEM) of a group for each time point (n = 3–6 per timepoint for the ad libitum condition, and n = 5–9 for the restricted fed condition). Ad libitum expression profiles of Nocturnin shown in 3A are the same data as shown in Figure 1A, and is regraphed here for convenient comparison. (B) CircWave analysis of eWAT gene expression data from (A) from ad libitum (left panel) and restricted fed mice (RF, right panel). The graphs are as described in Figure 1A. For all gene expression values, except those graphs marked with ‡, p-values <0.002. ‡ marks the data for which no curve could be fit. Note that the Nocturnin ad libitum data from liver and eWAT in this figure are the same data as shown in Figure 1A and are shown again here for direct comparison.
We found that the effect of restricted feeding was tissue- and gene-dependent. When mice were placed on the restricted feeding paradigm, Nocturnin's expression pattern in the liver shifted from a peak in the early night to a peak at ZT4 (Figure 3A, right panel). Though Nocturnin's expression pattern was arrhythmic in the white adipose tissue of ad libitum fed mice, restricted feeding resulted in the rhythmic expression of Nocturnin, which displayed a peak at ZT10, in this tissue (Figure 3B, right panel). Also in the white adipose tissue, the expression patterns of Bmal1 and Dbp1 shifted to resemble the profiles seen in the liver (Figure 3A, right panel), while the Per1 and Per2 expression patterns were arrhythmic in restricted-fed mice, unlike the expression profile of these genes in the liver (Figure 3A, right panel and Figure 3B, right panel).
Nocturnin expression is induced by the absence of the meal in the white adipose tissue of restricted-fed mice
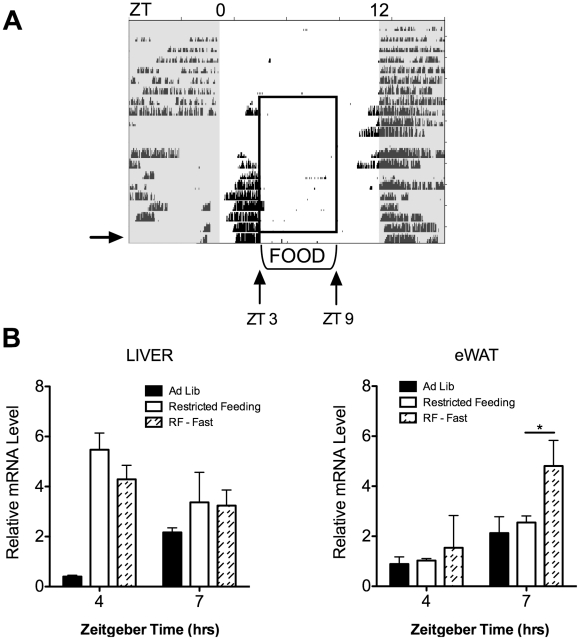
In restricted-fed mice, the peak expression of Nocturnin in liver and in eWAT occurs at ZT4 and ZT10, respectively. To test whether peak expression was dependent upon the presence of the expected meal in this paradigm, we placed animals on a restricted feeding schedule for 12 days and on the thirteenth day no meal was presented at ZT3 (Figure 4A). Tissues were harvested both at ZT4 and at ZT7. Gene expression analysis revealed that Nocturnin was unaffected by the absence of the meal in the liver (Figure 4B, left panel). However, in the white adipose tissue of restricted fed mice, Nocturnin expression was induced approximately two-fold at ZT7 in response to the fast (Figure 4B, right panel).
Figure 4. Nocturnin is induced by fasting in eWAT tissue of restricted fed mice.
(A) Schematic representation of feeding paradigm and absence of meal on the thirteenth day of restricted feeding. Actogram is as described in Figure 2, and is a representative wheel running actogram of restricted fed mice. Mice were habituated to a 12∶12 LD cycle for seven days, restricted feeding began on Day 8. On the 13th day of restricted feeding (indicated by the arrow), mice were not given the expected meal at ZT3, the condition labeled as RF-Fast. (B) Gene expression analysis of the liver (left panel) and the epididymal white adipose tissue (eWAT, right panel) reveal that Nocturnin is induced by the absence of the meal in eWAT only. The data from the ad libitum control group and from the restricted fed group (RF) are the same as from Figure 2 and has been re-graphed here for comparison. Data represent the mean of the group (±SEM); for each time point, n = 3 for the ad libitum group, n = 6 for the restricted-fed group, and n = 4 for the RF-fasted group. * = t-test, p-value = 0.019.
