Abstract
Cell population can be viewed as a quantum system, which like Schrödinger’s cat exists as a combination of survival- and death-allowing states. Tracking and understanding cell-to-cell variability in processes of high spatio-temporal complexity such as cell death is at the core of current systems biology approaches. As probabilistic modeling tools attempt to impute information inaccessible by current experimental approaches, advances in technologies for single-cell imaging and omics (proteomics, genomics, metabolomics) should go hand in hand with the computational efforts. Over the last few years we have made exciting technological advances that allow studies of cell death dynamically in real-time and with the unprecedented accuracy. These approaches are based on innovative fluorescent assays and recombinant proteins, bioelectrical properties of cells, and more recently also on state-of-the-art optical spectroscopy. Here, we review current status of the most innovative analytical technologies for dynamic tracking of cell death, and address the interdisciplinary promises and future challenges of these methods.
Keywords: programmed cell death, real-time assay, fluorescent probe, spectroscopy, lab-on-a-chip
The qualitative characteristics of programmed cell death (PCD), which is initiated and executed through multiple interconnected signaling cascades, differ not only between cell and stress types, but also within the clonal cell population encountering the same stress. The regulation of apoptosis involves a switch between the gradual (rheostat-type) and binary (all-or-none) events, which often establishes the threshold sensitivity for life-death decisions.1,2 Such level of complexity, with multiple variables concurrently expressed, requires an in-depth investigation of cell populations at a single cell level, kinetically, and with multivariate analytical capability.
The techniques applied to study PCD encompass a variety of molecular markers, such as fluorescent functional probes or genetically encoded fluorescently-tagged proteins, as well as phenotypic analysis of structural and bulk biochemical characteristics of dying cells. The most commonly used approach is a static assessment of cell status at a single time point. The very nature of cell death is, however, based on the multiple switches between slow but also variable in time decision-making processes, involving gradual accumulation of pro-apoptotic molecules (e.g., tBid, Bax complexes), followed by snap-action rapid activation of caspases, and variable kinetics and specificity of proteolytic degradation of endogenous targets. The cell-to-cell variability in time between the varying stages of cell death arises from the subtle fluctuations in the concentrations or states of regulatory proteins,3 protein oscillations, the induction of multiple compensatory mechanisms (e.g., autophagy), or ‘molecular noise’ originating from low copy number components.4 Therefore, the possibility to continuously track individual cells from the time of encountering stress signal, through the decision-making and execution phases of cell death, up to the point of complete demise, could provide previously inaccessible information of how complex biological systems progress from life-maintaining to death-allowing steady-states. This can be represented by a three-dimensional cubic (lattice) box which mimics the dynamics (time) of cellular changes (output) over encountered signals (input) (Fig. 1). In addition, any state space can represent multiple inputs-outputs (that is, y and z have more then one coordinate). The inherent complexity of cellular systems requires massive experimental parallelization, as snap-shot static analysis cannot reveal the full biological intricacy of the events.
Figure 1.
Rationale behind real-time analysis of living single cells. Each cell within a population is likely to respond to certain stimuli (inputs) with varying kinetics and outcomes (outputs). Steady-state measurement can mask this inherent variability. Dynamic labeling allows not only kinetic analysis but also enormous reduction of sample processing steps. This is important for the preservation of fragile apoptotic cells and allows obtaining content-rich kinetic data sets.
It should be noted that the intercellular variability in passing the “point of no return” during apoptotic process provides flexibility for cell populations to respond to stresses of different intensity by granting survival for the most resistant cells. In the case of cancer chemo- or radio-therapy such cells are of most interest because they escape the treatment and are responsible for cancer recurrence. The assessment of cells heterogeneity including identification of the stress resistant or sensitive cells for their further characterization at molecular level requires thus cytometric approach i.e., quantitative analysis of constituents in individual cells.
