Abstract
Coronary heart disease (CHD) is the leading cause of mortality in African Americans. To identify common genetic polymorphisms associated with CHD and its risk factors (LDL- and HDL-cholesterol (LDL-C and HDL-C), hypertension, smoking, and type-2 diabetes) in individuals of African ancestry, we performed a genome-wide association study (GWAS) in 8,090 African Americans from five population-based cohorts. We replicated 17 loci previously associated with CHD or its risk factors in Caucasians. For five of these regions (CHD: CDKN2A/CDKN2B; HDL-C: FADS1-3, PLTP, LPL, and ABCA1), we could leverage the distinct linkage disequilibrium (LD) patterns in African Americans to identify DNA polymorphisms more strongly associated with the phenotypes than the previously reported index SNPs found in Caucasian populations. We also developed a new approach for association testing in admixed populations that uses allelic and local ancestry variation. Using this method, we discovered several loci that would have been missed using the basic allelic and global ancestry information only. Our conclusions suggest that no major loci uniquely explain the high prevalence of CHD in African Americans. Our project has developed resources and methods that address both admixture- and SNP-association to maximize power for genetic discovery in even larger African-American consortia.
Author Summary
To date, most large-scale genome-wide association studies (GWAS) carried out to identify risk factors for complex human diseases and traits have focused on population of European ancestry. It is currently unknown whether the same loci associated with complex diseases and traits in Caucasians will replicate in population of African ancestry. Here, we conducted a large GWAS to identify common DNA polymorphisms associated with coronary heart disease (CHD) and its risk factors (type-2 diabetes, hypertension, smoking status, and LDL- and HDL-cholesterol) in 8,090 African Americans as part of the NHLBI Candidate gene Association Resource (CARe) Project. We replicated 17 associations previously reported in Caucasians, suggesting that the same loci carry common DNA sequence variants associated with CHD and its risk factors in Caucasians and African Americans. At five of these 17 loci, we used the different patterns of linkage disequilibrium between populations of European and African ancestry to identify DNA sequence variants more strongly associated with phenotypes than the index SNPs found in Caucasians, suggesting smaller genomic intervals to search for causal alleles. We also used the CARe data to develop new statistical methods to perform association studies in admixed populations. The CARe Project data represent an extraordinary resource to expand our understanding of the genetics of complex diseases and traits in non-European-derived populations.
Introduction
Coronary heart disease (CHD) is the leading cause of mortality in African-American men and women [1]. The risk factors for CHD in African Americans are similar to those reported in Caucasians, but their relative impact varies between the two ethnic groups. Multiple studies have reported that smoking, type-2 diabetes (T2D), hypertension, and LDL- and HDL-cholesterol (LDL-C and HDL-C) are significant independent risk factors for CHD in African Americans [2]–[5]. In general, hypertension and LDL-C have a larger and smaller impact on CHD risk, respectively, in African Americans compared with Caucasians [3]. There is also extensive evidence of the role of genetic factors in the familial aggregation of CHD and its predictors in African Americans [6]. However, the underlying genes remain largely unknown.
Recent advances in genome-wide association studies (GWAS) have made spectacular advances in identifying genes contributing to numerous common chronic diseases in Europeans and European Americans [7]. There are multiple loci convincingly associated with CHD risk in Caucasians, including many genes involved in lipid metabolism, as well as novel chromosomal regions that do not appear to contribute to risk through traditional risk factors [7]–[14]. However, there have been no large-scale GWAS for CHD and its risk factors in African Americans. GWAS in African Americans is important because new genes may be identified as a result of genetic variation private to populations of African-descent, differences in allele frequencies and in patterns of linkage disequilibrium (LD), differences in the relative impact of risk factors to disease, or differences in gene-environment interactions. Here we report a large (and for most phenotypes first) GWAS for CHD, type-2 diabetes (T2D), hypertension, LDL-C and HDL-C, and smoking in 8,090 African Americans as part of the National Heart, Lung, and Blood Institute (NHLBI)-sponsored Candidate gene Association Resource (CARe) Project [15].
Results
We genotyped 909,622 single nucleotide polymorphisms in 9,119 African Americans from the ARIC (N = 3,269), CARDIA (N = 1,209), CFS (N = 704), JHS (N = 2,200), and MESA (N = 1,737) population-based cohorts, on the Affymetrix Genome-Wide Human SNP Array 6.0 platform. Genotypes were called using Birdseed v1.33 [16], and stringent quality-control filters were applied (Tables S1 and S2). For samples that passed quality control (N = 8,100), principal component analysis (PCA) using EIGENSTRAT [17] revealed only ten population outliers across all cohorts; these samples were also excluded from the analysis (Text S1 and Figure S1). Overall, a total of 8,090 African Americans with very high genotype quality (average genotype success rate of 99.65%) were available for analysis. The demographics of these participants by cohort are shown in Table 1. To increase our coverage of common genetic variation and statistical power, and to facilitate comparisons across genotyping platforms, we imputed genotypes in the CARe African-American populations using MACH taking into account the admixed nature of the population (Text S1) [18], [19].
Table 1. Demographics of the CARe and replication African-American cohorts.