Nocturnin expression is induced by a rise in cAMP levels in adipocytes
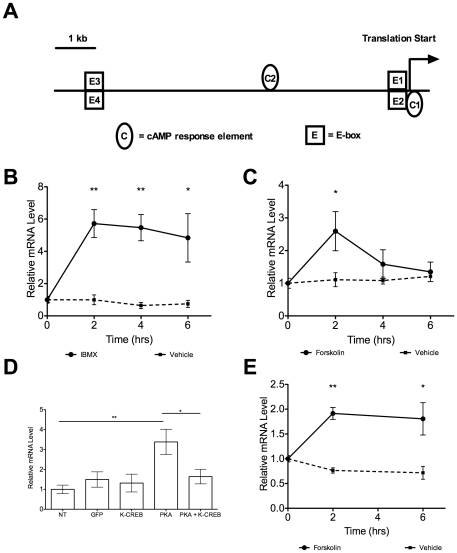
During fasting, white adipose tissue undergoes lipolysis mediated by catecholamine stimulated beta-adrenergic receptors which causes a rise in cAMP levels, leading to the activation of protein kinase A (PKA) which results in the phosphorylation of hormone sensitive lipase (HSL), perilipin A, and cAMP response element binding protein (CREB), and ultimately the breakdown of triglyceride-storing lipid droplets [24], [25], [26], [27]. It has been shown in Xenopus laevis that Nocturnin rhythmicity is regulated by p-CREB, but the activation of Nocturnin by p-CREB has not been demonstrated in mice [28]. There are conserved putative CREs within the 500 bp region upstream of Nocturnin's first exon (Figure 5A), and Nocturnin is acutely induced by 3-isobutyl-1-methylxanthine (IBMX) and forskolin in NIH3T3 fibroblasts (Figure 5B,C). Overexpression of PKA in NIH-3T3 fibroblasts resulted in an increase of Nocturnin expression, whereas cells co-transfected with PKA and a dominant negative CREB construct failed to exhibit the increase in Nocturnin expression, demonstrating that mammalian Nocturnin mRNA expression is induced by CREB activation (Figure 5D). We next examined whether Nocturnin could be induced by a rise in cAMP levels in mature 3T3-L1 adipocytes. Treatment of these cells with forskolin also resulted in an acute induction of Nocturnin expression (Figure 5E) showing that Nocturnin is regulated by the signaling cascade which occurs during lipolysis in white adipose tissue.
Figure 5. Nocturnin is induced by a rise in cAMP in NIH-3T3 fibroblasts and in 3T3-L1 adipocytes.
(A) Schematic representation of highly conserved putative transcription factor binding sites in the 10 kb region upstream of Nocturnin's translation start site. C = cAMP response element binding protein (CRE) sites. E = E-box elements. Locations of the CRE sites are C1 −29 bp and C2+3,016 bp. (B) Nocturnin is induced approximately 4.5 fold by IBMX (solid line) in NIH3T3 fibroblasts. No induction of Nocturnin was observed in the vehicle treated cells (dashed line). Data represent mean (± SEM), normalized to Cyclophilin B. n = 4 for each time point. P-value determined by t-test assuming equal variances, two-way p-value reported. ** p-value <0.002; * p-value <0.03. (C) Nocturnin is induced approximately 2.5 fold by forskolin (solid line) in NIH3T3 fibroblasts. The dashed line represents data from vehicle treated cells. Data represent mean (± SEM), normalized to Cyclophilin B. n = 3 for each time point. P-value determined by t-test assuming equal variances, two-way p-value reported. * p-value <0.03. (D) Transient transfection assay of NIH3T3 fibroblasts showing Nocturnin is induced by the overexpression of PKA in a CREB-dependent manner. Data represent the mean (± SEM), n = 6 for each group. T-test assuming equal variances performed, p-value reported. ** = p-value <0.01; * = p-value <0.04. NT = non-transfected; GFP = GFP transfected; K-CREB = pCMV-KCREB (dominant negative) alone transfected; PKA = pCMV-PKA transfected; PKA + K-CREB = pCMV-PKA and dominant negative pCMV-KCREB were cotransfected. (E) Nocturnin is induced approximately 2 fold by forskolin (solid line) in mature 3T3-L1 adipocytes. The dashed line represents data from vehicle treated cells. Data represent the mean (± SEM), n = 3. ** = p-value 0.0009, and * = p-value 0.036. Data were normalized to Cyclophilin B.