Given the above, an ideal approach to monitor cell death would require to: (i) focus at the level of individual living cells; (ii) follow cells over extended period of time to allow analysis of cell-to-cell variability; (iii) induce minimal perturbations to structural and bio-physiological properties of the cells; (iv) allow multiparameter analysis in combination with other approaches of similar nature, to correlate multiple events at a single cell level over time with each other; and (v) be applicable to high-throughput formats, i.e., straightforward in use and adaptable for automation. The innovative approaches to study cell death can be divided into molecular, based on novel probes and recombinant sensors, and bioengineered, based on ongoing developments in the fields of micro- and nano-technologies and spectroscopy. Here we discuss the use of these methods and their potential to enhance current understanding of sensitive and robust cell death networks.
The Fluorescent Toolbox for Analyzing Apoptosis
Analysis of mitochondrial outer membrane permeability (MOMP) and its regulators
Mitochondria act as central sentinels of stress signals, both in the mitochondrial (intrinsic) pathway of apoptosis and, in type II cells, in the death receptor (extrinsic) pathway of apoptosis.5,6 MOMP is the critical event leading to the release of pro-apoptotic proteins, such as cytochrome c, Smac, Omi or AK2 from the intermembrane mitochondrial space to the cytosol.7 It is also the point at which a graded signal (e.g., formation of Bax pores or accumulation of truncated Bid) is transformed into an all-or-none snap-action signal.2 Traditionally, MOMP has been studied using bulk biochemical approaches, such as cellular fractionation and western blot-based detection of cytochrome c and other proteins released from mitochondria upon MOMP, flow cytometric analysis of the amount of cytochrome c retained by fixed and permeabilized cells,8 as well as native blue-PAGE to show formation of Bax complexes.9 Although these approaches can provide interesting new mechanistic insights, e.g., about processing of pro-caspase 9 in mitochondria,10 they generally lack temporal single-cell resolution and do not reveal rare intermediate cellular states, or events such as mitochondrial fusion and fragmentation.11 They also do not reveal a possible mechanistic relationship in terms of cause-effect link between the particular events.
As it has been suggested that multiple kinetics of cytochrome c and Smac release exist during apoptosis,12 and that the release of cytochrome c does not take place simultaneously in all mitochondria,13 multiparameter real-time imaging of MOMP and pre-MOMP events is of paramount importance to detect small spatio-temporal changes. Fluorescently-tagged recombinant proteins, such as mitochondrial intermembrane space reporter protein (IMS-RP),2 cytochrome c-GFP and dsRed-Bax,12,14,15 GFP-Bax or Smac/DIABLO-YFP16–18 have all proven useful for real-time monitoring of MOMP on a single-cell level. For example, the imaging of cells for early spatially-confined MOMP, with the use of cytochrome c-GFP, Smac-mCherry and markers of mitochondrial transmembrane potential (ΔΦm), revealed that the release of cytochrome c and Smac, and loss of ΔΦm, occur in a similar spatial and temporal pattern.13
After the discovery that it is possible to fuse two proteins that fluoresce at different wavelengths and monitor fluorescence resonance energy transfer (FRET),19–21 the recombinant FRET-based sensors have been deployed to study MOMP and up-stream regulatory proteins. For example, it has been shown, using time lapse fluorescence microscopy and FRET, that the translocation of Bax to mitochondria involves several dynamic stages and leads to MOMP upon formation of small (>100 molecules) Bax-Bak complexes.18 FRET-Bid recombinant protein, i.e., Bid fused to yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) at the N-terminus and C-terminus, respectively, has been used for real-time detection of caspase 8-mediated cleavage of Bid, as well as translocation of tBid to mitochondria, at a single cell level.22 Other members of the Bcl-2 family can also be cleaved during the process of apoptosis, and the cleavage products, such as p18 Bax, can significantly affect the kinetics of apoptotic events.23 Therefore, we envisage that FRET-based recombinant proteins can provide substantial new insights into the dynamic role of Bcl-2 family members in apoptosis.