| Phenotypes1 | ARIC | CARDIA | CFS | JHS | MESA | MEC-T2D | Cleveland Clinic | PennCATH | NHANESIII | Jamaica SPT | Jamaica GXE | Health ABC |
| Gender | 1045/1785 | 366/583 | 213/308 | 842/1302 | 745/901 | 835/1349 | 345/374 | 280/222 | 718/1002 | 674/1055 | 232/736 | 468/651 |
| Age | 53.3±5.8 | 24.4±3.8 | 45.7±16.2 | 50.0±12.2 | 62.2±10.1 | 60.4±8.5 | 60.0±11.1 | 58.0±10.6 | 40.8±16.7 | 46.1±13.9 | 39.7±8.3 | 73.4±2.9 |
| Coronary heart disease | 110/2580 | NA | 25/475 | 125/1998 | NA | NA | 220/400 | 157/334 | NA | NA | NA | 244/895 |
| Type-2 diabetes | 529/2150 | NA | 98/403 | 339/1777 | 298/1348 | 1070/1114 | NA | NA | 168/899 | NA | NA | 335/757 |
| Hypertension | 1612/1132 | 36/913 | 209/260 | 1193/918 | 1019/625 | NA | NA | NA | 501/1219 | NA | NA | 871/263 |
| LDL-C | 138.7±43.3 (2588) | 111.3±31.0 (940) | 99.0±33.9 (295) | 125.2±36.6 (2111) | 116.5±33.4 (1631) | NA | NA | NA | 121.1±40.4 (805) | 123.5±39.1 (928) | 136.6±38.5 (928) | 123.9±36.7 (1128) |
| HDL-C | 54.7±17.5 (2613) | 54.4±13.0 (940) | 46.4±15.0 (483) | 50.0±14.1 (2138) | 52.5±15.3 (1639) | NA | NA | NA | 54.1±17.2 (805) | 48.4±12.6 (1413) | 51.5±11.5 (967) | 57.1±17.6 (1138) |
| Smoking | 14.2±9.4 (799) | 10.8±7.6 (359) | 10.8±6.7 (178) | 15.1±11.6 (659) | 14.6±18.6 (873) | NA | NA | NA | NA | NA | NA | NA |
For gender, we reporter the number of males/females. For age, we report the mean ± standard deviation in years. For coronary heart disease, type-2 diabetes, and hypertension, we report the number of cases/controls. For LDL-C and HDL-C, we report the mean ± standard deviation in mg/dl (number of samples with phenotypes available). For smoking, we report the mean ± standard deviation in daily cigarettes, excluding non-smokers (number of samples with phenotypes available). The number of CHD cases with concomitant type-2 diabetes in the CARe cohorts is: 44 for ARIC, 14 for CFS, and 43 for JHS. NA; not available or not analyzed in this study.
For all cohorts except CFS, single marker genetic association tests were performed by study using PLINK v1.06 [20] under an additive genetic model. We used linear regression for quantitative traits (HDL-C, LDL-C, and smoking) and logistic regression for dichotomous phenotypes (CHD, hypertension, and T2D). For CFS, family structure was modeled using linear mixed effects (LME) models and generalized estimating equations (GEE) for quantitative and dichotomous phenotypes, respectively [21]. For all analyses, the first ten principal components were used as covariates to account for global admixture and population stratification. A detailed description of the analysis methods and the phenotypic definitions used can be found in Text S1. Power calculations for the different phenotypes analyzed are summarized in Table S3; we have excellent power to find strong signals, but low to modest power for variants with weak phenotypic effects. The inflation factors (λs) observed were all near unity (Table S4), suggesting that most confounders, including population stratification, were well-controlled.
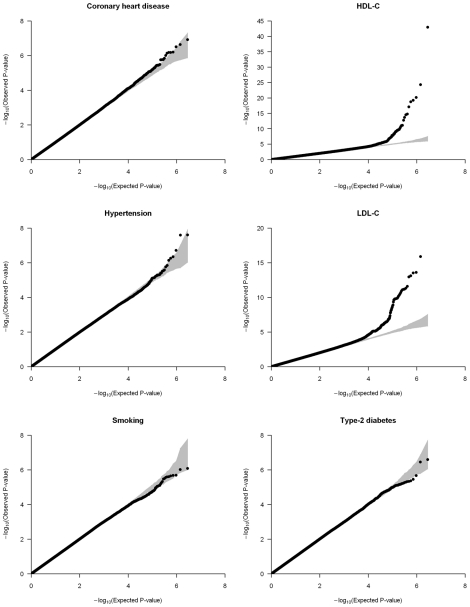
We applied genomic control to the individual cohorts' results and combined them using the inverse variance meta-analysis method [22]. Inflation factors of the meta-analysis results were modest and were again scaled using genomic control (Table S4). Quantile-quantile (QQ) plots of the six different meta-analyses after double genomic control corrections show that the test statistics follow the null expectations, except for the HDL-C and LDL-C meta-analyses, which show an upward departure from the null distributions at the lowest P-values (Figure 1). This departure is caused by known genetic variants with large effects on lipid levels (Figure S2).
Figure 1. Quantile-quantile (QQ) plots of the meta-analyses for coronary heart disease, HDL-C, hypertension, LDL-C, smoking, and type-2 diabetes analyzed in the CARe African-American samples (N = 8,090).
Each black circle represents an observed statistic for all genotypes and imputed SNPs (defined as the −log10(P-value)) against the corresponding expected statistic. The grey area corresponds to the 90% confidence intervals calculated empirically using permutations. The meta-analysis inflation factors are: coronary heart disease (λs = 0.991), HDL-C (λs = 1.030), hypertension (λs = 1.024), LDL-C (λs = 1.023), smoking (λs = 1.008), and type-2 diabetes (λs = 1.017). Data shown are genomic controlled before (for each study) and after the meta-analysis.
The main goal of this study was to identify new genetic risk factors for CHD and its predictors in African Americans. For five traits analyzed (we could not identify African-American replication cohorts for smoking), we identified SNPs with the strongest evidence of association in the CARe meta-analysis – SNPs were selected after accounting for LD to limit association signals redundancy – and sought replication using in silico data or direct genotyping in independent African-American cohorts (Table 1).
Combined results from a meta-analysis of the CARe and replication data are presented in Tables S5, S6, S7, S8, S9 and summarized in Table 2. We identified one novel locus that reached the generally accepted level for genome-wide significance (P≤5×10−8): SNP rs7801190 in the potassium/chloride transporter gene SLC12A9 and hypertension (OR = 1.31, combined P = 3.4×10−8). Despite reaching genome-wide significance, we are cautious in highlighting this association because it was identified using imputed genotypes (imputation quality r2_hat = 0.70) and the replication result, also obtained by imputation, was not statistically significant (P = 0.29). Indeed, when we directly assessed the quality of the imputation by directly genotyping rs7801190 in ARIC African-American samples (N = 2,572), we failed to validate the observed association with hypertension. This result suggests that the association between rs7801190 and hypertension status observed in the CARe African-American datasets is likely due to chance.