Discussion
Nocturnin −/− mice have no change in circadian locomotor activity rhythms or in the expression patterns of canonical clock genes in the liver suggesting that Nocturnin is an output of the circadian clock [12]. The observation that Nocturnin expression is not acutely induced by food nor by the absence of the expected meal in the livers of restricted fed mice suggests that the regulation of Nocturnin expression in the liver is likely the result of the local circadian clock, a food entrained oscillator, a systemic cue that is not dependent upon the daily presentation of food, or some combination thereof. Since Nocturnin shows robust rhythmicity in most tissues where it has been examined [13], our finding that Nocturnin is not strongly rhythmic in white adipose tissue under an ad libitum feeding paradigm was surprising. This finding suggests Nocturnin may be uncoupled from the regulation of the core clock machinery or influenced by other signals that are dominant to the clock in this situation.
A recent study demonstrated that the temporal pattern of feeding, as well as the availability of food, greatly affected the phase of the rhythmic expression of the transcriptome in the liver, and even is capable of partially restoring rhythmic gene expression patterns in the otherwise arrhythmic Cry1 −/−;Cry2 −/− mice [22]. Interestingly, Nocturnin becomes rhythmically expressed in the white adipose tissue of restricted fed mice, indicating that its rhythmic expression pattern is driven by the temporal availability of food. Nocturnin's peak expression time in the white adipose tissue of restricted fed mice is six hours later than the peak expression time in the liver, implying that different mechanisms modulate this effect. It has been shown that peripheral tissues respond to food entrainment differently, as the phase of peak Per1 expression between the lung and the liver differ, and the liver entrains to the feeding schedule faster than the lung. It was suggested that this difference could be due to the liver's important role in metabolism [2]. Our data contrasting the Nocturnin expression patterns in the liver and in the white adipose tissue of restricted fed mice suggest that even tissues directly involved in metabolic homeostasis have differing responses to food entrainment. Our data not only demonstrate a tissue-specific expression pattern of Nocturnin, but also demonstrate that the expression patterns of some core clock genes are tissue-specific.
In a restricted feeding paradigm, we have shown that Per1 and Per2 expression patterns become highly variable in eWAT and are expressed at mid-range levels when compared to the expression patterns in the white adipose tissue of ad libitum fed animals, unlike the liver. In contrast to our results, one previous study has reported rhythmic expression patterns of Per1 and Per2 in eWAT of restricted-fed animals, but perhaps the difference results from the manner in which the studies were conducted [20]. The restricted feeding regimen used in Zvonic et al. was less severe (12 hour food availability as compared to our 6 hour feeding window), fewer time points were used within a 24 hour period than were used in our study, and Zvonic et al. placed animals on restricted feeding for seven days instead of twelve to fourteen [20]. We have also shown that Bmal1 peak expression occurs three hours earlier in eWAT than in the liver on restricted feeding. Bmal1 expression levels are approximately three-fold higher in the eWAT of restricted-fed animals than in the eWAT of ad libitum fed animals, whereas restricted feeding results in a decrease of peak expression of Bmal1 in the liver. The tissue specific expression patterns of core clock genes in restricted-fed animals raises the possibility that the regulation of their expression may be achieved through unknown mechanisms or may be the result of different entraining signals.
The observation that Nocturnin is induced by fasting in restricted fed mice is the first example of Nocturnin's acute induction in the mouse. In Drosophila melanogaster it has been shown that curled, the homolog of Nocturnin, is induced by starvation further supporting a role for Nocturnin in fasting [29]. Our studies revealed that Nocturnin's induction is tissue specific and feeding regimen specific, as it only occurs in the white adipose tissue, and not in the liver, under a restricted feeding paradigm. Several possibilities could explain the feeding regimen specific induction of Nocturnin in eWAT. Gene expression in the rat salivary gland entrains to a feeding schedule after a sympathectomy has been performed, indicating that sympathetic input to the tissue modulates food entrainment and perhaps inhibits food entrainment in this tissue [30]. Nocturnin's inducibility in white adipose tissue may be suppressed when the phase of the tissue is entrained to the light-dark cycle, and this suppression could be relieved when the tissue phase-shifts in response to a feeding schedule.