The main obstacle to successful imaging of early apoptotic events, such as initiation of MOMP, is the variable time to onset and subsequent speed of these events. This intrinsic feature of apoptotic signaling makes it difficult to image single cells at high temporal resolution without causing photodamage and photobleaching by high power lasers of conventional laser scanning microscopy (LSM). Moreover, the speed of beam scanning movements and scan acquisition in single-beam LSM is usually not fast enough to capture video rate imaging of rapid cellular reactions, such as mitochondrial membrane depolarization.24 The expansion of multi-beam confocal microscopy (MBCM) and video-rate confocal microscopy (VRCM), based on spinning disc principle, provides new opportunities to study single living cells at a high temporal resolution, confocal sectioning and relatively low phototoxicity. In contrast to conventional single beam and single pinhole confocal microscopy, the spinning disc confocal systems utilize a disk that contains numerous pinholes or slit opening, and rotates allowing the pattern of pinholes to cover the entire specimen. Although spinning disc imaging compromises somewhat the spatial resolution as compared to LSM, it achieves much better florescence transmission and image acquisition rates. It is thus preferable for 4D (time-lapse) multi-colour imaging of living specimens that require enhanced spatial resolution, full frame imaging at high frame rates (up to 15 frames per second) and reasonable sensitivity with low power lasers or mercury arc lamps.24
Probing Caspase Activation
Activation of caspases is critical for execution of apoptosis.25 Real-time monitoring of caspase activity at a single cell level is crucial to the understanding of concepts such as bistability, variable-delay caspase activation, and integration of gradual signals into all-or-none decision of apoptosis execution.2
Several techniques have been recently used to address this issue, including probes based on cell-permeable organic fluorogenic caspase substrates. Non-fluorescent substrates with incorporated lipophilic moieties pass freely through the cellular membranes, become fluorescent upon caspase-mediated cleavage, and are retained within the cell, usually by the charge of the fluorochromes. For example, a hydrophobic red fluorescent dye, cresyl violet, linked to the bi-substituted target peptide aspartate-glutamate-valine-aspartate (z-DEVD)2, can detect active caspase 3/7 within living cells without the requirement of washing steps.26 Further advantages of this probe, known under the trademark Magic Red® (ICN Biomedicals and Immunochemistry Technologies), include low autofluorescence and reduced phototoxicity, both important in live cell imaging.