Table 2. Novel genetic associations (P≤1×10−6) between SNPs and coronary heart disease or its risk factors in African Americans.
| Trait | SNP | CHR (POS)1 | Reference allele (reference allele frequency)2 | CARe meta-analysis | Replication | Combined | Locus | |||
| OR [95% CI] or Beta (SE)3 | P-value4 | OR [95% CI] or Beta (SE)3 | P-value | OR [95% CI] or Beta (SE)3 | P-value | |||||
| HDL-C | rs7323893 | 13 (87502707) | T (0.91) | −0.138 (0.030) | 5.7×10−6 | −0.131 (0.047) | 0.0053 | −0.136 (0.025) | 1.3×10−7 | |
| rs937254 | 15 (55697456) | A (0.57) | 0.077 (0.017) | 5.4×10−6 | 0.078 (0.043) | 0.067 | 0.077 (0.016) | 1.0×10−6 | GCOM1 | |
| Hypertension | rs7801190 | 7 (100296029) | C (0.73) | 1.35 [1.22–1.50] | 2.5×10−8 | 1.13 [0.90–1.44] | 0.29 | 1.31 [1.19–1.44] | 3.4×10−8 | SLC12A9 |
| LDL-C | rs13161895 | 5 (179403807) | T (0.08) | 0.151 (0.035) | 2.3×10−5 | 0.139 (0.052) | 0.0077 | 0.147 (0.029) | 5.8×10−7 | RNF130 |
Coordinates are on NCBI build 36.1.
Average frequency for the reference allele across all available African-American CARe samples.
Direction of the effect given for the reference allele; OR, odds ratio; CI, confidence interval; SE, standard error.
P-values are scaled using genomic control.
To validate our phenotype modeling and analytical strategy, we sought to replicate in the CARe meta-analyses genetic associations previously reported in populations of European ancestry. We retrieved all index SNPs associated at genome-wide significance level with CHD, T2D, hypertension, HDL-C, LDL-C, and smoking in Caucasians as well as their proxy SNPs (defined as markers with an r2≥0.5 with the index SNPs in HapMap samples of European ancestry (CEU)) (Table S10) [23]. We then determined whether there was also evidence of association for the same signals in this large sample of African Americans. We detected modest to strong evidence of replication for one locus associated with CHD, one locus with T2D, nine with HDL-C, and six with LDL-C (Table 3 and Table S11). We did not replicate signals associated with smoking or hypertension. Furthermore, the top ten associated SNPs in a recent hypertension GWAS performed in African Americans [24] were not associated with hypertension in the CARe meta-analysis (different direction of effect and/or P>0.05). Since these hypertension association signals did not replicate in the original publication, non-replication here may result from their being falsely positive in the original report. Although replication of some of the above loci in African-derived populations had been reported previously [25], for most of them, the CARe results represent the first replication in populations of African ancestry.
Table 3. Replication of associations previously reported in Caucasians in the CARe African-American meta-analyses.
| Trait | Locus | Chr. | CARe SNPa | Positionb | Effect allelec | Average effect allele frequency in CARe (SE) | Odds ratio/Betad | 95% CI/SEd | P-valuee | Reference |
| Coronary heart disease | CDKN2A, CDKN2B | 9 | rs4977574 | 22088574 | G | 0.177 (0.004) | 1.18 (OR) | [0.93–1.49] | 0.17 | [10] |
| 9 | rs6475606 (p) | 22071850 | C | 0.109 (0.003) | 2.00 (OR) | [1.34–2.96] | 6.4×10−4 | |||
| Type-2 diabetes | TCF7L2 | 10 | rs7903146 | 114748339 | T | 0.291 (0.005) | 1.33 (OR) | [1.19–1.48] | 3.5×10−7 | [29] |
| HDL-C | GALNT2 | 1 | rs2144300 | 228361539 | T | 0.143 (0.012) | 0.092 (BETA) | 0.029 | 0.0015 | [13] |
| PPP1R3B | 8 | rs9987289 | 9220768 | A | 0.191 (0.005) | −0.090 (BETA) | 0.022 | 4.3×10−5 | [30] | |
| LPL | 8 | rs10503669 | 19891970 | A | 0.059 (0.006) | 0.137 (BETA) | 0.035 | 7.2×10−5 | [13] | |
| rs10096633 (p) | 19875201 | T | 0.430 (0.039) | 0.101 (BETA) | 0.017 | 1.5×10−9 | ||||
| ABCA1 | 9 | rs3905000 | 106696891 | A | 0.161 (0.011) | −0.043 (BETA) | 0.022 | 0.054 | [31] | |
| rs13284054 (p) | 106708894 | C | 0.850 (0.005) | 0.090 (BETA) | 0.027 | 0.0011 | ||||
| FADS1, FADS2, FADS3 | 11 | rs174547 | 61327359 | C | 0.092 (0.009) | −0.055 (BETA) | 0.030 | 0.068 | [26] | |
| 11 | rs1535 (p) | 61354548 | A | 0.820 (0.009) | −0.102 (BETA) | 0.025 | 6.7×10−5 | |||
| LIPC | 15 | rs1800588 | 56510967 | T | 0.497 (0.017) | 0.102 (BETA) | 0.018 | 1.5×10−8 | [32] | |
| rs8034802 (p) | 56512084 | A | 0.362 (0.010) | 0.104 (BETA) | 0.017 | 1.3×10−9 | ||||
| CETP | 16 | rs3764261 | 55550825 | A | 0.305 (0.010) | 0.203 (BETA) | 0.023 | 8.6×10−18 | [13] | |
| rs247617 (n) | 55548217 | A | 0.258 (0.006) | 0.260 (BETA) | 0.019 | 1.2×10−43 | ||||
| LCAT | 16 | rs255052 | 66582496 | A | 0.218 (0.009) | 0.132 (BETA) | 0.020 | 6.6×10−11 | [13] | |
| PLTP | 20 | rs7679 | 44009909 | T | 0.958 (0.005) | 0.052 (BETA) | 0.041 | 0.22 | [26] | |
| rs6065904 (p) | 43968058 | A | 0.202 (0.010) | −0.0904 (BETA) | 0.023 | 7.4×10−5 | ||||
| LDL-C | DOCK7 | 1 | rs10889353 | 62890784 | A | 0.618 (0.012) | 0.049 (BETA) | 0.017 | 0.0040 | [31] |
| 1 | rs10889335 (p) | 62732689 | A | 0.606 (0.018) | 0.068 (BETA) | 0.018 | 1.2×10−4 | |||
| CELSR2, PSRC1, SORT1 | 1 | rs12740374 | 109619113 | T | 0.235 (0.003) | −0.174 (BETA) | 0.021 | 1.3×10−16 | [26] | |
| PCSK9 | 1 | rs10493178 (n) | 55369655 | A | 0.878 (0.007) | 0.177 (BETA) | 0.025 | 4.7×10−12 | ||
| APOB | 2 | rs562338 | 21141826 | A | 0.598 (0.009) | −0.089 (BETA) | 0.017 | 3.1×10−7 | [32] | |
| rs503662 (p) | 21267647 | T | 0.652 (0.010) | −0.110 (BETA) | 0.018 | 2.5×10−9 | ||||
| LDLR | 19 | rs6511720 | 11063306 | T | 0.144 (0.002) | −0.208 (BETA) | 0.038 | 7.2×10−8 | [26] | |
| APOE, APOC1, APOC4, APOC2 | 19 | rs1160985 (n) | 50095252 | T | 0.635 (0.011) | −0.166 (BETA) | 0.017 | 7.2×10−21 |
To be included in this table, we require a two-tailed P≤0.05 after Bonferroni correction for the number of independent loci reported in the NHGRI database (Table S5). We report the association results for the published (index) SNP, unless it is not available. In that case, we report results for a proxy SNP (r2≥0.5 with original SNP in HapMap CEU; see Table S5 for additional details).