Another potential explanation for the restricted-feeding specific induction of Nocturnin in response to fasting may be found in the observation that there are different methods of energy utilization during the different stages of food deprivation. The early fasting stage is categorically centered on glycogenolysis, and after glycogen stores in the liver are depleted, fuel sources are produced by gluconeogenesis and by lipolysis of white adipose tissue [31]. The difference in metabolic states between mice receiving one 16 hr fast and that of mice receiving daily 18 hr fasts may account for the induction of Nocturnin seen in restricted fed mice.
Perhaps another reason for the feeding paradigm-specific induction of Nocturnin is the fact that lipolysis in response to an acute stress, such as that presented to the restricted fed mice, is robust, transitory, and predominantly mediated by catecholamines [32]. Our data show that Nocturnin is induced by a rise in cAMP levels, a central step in the well-studied hormone-induced lipolysis cascade. We show that Nocturnin expression increases in response to forskolin, to IBMX, and to the overexpression of PKA in NIH3T3 fibroblasts (Figure 5). A previous study reported that Nocturnin is not induced by forskolin in NIH3T3 fibroblasts [14]. This seeming contradiction is likely the result of the differing treatment of the cells with forskolin. The previous study treated NIH3T3 fibroblasts with forskolin for 15 minutes, whereas our present study harvested NIH3T3 fibroblasts treated with forskolin for 2, 4, or 6 hours (Figure 5C). Our data also show that Nocturnin is induced by forskolin in mature 3T3-L1 adipocytes, further supporting the finding that Nocturnin expression increases in response to a rise in cAMP. The finding that Nocturnin is induced by a fast in white adipose tissue of restricted fed mice is contrary to the original hypothesis that Nocturnin would be induced by feeding. The fact that Nocturnin is not induced by feeding in liver or in white adipose tissue, while it is induced by insulin in 3T3-L1 preadipocytes [15], further highlights the tissue specificity and developmental specificity of Nocturnin's inducibility, positioning it to regulate a diverse set of transcripts throughout the organism in response to a variety of stimuli.
Mammalian Nocturnin has been shown to exhibit deadenylase activity in vitro, suggesting that its function is to control mRNA stability or translatability. However, direct mRNA targets of Nocturnin's post-transcriptional regulation have yet to be identified. How this function contributes to the fasting response in eWAT is unclear but many interesting possibilities exist. Perhaps Nocturnin contributes to the post-transcriptional control of mRNAs involved in lipolysis or adipocyte signaling [33], [34], [35]. The recent study showing Nocturnin promotes adipogenesis by increasing the nuclear translocation of PPARγ suggests that Nocturnin may have other functions in addition to deadenylase activity [15]. The observation that Nocturnin plays a role in adipogenesis, exhibits a food entrainable circadian expression profile in the liver (Figure 3A), and contains a leucine zipper motif in the Xenopus laevis model is reminiscent of another leucine zipper containing protein, glucocorticoid induced leucine zipper (GILZ) [15], [36], [37]. Perhaps the functions of GILZ may provide new insights into other roles of Nocturnin. Future studies investigating the role that Nocturnin plays in white adipose tissue, and how that differs from the role Nocturnin plays in the liver, will add to our knowledge regarding the crosstalk between circadian clocks and metabolism.
Future studies of Nocturnin should also investigate different white adipose tissue depots, as it has been shown that qualitative differences exist between different depots. For example, human visceral white adipose tissue exhibits a stronger lipolytic response to catecholamines than subcutaneous adipose tissue, and the anti-lipolytic affect of insulin is stronger in subcutaneous adipose tissue than visceral adipose tissue [38], [39]. Also, the expression of leptin is higher in epididymal white adipose tissue than in subcutaneous adipose tissue [40]. These differences among depots of white adipose tissue may also extend to the expression patterns of core clock genes and of Nocturnin, as well as the regulatory mechanisms behind the patterns.