Another promising fluorogenic system utilizes xanthenes dyes forming H-type dimers with spectral characteristics described under exciton theory.27 Upon formation, xanthene H-type dimers exhibit fluorescence quenching, a feature utilized by inserting the linker between two rhodamine fluorophores. The linker is an 18-amino acid peptide constituting the recognition and cleavage sequence from CPP32 (PARP-1), a physiological target for caspase 3. As a result, proximity-enforced formation of H-type rhodamine dimers generates quenched and cell-permeable caspase substrate, known under the trademark PhiPhiLux (OncoImunin, Inc., USA). When activated caspases cleave the liker, the rhodamine molecules are released and become highly fluorescent.28–30 Importantly, cleaved fluorescent fragments are well retained on the side of the membrane where the cleavage took place, allowing detection of low-level, compartmentalized caspase activity in living cells.29,31
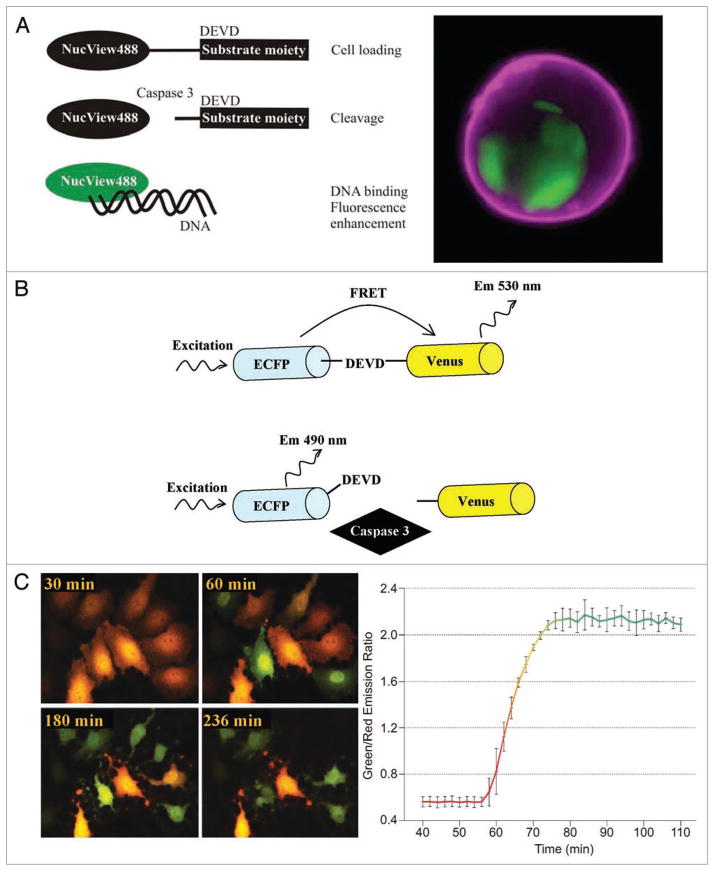
In another twist to fluorogenic caspase substrate design, Can et al. directly linked a DNA-binding dye, NucView488, to the caspase-3 recognition peptide DEVD (Fig. 2A).32,33 NucView488 (2-[(1-(5-((2-aminoethylamino)carbonyl)pentyl)quinolinium-4(1H)-ylidene)methyl]-3-methylbenzo[d]thiazolium iodide) is a DNA binding probe, derivative of thiazole orange. When attached to a highly negatively charged group of Ac-DEVD, its ability to electrostaticaly bind to DNA is inactivated. Substrate cleavage by active caspase 3 initiates dye release, restoration of its high positive charge, and translocation to the nucleus. Upon binding to DNA NucView488 becomes highly fluorescent34 (Fig. 2A). DEVD-NucView488 probe is reportedly not cytotoxic and does not affect cell fate decisions even during long-term incubations.32,33
Figure 2.
Dynamic analysis of caspase activation. (A) Left, principle of the DEVD-NucView 488 probe. Right, confocal image of apoptosing Jurkat T cell, stained with DEVD-NucView 488 (green) and CF647-Annexin V (violet) probes. Note the nuclear localization of the NucView488 probe following cleavage of the DEVD substrate by caspase 3. NucView 488 staining reveals also characteristic apoptotic fragmentation of the nucleus. Data courtesy of Biotium Inc., (Hayward, CA, USA). (B) Diagram of a FRET sensor. The fluorescence resonance energy transfer (FRET) that occurs between the pairs of fluorescent proteins when they are linked by the peptide is abruptly terminated upon caspase-mediated cleavage of the peptide linker. (C) Caspase activation visualized by a loss of FRET between TagGFP (green, Ex/Em 482/505 nm) and TagRFP (red, Ex/Em 555/584 nm). HeLa cells were transiently transfected with Casper3-GR, and then treated Staurosporine (2 μM). Left, time-lapse images were acquired using 520 nm band-pass filter for TagGFP and 580 nm for TagRFP. Right, green to red fluorescence signal ratio after stimulation of cells with Starosporine indicates activation of caspase 3. Data courtesy of Evrogen JSC (Moscow, Russia).