Proxy SNPs are marked with (p). SNPs that have a strong association signal but are not in LD with the published SNP are marked with (n) as potentially novel.
Position on NCBI build 36.1.
Effect alleles are given on the forward strand. For proxy SNPs, we phased HapMap CEU genotypes for the index and proxy SNPs to determine haplotypes, to be able to assess consistency of the direction of effect.
For dichotomous phenotypes, we report odds ratio (OR) and 95% confidence interval (CI); for quantitative traits, we report effect size (beta, in standard deviation units) and standard error (SE).
P-values are corrected using genomic control.
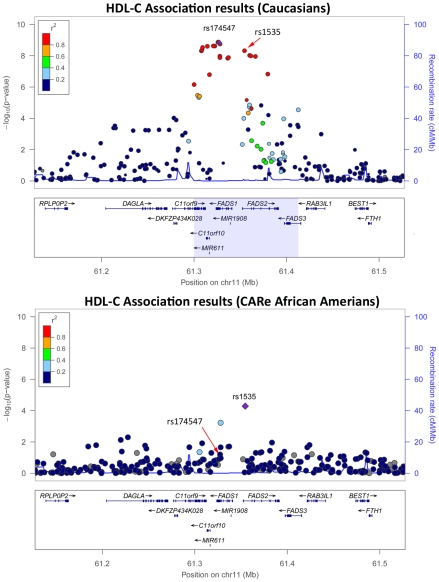
Taking advantage of the LD patterns in African Americans (LD breakdown over shorter distances compared with Caucasians), we assessed whether we could fine-map some of the associations previously reported in Caucasians. For this, we evaluated SNPs that were correlated with the index SNP in HapMap CEU (r2≥0.5), but largely uncorrelated with it in HapMap samples of African descent (YRI)(r2≤0.1). In most cases, the same signals were responsible for the associations in Caucasians and African Americans (Table 3 and Table S11). However, we found five examples where the predominant association signals were at SNPs strongly correlated with the index SNPs in HapMap CEU but weakly or not correlated with the index SNPs in HapMap YRI: the CDKN2A/CDKN2B locus for CHD and the FADS1-3, PLTP, LPL, and ABCA1 loci for HDL-C (Table S12). Using available genetic association results for myocardial infarction [10] and HDL-C [26] in Caucasians, we illustrate in Figure 2 and Figure S3 how our results in African Americans can help refine association signals. For instance, for the FADS locus, the index SNP in Caucasians (rs174547) is in strong LD with the top SNP in the CARe African-American meta-analysis (rs1535) in HapMap CEU (r2 = 1) but not in HapMap YRI (r2 = 0.09). The region of strong LD around rs174547 in HapMap CEU is 113 kb wide and includes the three FADS genes, whereas rs1535, located in an intron of FADS2, is in strong LD with no other markers in HapMap YRI (Figure 2). Comparison of association signals regionally in African Americans and European-derived individuals can thus be useful in two ways: (1) they may suggest smaller chromosomal regions for re-sequencing experiments to attempt to identify causal variant(s) that underlie shared signals between African- and European-derived chromosomes or (2) they may indicate that the index SNPs for African and European populations are linked to distinct causal variants. A third potentially interesting result from trans-ethnic comparison of association results is the identification of ethnic-specific association signals. For instance, at the ABCA1 locus, three SNPs in LD (rs4743763, rs4149310, and rs2515629) are associated with HDL-C in CARe African Americans (P<1×10−5), but not in Caucasians (Figure S3D).
Figure 2. HDL-C association results in Caucasians (top panel) [26] and in the CARe African Americans (bottom panel) at the FADS locus.
Plots were generated using LocusZoom [33]. Under the top panel, the light blue box corresponds to the chromosomal interval flanked by the leftmost and rightmost SNPs with a r2≥0.3 with the index SNP rs174547 in HapMap CEU. In the bottom panel, there is no light blue box because the top CARe SNPs at this locus, rs1535, is not in strong LD with any markers in HapMap YRI.
The optimal analytical strategy for GWAS in recently admixed populations has not been established. In African Americans, an ideal test statistic would incorporate both genotype information as traditionally used in GWAS, but also, at each locus, the probability that a given individual has zero, one, or two copies of a European (or African) chromosomal segment. This method would be particularly informative in a case where, for example, the causal allele is not in LD with any markers on the genotyping array, but is at higher frequency on one ancestral background. To explore the benefits of such a statistical framework, we designed and applied a novel method that combines evidence of association from genotypes and local ancestry estimates; the method is described in details in Text S1. Briefly, we use a panel of ancestry informative markers across the genome and a new implementation of the software ANCESTRYMAP [27] to estimate, for each of the CARe African Americans genotyped, the probabilistic proportion of European ancestry (0–100%) at the locus for each of the ∼900,000 SNPs genotyped on the Affy6.0 platform. For each SNP, we can then compute association between the phenotype and both the SNP genotype and the SNP estimate of local ancestry to generate a combined score that summarizes allelic variation and admixture. This method was used to produce the association data presented in Table 4.