It is interesting to entertain the possibility that white adipose tissue could play a prominent role in food entrainment. Given its ability to dynamically respond to food or the lack thereof, its ability to integrate a variety of systemic signals both neural and humoral, and its function as an endocrine organ, it is well positioned to aid the adaptation of the animal to the timing of food availability. Our studies suggest a role for Nocturnin in the white adipose tissue's response to fasting in restricted fed mice, and highlight the usefulness of Nocturnin and white adipose tissue as tools with which to investigate the intersection of circadian timing and metabolism.
Materials and Methods
Animals
All experiments were conducted under the guidelines set forth by the Institutional Animal Care and Use Committee at the University of Virginia. For all experiments, C57BL/6 male mice aged 4–6 months were used. Mice were housed in 12∶12 LD with ad libitum feeding to establish whether Nocturnin is rhythmically expressed in white adipose tissue, and to compare that expression profile to the liver as a control. All studies thereafter were conducted in 12∶12 LD to keep the photic stimuli consistent with the experiment conducted in Figure 1A. For all timepoints, mice were euthanized via CO2 followed by cervical dislocation. For time points conducted during the dark hours, mice were euthanized in the dark, and tissues were collected in the light. Specifically, for studies testing the dependence of Nocturnin's expression on food, mice were housed in 12∶12 LD, with running wheels, and were fed standard chow ad libitum for 7 days. Then food was removed at ZT12 (onset of darkness), and mice were euthanized 25 hours later, at ZT13. Tissues were harvested, frozen in liquid nitrogen, and stored at -80°C until further use. For studies testing the induction of Nocturnin expression in response to feeding after a fast, mice were habituated for 7 days to 12∶12 LD, with running wheels, and with ad libitum access to standard chow. On the eighth day, food was removed at ZT12. On day nine, the cohort was split and one group remained without food while the other group received a meal at ZT2. All mice were euthanized at ZT4, tissues were harvested and frozen in liquid nitrogen, and stored at −80°C until further use. For the restricted feeding experiments, mice were habituated to 12∶12 LD, with running wheels, and with ad libitum access to standard chow for 7 days. At ZT9 on the seventh day, food was removed. Each day, from this day on, food was placed into food bowls in the bottom of the cages at ZT3 and was removed at ZT9. Ad libitum fed control mice had free access to food, but also received the food bowls at the same time and in the same manner as the restricted fed mice. Mice were on restricted feeding for 12-14 days before harvesting tissues every three hours. Tissues were frozen in liquid nitrogen and were stored at -80°C until further use. To test the affect the absence of the expected meal would have on Nocturnin expression, mice were habituated for 7 days in cages with running wheels placed in 12∶12 LD with ad libitum access to standard chow. They were placed on restricted feeding, as described above, for 12 days. On the 13th day, no food was given at ZT3. Tissues were harvested at ZT4 and at ZT7, frozen on liquid nitrogen, and stored at -80°C until further use.
RNA isolation and cDNA Synthesis
Tissues were homogenized in Trizol (Invitrogen, Carlsbad, CA) using Lysing Matrix D (QBiogene, Carlsbad, CA) and the FastPrep homogenizer (QBiogene, Carlsbad, CA). RNA isolation was conducted according to the Trizol manufacturer's instructions. Total RNA was DNAse-treated (Sigma Aldrich, St. Louis, MO), extracted by acidic phenol chloroform extraction (Ambion, Austin, TX) and precipitated with ammonium acetate (Ambion, Austin, TX) and ethanol. 500 ng of total RNA was used to synthesize cDNA using the iScript cDNA Synthesis kit (BioRad, Hercules, CA), according to manufacturer's instructions.
Real-time PCR
Real-time PCR was conducted with the Single Color MyiQ Detection System and iQ SYBR Green Supermix (BioRad, Hercules, CA). See Table S1 for a list of the primer sequences used. Data acquired from the restricted fed mice, ad libitum fed mice, and the fasted restricted fed mice were normalized to acidic ribosomal phosphoprotein PO (36B4). The data are graphed and reported as the mean fold change above 36B4 levels (±SEM). CircWave v1.4 software (courtesy of Dr. Roelof Hut, http://www.euclock.org/) was used to analyze the rhythmicity of gene expression patterns. p-values reported are the result of an f-test, user defined alpha = 0.5. Graphs marked with ‡ represent data sets to which no curve could be fit, and all other graphs have a p-value ≤0.05. Forskolin induction data were normalized to Cyclophilin B. Points represent mean and error bars represent the standard error of the mean (±SEM). T-tests assuming equal variances were performed (alpha = 0.05) and the two-way p-values were reported.