Activation of caspases can also be assayed by using genetically-encoded recombinant caspase substrates. Recombinant FRET probes based on GFP variants have been successfully applied in single-cell kinetic analysis of caspase activity.34–37 Recently, several new assays have been designed using the tandem molecules of green fluorescent protein (GFP), blue fluorescent protein (BFP), enhanced cyan-(ECFP) or yellow-fluorescent protein (YFP), or improved version of YFP called Venus, covalently linked by a short caspase-cleavable peptide.38–42 FRET that occurs between the pairs of these fluorescent proteins when they are linked by the peptide is abruptly terminated upon cleavage of the peptide linker (Fig. 2B). Caspase activation, thus, manifests by loss of FRET. The drawback of FRET probes is their poor sensitivity due to spectral overlap between cyan and yellow FRET partners, and often the low dynamic range of FRET sensors.43 As it is critical to use FRET probes with high signal to noise ratio in order to facilitate the detection of low caspase activities, new FRET sensors have been recently developed, including an optimized CFP-YFP pair called CyPet-YPet,44 as well as bright monomeric fluorescent TagGFP and TagRFP pair.43 The TagGFP-DEVD-TagRFP sensor shows increased dynamic range, bright fluorescence and enhanced pH and photo-stability (Fig. 2C). Finally, a FRET probe for simultaneous measurement of caspase 3 and caspase 6 activity has been developed, based on CFP-YFP-mRFP fusion protein containing caspase 3 cleavage site between CFP and YFP, and caspase 6 cleavage site between YFP and mRFP.45 Noteworthy, the major disadvantage of all FRET methods is the transfection step which lacks versatility, as not all cells are equally susceptible to gene delivery, is difficult to adopt during automated dispensing, increases costs, and potentially interferes with the robustness of cell physiology.33
Apoptotic Changes within Plasma Membrane
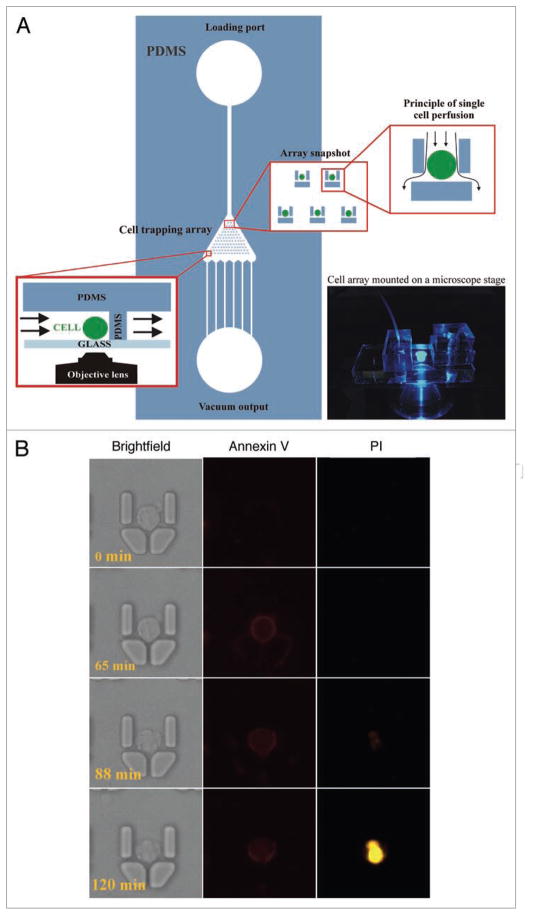
A characteristic asymmetric distribution of plasma membrane phospholipids between inner and outer leaflets is lost during caspase-dependent apoptosis when phosphatidylserine (PS) becomes exposed on the outer leaflet.46–48 The detection of exposed PS residues with a fluorochrome-tagged 36 kDa anticoagulant protein Annexin V, which reversibly binds to PS residues in the presence of milimolar concentration of Ca2+ ions, allows detection of apoptotic cells.47 Interestingly, fluorescent conjugates of Annexin V are typically used in end-point assays to quantify apoptotic frequency in vitro. This is somehow contradictory to an abundant number of reports showing non-invasive methods with radio-labeled Annexin V in dynamic analysis of apoptosis in vivo.49–51 Indeed, in our recent work we have shown that a dynamic tracking of apoptosis on state-of-the-art microfluidic cell arrays is feasible using fluorescent Annexin V conjugates continuously present in perfusion culture medium (Fig. 3).52 The only consideration for such assays is the necessity to increase concentration of calcium ions in culture medium. Moreover, Annexin V conjugated to inorganic, semiconductor nanocrystals (Quantum Dots; QDs)53,54 has increased photostability and narrow emission spectra. Recently polarity-sensitive indicator of viability and apoptosis (pSIVA), based on Annexin B12, has been developed and successfully applied in live-cell imaging of etoposide-treated cells and nerve growth factor (NGF)-deprived neurons.55 Annexin V functionalized quantum dots and polarity-sensitive annexin-based biosensors are feasible for real-time and multiparameter studies of apoptosis,55–58 and could be used for high-throughput screening on microfluidic platforms59 or with the use of MRI.49
Figure 3.