Table 4. Top novel associations (P≤1×10−6) identified using SNP genotype and estimate of local African versus European ancestry.
| TRAIT | SNP | CHR (POS)1 | Reference allele | Reference allele frequency | SNP-only | SNP+Estimate of local ancestry | Closest genes7 | |||||
| CARe2 | CEU | YRI | Beta (SE)3 | P-value4 | Zgeno5 | Zloc.anc6 | P-value4 | |||||
| Coronary heart disease | rs6674681 | 1 (79493711) | T | 0.75 | 0.23 | 0.88 | 0.0892 (0.1151) | 0.44 | 3.524 | −4.14 | 3.8×10−7 | |
| rs6753112 | 2 (231895399) | T | 0.87 | 0.31 | 0.91 | −0.2459 (0.1357) | 0.07 | −3.977 | 3.625 | 5.2×10−7 | ARMC9 | |
| HDL-C | rs8078633 | 17 (559286) | C | 0.31 | 1.00 | 0.18 | −0.0006 (0.0186) | 0.98 | −3.893 | 4.723 | 3.6×10−7 | APPBP2 |
| Hypertension | rs10218356 | 23 (19168233) | A | 0.20 | 0.94 | 0.04 | −0.0915 (0.0523) | 0.08 | −4.056 | 3.891 | 6.5×10−7 | |
| LDL-C | rs17441606 | 2 (19431916) | A | 0.17 | 0.32 | 0.11 | −0.0759 (0.0219) | 6.2×10−4 | −4.234 | 4.021 | 4.0×10−8 | OSR1 |
| rs9306885 | 2 (19852313) | T | 0.26 | 0.72 | 0.16 | −0.0329 (0.0197) | 0.10 | −4.144 | 4.321 | 1.7×10−8 | ||
| rs6728440 | 2 (19862827) | A | 0.96 | 0.87 | 1.00 | 0.0978 (0.0446) | 0.03 | 3.527 | −4.029 | 5.9×10−8 | TTC32 | |
| rs7560236 | 2 (22930288) | T | 0.06 | 0 | 0.08 | 0.1493 (0.0366) | 5.5×10−5 | 4.568 | 3.804 | 2.1×10−8 | ||
| rs6748157 | 2 (28586865) | A | 0.86 | 0.51 | 0.98 | 0.03 (0.0247) | 0.23 | 3.592 | −4.062 | 4.1×10−8 | PLB1 | |
| Smoking | rs7075036 | 10 (16904816) | T | 0.69 | 0.32 | 0.80 | −0.1326 (0.0296) | 8.4×10−6 | −5.036 | 2.054 | 8.0×10−7 | RSU1, CUBN |
| rs11088655 | 21 (18128360) | T | 0.41 | 0.22 | 0.52 | 0.0938 (0.0275) | 6.9×10−4 | 4.347 | 3.637 | 2.4×10−7 | C21orf91 | |
| rs16982414 | 21 (28711411) | T | 0.90 | 0.99 | 0.84 | −0.1646 (0.0432) | 1.5×10−4 | −4.375 | −3.327 | 6.0×10−7 | ||
Global ancestry is included in the model for both methods.
Coordinates are on NCBI build 36.1.
Average frequency for the reference allele across all available African American CARe samples.
Direction of the effect given for the reference allele; SE, standard error.
P-values are scaled using genomic control.
Z-score for the SNP genotype information. A Z-score >0 means that the trait (or the risk to develop the disease) increases with the number of copies of reference alleles.
Z-score for the local ancestry estimate information. A Z-score >0 means that the trait (or the risk to develop the disease) increases with the number of copies of European chromosomes.
Genes in a 200 kb window.
Our method to assess combined SNP- and ancestry-association was tested explicitly on CHD and its risk factors in the CARe African-American samples (Figures S4, S5). For each SNP, we compared the test statistic obtained using the SNP-alone or the SNP+admixture information (in both methods, global ancestry is included as a covariate), focusing on markers that would not have been prioritized for follow-up replication when considering only SNP genotype association results (Figure S6). Across the six phenotypes, we identified 12 SNPs outside the previously known loci with a P≤1×10−6 in this SNP+admixture test statistic (Table 4). Most of these SNPs have a large allele frequency difference between the HapMap CEU and YRI individuals, suggesting that local ancestry might confound simple SNP association testing. For instance, the frequency of the C-allele at rs8078633 near the APPBP2 gene is 100% and 18% in CEU and YRI, respectively. The association between this SNP and HDL-C levels is weak when considering only allelic variation (P = 0.98) but becomes highly significant when evidence from the genotype and the estimate of local ancestry is combined (P = 3.6×10−7) (Table 4). This composite approach also identified a SNP near the phospholipase B1 gene (PLB1) that is strongly associated with LDL-C levels (P = 4.1×10−8), but that would not have been noticed using traditional genotype-only association testing (P = 0.23) (Table 4). As more large-scale GWAS in individuals of African ancestry are completed, it will be important to replicate these results.
Discussion
Most large-scale genetic efforts to identify risk factors for CHD have focused so far on populations of European ancestry. Given the prevalence of the disease in African Americans, and the development of better genotyping platforms that more completely survey common genetic variation in African-derived genomes [16], it is now both pertinent and timely to investigate the genetics of CHD in populations of African ancestry. The CARe Project was launched four years ago with the specific goal to create a resource for association studies of various heart-, lung-, and blood-related phenotypes across different ethnic groups [15]. In this article, we present results from the largest GWAS to date for CHD and its risk factors in African Americans. Despite being the largest, the size of our GWAS is modest compared to that of some European-derived consortia. As a consequence, we had limited discovery power and did not identify novel loci specifically associated with CHD or its risk factors that reach genome-wide significance in our African-American dataset.