Cell culture
NIH3T3 fibroblasts were cultured in high glucose-L-glutamine DMEM (Invitrogen, Carlsbad, CA) with 10% Fetal Bovine Serum-Premium Select (Atlanta Biologicals, Lawrenceville, GA) at 37°C with 5% CO2. For induction experiments in NIH3T3 fibroblasts, cells were seeded and grown to full confluence before adding IBMX (to a final concentration of 0.5 mM) or forskolin (to a final concentration of 10 µM). The vehicle used to dissolve both forskolin and IBMX was ethanol. Cells were harvested 0, 2, 4, or 6 hours post-treatment by washing the cells once with cold phosphate buffered saline, scraping and lysing with Trizol (Invitrogen, Carlsbad, CA), and storing the lysates at −80°C until further use. Transient transfections were conducted using FuGene6 (Roche Applied Sciences, Indianapolis, IN) and plasmids pCMV-PKA or dominant negative pCMV-KCREB (Clonetech, Mountain View, CA). Cells were collected approximately 24 hours after transfection in the same manner as described before. 3T3-L1 pre-adipocytes were cultured in high glucose-L-glutamine DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% Fetal Bovine Serum-Premium Select (Atlanta Biologicals, Lawrenceville, GA) at 37°C with 5% CO2. Pre-adipocytes were differentiated by growing to confluence and adding Differentiation Media: DMEM-hg-L-glutamine, 10% FBS, 10 µg/mL insulin (Sigma Aldrich, St. Louis, MO), 1 µM Dexamethasone (Sigma Aldrich, St.Louis, MO), 0.5 mM 3-Isobutyl-1-methylxanthine (Sigma Aldrich, St.Louis, MO), and 1 µM Rosiglitazone (Cayman Chemicals, Ann Arbor, MI), set as Day 0. Forty-eight hours later, media was replaced with DMEM-hg-L-glutamine, 10%FBS, 10 µg/mL insulin, 1 µM Rosiglitazone. Every 48 hours media was changed with standard culture media (DMEM-hg-L-glutamine 10%FBS) and 10 µg/mL insulin. Differentiation was confirmed by visual observation of cell morphology and forskolin induction experiments were conducted on Day 10 post-differentiation. For induction studies, 3T3-L1 adipocytes were first serum starved by adding serum free DMEM-hg-L-glutamine for two hours. Then 10 µM forskolin (Sigma Aldrich, St. Louis, MO) or vehicle (ethanol) was added to the cells and cells were collected 0, 2, and 6 hours post-treatment. Samples were collected as above and frozen at −80°C until further use.
Promoter analysis
Mouse (AC118601) and human (AC093602) Nocturnin sequences were aligned, and the 10 kb region upstream of the translation start site was identified. These sequences were analyzed with MatInspector. Putative sites reported here have 100% conservation of the core sequence and 75% conservation of the matrix sequence.
Supporting Information
Primer sequences for quantitative RT-PCR.