Real-time, dynamic analysis of apoptosis using fluorescent Annexin V conjugate on a microfluidic live-cell array. (A) Overview of the microfluidic live-cell array. Note the array of micromechanical traps that allow single cell positioning and perfusion. (B) Typical, time-resolved images of a single HL-60 cell undergoing apoptosis after perfusion with Staurosporine (2 μM), with time points indicated at the lower left corner. Cells were stimulated in the presence of Annexin V-APC and plasma membrane permeability marker propidium iodide (PI). Time-lapse images were collected every minute. Note the stochastic nature of cell death with the gradual exposure of the phosphatidylserine and increase in plasma membrane permeability to PI during apoptosis.
Plasma membrane permeability, although not specific to apoptotic cell death, is a common marker of dead cells. It is detected using charged fluorescent probes, e.g., propidium iodide (PI), 7-aminoactinomycin D (7-AAD), SYTOX or YO-PRO1, which enter cells and stain nucleic acids only upon permeabilization of the plasma membrane.60,61 In contrary to the commonly suggested toxicity of these probes, we have recently described their applicability to dynamically trace cell death in real-time.60,62,63 The proliferation rates, viability, and even quantitative responses to cytotoxic and DNA damaging agents were not significantly affected by a continuous growth of cells in the presence of these fluorochromes.60,62,63 While some increase in fluorescence intensity of live cells growing in their presence was seen, the intensity of fluorescence was many-fold enhanced upon cell death. The finding that charged cationic fluorescent markers do not affect cell growth or DNA integrity in any significant degree implies a possibility of their use in real-time single-cell analysis.
Autophagic Flux
Macroautophagy (hereafter referred to as autophagy) is an intra-cellular bulk degradation system for long-lived proteins and whole organelles. Emerging evidence suggests that autophagy promotes survival of cells exposed to various stress, but can also contribute to cell death when induced above an acceptable for cellular homeostasis threshold.64,65 The analysis of the kinetics of autophagy could thus reveal new aspects related to the robustness of cellular physiology. To date only a handful of methods have been introduced to quantify autophagy. Fluorescent microscopy (using GFP tagged MAP1-LC3 protein) as well as transmission electron microscopy are used to follow steady-state accumulation of autophagosomes, whereas bafilomycin A1-inhibitable half-life assessments of long-lived proteins is applied to access the catabolic autophagic activity.66,67 Current methods designed to quantify autophagic activity are, however, time consuming, labor intensive, and not feasible for measuring the dynamics of autophagic flux. Recently, the elegant solution has been proposed by Farkas and colleagues68 who designed a luciferase-based reporter assay (RLuc-LC3) to measure the autophagic flux in real-time. Particular advantage of the RLuc-LC3 assay lies in a broad dynamic range and applicability to a dynamic analysis of cell population. The assay has been validated by screening a library of small-molecule kinase inhibitors, proving its applicability for tracking dose- and stimulus-dependent differences in autophagy kinetics.68
Metal Nanoparticles–Because Stochastic Reactions Need Single Molecule Tracking
Advances in organic chemistry, materials science and molecular biology have recently created new classes of probes for cell imaging, such as functionalized gold or silver nanoparticles. The binding of proteins to such nanoparticles changes the wavelength of plasmon resonance, which can be followed by dark-field microscopy. Moreover, the light-scattering intensity of a pair of nanoparticles is greater then for two separate particles, and increases even further for multi-particle assemblies. This feature has been recently utilized to study activation of caspase 3. To this aim peptides with caspase 3 cleavage sequence DEVD have been linked to crown nanoparticle probes composed of a NeutrAvidin-coated gold-core nano-particle with 5 biotinylated gold satellite nanoparticles.69 Although the intracellular movement of crown nanoparticles is limited due to their large size, small cleaved satellite nanoparticles can move and allow analysis of single molecule