We also attempted to replicate in the CARe African-American participants genetic associations to CHD and its risk factors previously identified in Caucasians. We could replicate 17 of those associations; for many of them, this was the first replication in a non-European-derived population (Table 3). For five of these 17 associations, we showed how cross-ethnic comparisons of genetic association results may help refine genomic intervals carrying causal alleles (Figure 2 and Figure S3). There were, however, a large number of loci originally found in Caucasians that were not replicated in the CARe meta-analyses presented in this manuscript (Table S11). Because our sample size was relatively modest, that we used stringent statistical thresholds to declare replication in order to control our false positive rate, and that effect sizes could be weaker for given loci across different ethnic groups, our limited power probably explains why many loci did not replicate in the CARe African Americans. Alternatively, some of these non-replications could be explained by the absence of variants within these loci associated with these traits in African Americans. Our data does not allow us to distinguish these two possibilities, and larger replication studies in African-American cohorts will be needed to draw informative conclusions.
Taken together, our results suggest that CHD risk in African Americans is not influenced by loci with major phenotypic effect on disease risk, but rather by multiple variants of weak effect, as we have observed for CHD and other traits in Caucasians. Because opportunities for replication and meta-analysis with other African-American cohorts are evolving rapidly, the CARe dataset is an outstanding public resource that provides a strong base for discovery of genetic contributors to CHD in non-European-derived populations.
Materials and Methods
Ethics statement
All participants gave informed written consent. The CARe project is approved by the ethic committees of the participating studies and of the Massachusetts Institute of Technology.
Studies
African-American participants for the GWAS were drawn from five population-based studies: Atherosclerosis Risk in Communities (ARIC; N = 3,269), Coronary Artery Risk Development in young Adults (CARDIA; N = 1,209), Cleveland Family Study (CFS; N = 704), Jackson Heart Study (JHS; N = 2,200), and Multi-Ethnic Study of Atherosclerosis (MESA; N = 1,737). Although longitudinal data is available for most participants, only information collected at recruitment was considered in this GWAS. Replication results for top SNP associations were obtained using in silico or de novo genotyping from four African-American and African-Caribbean population-based cohorts (Health, Aging, and Body Composition Study (Health ABC; N = 1,119), National Health and Nutrition Examination Survey III (NHANES III, N = 1,720), Jamaica Spanish Town (SPT, N = 1,746) and Jamaica GXE (N = 969), one nested case-control panel from the population-based Multiethnic Cohort (MEC, N = 2,184), and two case-control panels (Cleveland Clinic, N = 620, and PennCATH, N = 491). A detailed description of all cohorts and phenotype definitions used in this study is provided in Text S1.
Genotyping and quality controls
All discovery samples (GWAS) were genotyped on the Affymetrix Genome-Wide Human SNP array 6.0 according to the manufacturer's protocol. For replication, the MEC samples were genotyped by Taqman, and the NHANES III, Jamaica SPT, Jamaica GXE, Cleveland, and UPENN samples were genotyped using Illumina's Oligos Pool All (OPA) technology. The Health ABC samples were genotyped on the Illumina Human1M-Duo BeadChip array as part of an independent GWAS; SNP results for the replication of the CARe findings were extracted and analyzed. Several quality control (QC) filters were applied to the genome-wide genotype data: DNA concordance checks; sample and SNP genotyping success rate (>95%, minor allele frequency ≥1%); sample heterozygosity rate, identity-by-descent analysis to identify population outliers (Figure S1), problematic samples, and cryptic relatedness; Mendel errors rate in CFS and JHS, and SNP association with chemistry plates. For replication, SNPs and samples with genotyping success rate <90% were excluded. Because of the admixed nature of the participants, SNPs were not removed solely because they departed from Hardy-Weinberg equilibrium. A detailed description of the quality control checks applied to the discovery (GWAS) and replication genotyping data can be found in Text S1.
SNP imputation
To increase coverage and facilitate comparison with other datasets, we imputed genotype data using MACH v1.0.16 [19]. We built a panel of reference haplotypes using HapMap phase II CEU and YRI data, and imputed SNP genotypes using all Affymetrix 6.0 SNPs that passed the QC steps described above. Using and independent dataset of ∼12,000 SNPs genotyped on the same DNA but with a different platform, we estimated an allelic concordance rate of 95.6% (Text S1).
Association analyses
SNP-only based genetic association analysis of quantitative (HDL-C, LDL-C, smoking) and dichotomous (coronary heart disease, type-2 diabetes, hypertension) traits were carried out using linear and logistic statistical framework, respectively, in PLINK (unrelated cohorts: ARIC, CARDIA, JHS, MESA, UPENN, Cleveland, MEC, NHANES III, and Health ABC) or using R scripts that model family structure (related cohort: CFS) [28]. For the cohorts with genome-wide genotyping data available, the first ten principal components were included in each analysis to account for population stratification and admixture. The method to estimate local ancestry was implemented in ANCESTRYMAP and is described in details in Text S1. To combine allelic and local ancestry information (Table 4), we calculated a chi-square statistic with two degrees-of-freedom. Association results were combined across cohorts using an inverse variance meta-analysis approach as implemented in metal.
URL
CARe: http://www.broadinstitute.org/gen_analysis/care/index.php/Main_Page; MACH: http://www.sph.umich.edu/csg/abecasis/MACH; METAL: http://www.sph.umich.edu/csg/abecasis/Metal/index.html; PLINK: http://pngu.mgh.harvard.edu/~purcell/plink.
Supporting Information
Plots of the two main principal components (PC) in the CARe African-American samples. European-Americans and Nigerians samples are used as reference populations. We note that the first principal component (PC1) captures European vs. African global ancestry. For CFS, outliers on PC2 all belong to the same large family.
(0.25 MB TIF)
Manhattan plots summarizing the meta-analysis results for the six phenotypes analyzed after double genomic control scaling. The dashed line highlights genome-wide significance (P-value = 5×10−8). The genome-wide significant loci include 8p23 (chr8), LPL (chr8), LIPC (chr15), LCAT (chr16), and CETP (chr16) for HDL-C, SLC12A9 (chr7) for hypertension, and PCSK9 (chr1), CELSR2-PSRC1-SORT1 (chr1), and APOE (chr19) for LDL-C.
(1.27 MB TIF)
Graphical representation of the information summarized in Table S12. Plots were drawn using LocusZoom [26]. CHD association results in Caucasians are from Kathiresan et al. [27]. HDL-C association results in Caucasians are from Kathiresan et al. [28]. Under each plot, the light blue box corresponds to the genomic intervals flanked by the leftmost and rightmost SNPs with and r2≥0.3 with the index SNPs (purple diamond). For the results in Caucasians, we used LD based on HapMap CEU, and for the results in African Americans, LD based on HapMap YRI. For the PLTP and ABCA1 loci, the CARe SNPs (respectively rs6065904 and rs13284054) define genomic intervals of 0.2 kb using the r2≥0.3 threshold, which appear as light blue lines on the plots.