(DOC)
Acknowledgments
We thank the members of the Green laboratory (especially Hyun Jeung Sung and Silvia LaRue for excellent technical assistance).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the National Institutes of Health R01 GM076626 and R01 MH090247 to CBG (http://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stephan FK. The “other” circadian system: food as a Zeitgeber. J Biol Rhythms. 2002;17:284–292. doi: 10.1177/074873040201700402. [DOI] [PubMed] [Google Scholar]
- 2.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 3.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 11.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, et al. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- 12.Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, et al. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001;1:9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, et al. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci U S A. 2010;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zechner R, Strauss JG, Haemmerle G, Lass A, Zimmermann R. Lipolysis: pathway under construction. Curr Opin Lipidol. 2005;16:333–340. doi: 10.1097/01.mol.0000169354.20395.1c. [DOI] [PubMed] [Google Scholar]
- 18.Havel PJ. Update on adipocyte hormones: regulation of energy balance and carbohydrate/lipid metabolism. Diabetes. 2004;53(Suppl 1):S143–151. doi: 10.2337/diabetes.53.2007.s143. [DOI] [PubMed] [Google Scholar]
- 19.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Downes M, Yu RT, Bookout AL, He W, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 22.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, et al. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belfrage P, Fredrikson G, Nilsson NO, Stralfors P. Regulation of adipose tissue lipolysis: phosphorylation of hormones sensitive lipase in intact rat adipocytes. FEBS Lett. 1980;111:120–124. doi: 10.1016/0014-5793(80)80775-7. [DOI] [PubMed] [Google Scholar]
- 25.Galitzky J, Carpene C, Bousquet-Melou A, Berlan M, Lafontan M. Differential activation of beta 1-, beta 2- and beta 3-adrenoceptors by catecholamines in white and brown adipocytes. Fundam Clin Pharmacol. 1995;9:324–331. doi: 10.1111/j.1472-8206.1995.tb00506.x. [DOI] [PubMed] [Google Scholar]
- 26.Kawamura M, Jensen DF, Wancewicz EV, Joy LL, Khoo JC, et al. Hormone-sensitive lipase in differentiated 3T3-L1 cells and its activation by cyclic AMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1981;78:732–736. doi: 10.1073/pnas.78.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stralfors P, Belfrage P. Phosphorylation of hormone-sensitive lipase by cyclic AMP-dependent protein kinase. J Biol Chem. 1983;258:15146–15152. [PubMed] [Google Scholar]
- 28.Liu X, Green CB. Circadian regulation of nocturnin transcription by phosphorylated CREB in Xenopus retinal photoreceptor cells. Mol Cell Biol. 2002;22:7501–7511. doi: 10.1128/MCB.22.21.7501-7511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gronke S, Bickmeyer I, Wunderlich R, Jackle H, Kuhnlein RP. Curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin. Genetics. 2009;183:219–232. doi: 10.1534/genetics.109.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vujovic N, Davidson AJ, Menaker M. Sympathetic input modulates, but does not determine, phase of peripheral circadian oscillators. Am J Physiol Regul Integr Comp Physiol. 2008;295:R355–360. doi: 10.1152/ajpregu.00498.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T, Hung CC, Randall DJ. The comparative physiology of food deprivation: from feast to famine. Annu Rev Physiol. 2006;68:223–251. doi: 10.1146/annurev.physiol.68.040104.105739. [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Soni KG, Semache M, Casavant S, Fortier M, et al. Lipolysis and the integrated physiology of lipid energy metabolism. Mol Genet Metab. 2008;95:117–126. doi: 10.1016/j.ymgme.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Ahima RS, Prabakaran D, Mantzoros C, Qu D, Lowell B, et al. Role of leptin in the neuroendocrine response to fasting. Nature. 1996;382:250–252. doi: 10.1038/382250a0. [DOI] [PubMed] [Google Scholar]
- 34.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 35.Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem. 2004;279:47066–47075. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 36.Green CB, Besharse JB. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc Natl Acad Sci USA. 1996;93:14884–14888. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimble JM, Ptitsyn AA, Goh BC, Hebert T, Yu G, et al. Delta sleep-inducing peptide and glucocorticoid-induced leucine zipper: potential links between circadian mechanisms and obesity? Obes Rev. 2009;10(Suppl 2):46–51. doi: 10.1111/j.1467-789X.2009.00661.x. [DOI] [PubMed] [Google Scholar]
- 38.Mauriege P, Galitzky J, Berlan M, Lafontan M. Heterogeneous distribution of beta and alpha-2 adrenoceptor binding sites in human fat cells from various fat deposits: functional consequences. Eur J Clin Invest. 1987;17:156–165. doi: 10.1111/j.1365-2362.1987.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 39.Bolinder J, Engfeldt P, Ostman J, Arner P. Site differences in insulin receptor binding and insulin action in subcutaneous fat of obese females. J Clin Endocrinol Metab. 1983;57:455–461. doi: 10.1210/jcem-57-3-455. [DOI] [PubMed] [Google Scholar]
- 40.Trayhurn P, Thomas ME, Duncan JS, Rayner DV. Effects of fasting and refeeding on ob gene expression in white adipose tissue of lean and obese (oblob) mice. FEBS Lett. 1995;368:488–490. doi: 10.1016/0014-5793(95)00719-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences for quantitative RT-PCR.
(DOC)