trajectories. Briefly, a scattering intensity map of the crown nanoparticle-labelled cell is obtained, and once the apoptotic stimulus is administered the time trace of single probes is analyzed.69 Such an approach allows detection of early and/or spatially confined caspase activation. In the future crown nanoparticles could be used to study the activity of other caspases or protein conformational changes (e.g., during mitochondrial outer membrane pore formation), and potentially to assess the kinetics of reactions that, due to the spatial location of protein components and their diffusion rates, constitute low probability stochastic events (e.g., apoptosome assembly). Similar technology, referred to as SNaPT (Single Nanoparticle Photothermal Tracking), has also been used to quantify conformational changes of mitochondria under selected pro-apoptotic stimuli in live COS-7 cells.70
Label-Free Approaches
Kinetic tracking of cell death based on labeling approaches is a compelling prospect, hampered, however, by possible adverse effects of ectopic probes on processes within the living cells. For example, photoexcitation of TMRM can trigger permeability transition pore (PTP), release of cytochrome c and relocation of Bax.71 Therefore, technologies for measuring functional responses in living cells without the use of probes or recombinant proteins are gaining increasing interest.72
Capacitance Spectroscopy
Capacitance cytometry represents a pioneering approach with prospective applications in the analysis of cell cycle and fractional DNA content (sub-G1).73 This technology is based on a linear relationship between the DNA content of eukaryotic cell and the change in capacitance that is evoked by the passage of individual cell across a 1-kHz electric field.73 As capacitance measurements do not require cell processing and can be repeatedly performed on the same population of living cells, they are particularly useful in the real-time monitoring of changes in cell cycle distribution.73
Alternating Current (AC) Impedance Spectroscopy
Microelectronic alternating current (AC) impedance spectroscopy is another up-and-coming technology for monitoring of cell death, developed and commercialized by Roche Applied Science in partnership with ACEA Bioscience under the trademark of xCEL-Ligence system. Impendence spectroscopy allows label-free analysis of adherent cells cultured on proprietary micro-electrode plates. It relies on the local ionic environment at the electrode-solution interface, which affects the impedance value of the electrode. The more cells attach to an array of inter-digitated electrodes, the larger the increase in total impedance. Impedance spectroscopy can be used to monitor not only cell viability, but also cell proliferation and degree of cell adhesion. The technology offers several advantages, including the lack of labeling, real-time monitoring and potential for automation. The main disadvantages of impedance spectroscopy are (i) its applicability to adherent cells only; and (ii) lack of specificity for cell death versus cell detachment or decrease in proliferation rate. The latter is a serious obstacle to characterization and quantification of cell death. When apoptotic signaling is triggered cells often start to detach from the surface leading to a rapid loss of impedance values. Primary necrotic and late apoptotic/secondary necrotic cells will, however, also lose contact with the substrate, yielding in principle the same results. Moreover, cells that respond by detachment may still maintain viability for a long-time before committing to cell death. Therefore, a drop in impedance value can be observed irrespectively of the type, and even in the absence, of cell death. Finally, impedance spectroscopy is a bulk technique that measures the average impedance over the total area covered by micro-electrode array, thus lacking single-cell specificity. Recent developments can increase the specificity of impedance cytometry, and allow quantitative analysis of the results. For example, Tong and co-authors have developed novel biosensor which incorporates electrochemical impedance measurements with the specificity of Annexin V.74 Overall, impedance spectroscopy can provide a valuable complement to the flow cytometry and imaging methods, especially during screening of compounds that induce irreversible or reversible cell cycle arrest leading to e.g., senescence.