(1.59 MB TIF)
Quantile-quantile (QQ) plots of the meta-analyses results that take into account local ancestry estimates and SNP genotypes (Chi-square with two degrees-of-freedom (N = 8,090). Each black circle represents an observed statistic for genotyped SNPs only (defined as the −log10(P-value)) against the corresponding expected statistic. The grey area corresponds to the 90% confidence intervals calculated empirically using permutations. The meta-analysis inflation factors are: coronary heart disease (λs = 0.923), HDL-C (λs = 1.030), hypertension (λs = 1.121), LDL-C (λs = 1.290), smoking (λs = 1.060), and type-2 diabetes (λs = 1.109). Data shown is genomic controlled before (for each study) and after the meta-analysis.
(0.26 MB TIF)
Manhattan plots summarizing the meta-analysis results that take into account local ancestry estimates and SNP genotypes (two degrees-of-freedom). Results are shown for the six phenotypes analyzed after double genomic control scaling. The dashed line highlights genome-wide significance (P-value = 5×10−8). The genome-wide significant loci include LCAT (chr16) and CETP (chr16) for HDL-C and PCSK9 (chr1), CELSR2-PSRC1-SORT1 (chr1), 2p24 (chr2), and APOE (chr19) for LDL-C.
(1.23 MB TIF)
Comparison of P-values (-log10 scale) for the meta-analysis results obtained using SNP genotype-only (x-axis) or SNP genotype+estimate of local ancestry (y-axis) to compute the test statistics. Each black circle corresponds to a SNP. In total, results from ∼885,000 genotyped SNPs were available for each method. The gray line represents perfect correlation (x = y). The horizontal and vertical dashed lines represent the pre-defined threshold for genome-wide significance (P-value = 5×10−8).
(0.45 MB TIF)
Sample exclusion for the CARe Affy6.0 datasets.
(0.05 MB DOC)
SNP exclusion for the CARe Affy6.0 datasets.
(0.04 MB DOC)
Power calculations for the different phenotypes analyzed by the CARe Project in this study. The assumptions are: an additive inheritance model, an effect allele frequency of 10%, and an alpha threshold of 1×10−6 (which was arbitrary selected to promote SNPs for replication). We also assume that all samples belong to a single study; in practice, there is a small loss of power because of the meta-analysis. For case-control, power calculations were done using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/). For quantitative traits, power calculations were done using Quanto (http://hydra.usc.edu/gxe/).
(0.04 MB DOC)
Inflation factors (λs) for the different phenotypes analyzed, by cohort and for the corresponding meta-analyses.
(0.04 MB DOC)
Coronary heart disease replication results for SNPs from the CARe meta-analysis.
(0.25 MB DOC)
HDL-C replication results for SNPs from the CARe meta-analysis.
(0.21 MB DOC)
Hypertension replication results for SNPs from the CARe meta-analysis.
(0.21 MB DOC)
LDL-C replication results for SNPs from the CARe meta-analysis.
(0.22 MB DOC)
Type-2 diabetes replication results for SNPs from the CARe meta-analysis.
(0.10 MB DOC)
Loci associated at genome-wide significance level with CHD or its risk factors in Caucasians (data retrieved from the NHGRI database; http://www.genome.gov/26525384).
(0.30 MB DOC)
CARe meta-analysis association results (including data from ARIC, CARDIA, CFS, JHS, and MESA) for the SNPs associated with coronary heart disease and its risk factors in Caucasians. In yellow, we highlight the five loci where association signals in African Americans define smaller genomic intervals to search for causal allele(s).
(0.32 MB DOC)
Linkage disequilibrium-based genomic intervals of loci previously associated with coronary heart disease and HDL-C in Caucasians using the CARe datasets.
(0.05 MB DOC)
Supporting information.
(0.27 MB DOC)
Acknowledgments
The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the research institutions, study investigators, field staff, and study participants in creating this resource for biomedical research.
Footnotes
JBM has a research grant from GSK and a consulting agreement with Interleukin Genetics, SLH reports being listed as co-inventor on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics. SLH reports having been paid as a consultant or speaker for the following companies: AstraZeneca Pharmaceuticals LP, BG Medicine, Merck & Co., Pfizer Takeda, Esperion, and Cleveland Heart Lab. SLH reports receiving research funds from Abbott, Liposcience, and Cleveland Heart Lab. WHWT reports receiving research grant support from Abbott Laboratories.
The grants and contracts that have supported CARe are listed at http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx, including HHSN268200625226C (ADB No. N01-HC-65226). Additional support for this work was provided by: the Fondation de l'Institut de Cardiologie de Montreal and the Centre of Excellence in Personalized Medicine (CEPMed) (to GL), NIDDK K24 DK080140 (to JBM). The Health ABC study was supported by NIA contracts N01AG62101, N01AG62103, and N01AG62106. The Health ABC genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences, and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. The Health ABC research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. The Cleveland Clinic GeneBank study was supported through NIH grants 1P01HL098055-01, P01HL087018, P01HL076491, R01DK080732, and R01HL103931-01. A portion of analyses for the Cleveland Clinic GeneBank was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 (RR10600-01, CA62528-01, RR14514-01) from the National Center for Research Resources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.NHLBI. 2009. 2009 NHLBI Morbidity and Mortality Chart Book.
- 2.Thomas J, Thomas DJ, Pearson T, Klag M, Mead L. Cardiovascular disease in African American and white physicians: the Meharry Cohort and Meharry-Hopkins Cohort Studies. J Health Care Poor Underserved. 1997;8:270–283; discussion 284. doi: 10.1353/hpu.2010.0526. [DOI] [PubMed] [Google Scholar]
- 3.Jones DW, Chambless LE, Folsom AR, Heiss G, Hutchinson RG, et al. Risk factors for coronary heart disease in African Americans: the atherosclerosis risk in communities study, 1987–1997. Arch Intern Med. 2002;162:2565–2571. doi: 10.1001/archinte.162.22.2565. [DOI] [PubMed] [Google Scholar]
- 4.Cooper R, Rotimi C. Hypertension in blacks. Am J Hypertens. 1997;10:804–812. doi: 10.1016/s0895-7061(97)00211-2. [DOI] [PubMed] [Google Scholar]
- 5.Cooper RS, Zhu X. Racial differences and the genetics of hypertension. Curr Hypertens Rep. 2001;3:19–24. doi: 10.1007/s11906-001-0073-z. [DOI] [PubMed] [Google Scholar]
- 6.Katzmarzyk PT, Perusse L, Rice T, Gagnon J, Skinner JS, et al. Familial resemblance for coronary heart disease risk: the HERITAGE Family Study. Ethn Dis. 2000;10:138–147. [PubMed] [Google Scholar]
- 7.WTCCC. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 10.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 15.Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, et al. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang SJ, Chiang CW, Palmer CD, Tayo BO, Lettre G, et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 26.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plots of the two main principal components (PC) in the CARe African-American samples. European-Americans and Nigerians samples are used as reference populations. We note that the first principal component (PC1) captures European vs. African global ancestry. For CFS, outliers on PC2 all belong to the same large family.
(0.25 MB TIF)
Manhattan plots summarizing the meta-analysis results for the six phenotypes analyzed after double genomic control scaling. The dashed line highlights genome-wide significance (P-value = 5×10−8). The genome-wide significant loci include 8p23 (chr8), LPL (chr8), LIPC (chr15), LCAT (chr16), and CETP (chr16) for HDL-C, SLC12A9 (chr7) for hypertension, and PCSK9 (chr1), CELSR2-PSRC1-SORT1 (chr1), and APOE (chr19) for LDL-C.
(1.27 MB TIF)
Graphical representation of the information summarized in Table S12. Plots were drawn using LocusZoom [26]. CHD association results in Caucasians are from Kathiresan et al. [27]. HDL-C association results in Caucasians are from Kathiresan et al. [28]. Under each plot, the light blue box corresponds to the genomic intervals flanked by the leftmost and rightmost SNPs with and r2≥0.3 with the index SNPs (purple diamond). For the results in Caucasians, we used LD based on HapMap CEU, and for the results in African Americans, LD based on HapMap YRI. For the PLTP and ABCA1 loci, the CARe SNPs (respectively rs6065904 and rs13284054) define genomic intervals of 0.2 kb using the r2≥0.3 threshold, which appear as light blue lines on the plots.
(1.59 MB TIF)
Quantile-quantile (QQ) plots of the meta-analyses results that take into account local ancestry estimates and SNP genotypes (Chi-square with two degrees-of-freedom (N = 8,090). Each black circle represents an observed statistic for genotyped SNPs only (defined as the −log10(P-value)) against the corresponding expected statistic. The grey area corresponds to the 90% confidence intervals calculated empirically using permutations. The meta-analysis inflation factors are: coronary heart disease (λs = 0.923), HDL-C (λs = 1.030), hypertension (λs = 1.121), LDL-C (λs = 1.290), smoking (λs = 1.060), and type-2 diabetes (λs = 1.109). Data shown is genomic controlled before (for each study) and after the meta-analysis.
(0.26 MB TIF)
Manhattan plots summarizing the meta-analysis results that take into account local ancestry estimates and SNP genotypes (two degrees-of-freedom). Results are shown for the six phenotypes analyzed after double genomic control scaling. The dashed line highlights genome-wide significance (P-value = 5×10−8). The genome-wide significant loci include LCAT (chr16) and CETP (chr16) for HDL-C and PCSK9 (chr1), CELSR2-PSRC1-SORT1 (chr1), 2p24 (chr2), and APOE (chr19) for LDL-C.
(1.23 MB TIF)
Comparison of P-values (-log10 scale) for the meta-analysis results obtained using SNP genotype-only (x-axis) or SNP genotype+estimate of local ancestry (y-axis) to compute the test statistics. Each black circle corresponds to a SNP. In total, results from ∼885,000 genotyped SNPs were available for each method. The gray line represents perfect correlation (x = y). The horizontal and vertical dashed lines represent the pre-defined threshold for genome-wide significance (P-value = 5×10−8).
(0.45 MB TIF)
Sample exclusion for the CARe Affy6.0 datasets.
(0.05 MB DOC)
SNP exclusion for the CARe Affy6.0 datasets.
(0.04 MB DOC)
Power calculations for the different phenotypes analyzed by the CARe Project in this study. The assumptions are: an additive inheritance model, an effect allele frequency of 10%, and an alpha threshold of 1×10−6 (which was arbitrary selected to promote SNPs for replication). We also assume that all samples belong to a single study; in practice, there is a small loss of power because of the meta-analysis. For case-control, power calculations were done using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/). For quantitative traits, power calculations were done using Quanto (http://hydra.usc.edu/gxe/).
(0.04 MB DOC)
Inflation factors (λs) for the different phenotypes analyzed, by cohort and for the corresponding meta-analyses.
(0.04 MB DOC)
Coronary heart disease replication results for SNPs from the CARe meta-analysis.
(0.25 MB DOC)
HDL-C replication results for SNPs from the CARe meta-analysis.
(0.21 MB DOC)
Hypertension replication results for SNPs from the CARe meta-analysis.
(0.21 MB DOC)
LDL-C replication results for SNPs from the CARe meta-analysis.
(0.22 MB DOC)
Type-2 diabetes replication results for SNPs from the CARe meta-analysis.
(0.10 MB DOC)
Loci associated at genome-wide significance level with CHD or its risk factors in Caucasians (data retrieved from the NHGRI database; http://www.genome.gov/26525384).
(0.30 MB DOC)
CARe meta-analysis association results (including data from ARIC, CARDIA, CFS, JHS, and MESA) for the SNPs associated with coronary heart disease and its risk factors in Caucasians. In yellow, we highlight the five loci where association signals in African Americans define smaller genomic intervals to search for causal allele(s).
(0.32 MB DOC)
Linkage disequilibrium-based genomic intervals of loci previously associated with coronary heart disease and HDL-C in Caucasians using the CARe datasets.
(0.05 MB DOC)
Supporting information.
(0.27 MB DOC)