Surface Resonance Spectroscopy
Resonant plastic-based photonic crystal technology is an elegant, label-free method for detecting the proliferation and cell death in real-time.75 Photonic crystals are composed of a periodic arrangement of dielectric materials such as TiO2. Sensor measures changes in the reflected light wavelength within an evanescent region adjacent to the surface during cell attachment and spreading.74 Importantly, the resonant wavelength emitted by the crystal is measured after illumination with white light and reflected spectrum of each pixel is collected using an imaging spectrometer.74 This technology has been used to dynamically monitor proliferation of MCF-7 breast cancer cells and their detachment during drug-induced apoptosis.75
Thermal Lens Spectroscopy
Tamaki and colleagues have recently developed a revolutionary scanning thermal lens microscopy for detection of non-fluorescent biological substances with extremely high sensitivity and spatial resolution.76 This technology utilizes a photo-thermal effect of a non-radiative relaxation of photo-excited species in a very small volume coupled with a rapid thermal conduction between a solid-liquid interface.76 Compared to any of the currently available absorption methods thermal lens spectroscopy has a much higher sensitivity, which allows kinetic monitoring of rapid intracellular events associated with apoptosis.76 For example, high spatial resolution of thermal lens spectroscopy permitted single-cell quantitative analysis of cytochrome c release from mitochondria during apoptosis.76
Raman Spectroscopy
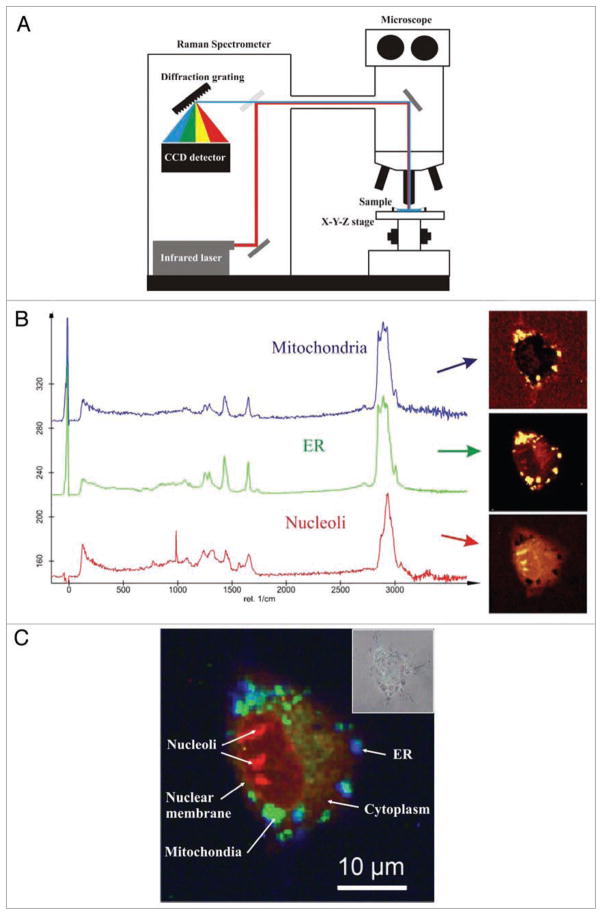
Non-destructive spectroscopies such as infrared micro-spectroscopy and Raman micro-spectroscopy attract an increasing interest for a label-free analysis at the single cell level.77,78 Raman spectroscopy is an optical technique based on inelastic scattering of photons by molecular vibrations of biopolymers79 (Fig. 4A). To avoid destruction of live specimens, it relies mostly on scattering of monochromatic light in the near-infrared spectrum. The wavelengths and intensities of the scattered light are collected from a very small volume and provide a chemical fingerprint of cells without the use of fluorescent labels.79 When applied in a hyperspectral imaging mode, thousands of Raman spectra is used to generate detailed Raman maps, which can provide content-rich information, with a very high spatial and temporal resolution, on molecular changes within living cells78,79 (Fig. 4B and C).
Figure 4.
Raman micro-spectroscopy in single-cell analysis of apoptosis. (A) Basic schematics of confocal Raman microscope. Scattered photons of light are collected and spectrally analyzed after passing through the diffraction grating and a CCD detector. Raman scattering occurs when photon of light collides with a molecule and interacts with its electron cloud and the chemical bonds. The inelastic interaction changes the vibration state of the molecule. Exited vibrational quantum stage of the molecule leads to an energy shift between the incident light and the back-scattered light. This energy shift is a unique chemical fingerprint of the molecule and can be used for qualitative and quantitative analysis of live cells. (B) Averaged basic Raman spectra obtained from different cellular compartments: mitochondria, endoplasmic reticulum (ER) and nucleoli, in a living HeLa cell (left). Basic spectra were constructed by averaging 10,000 spectral sets. Raman images were generated by the fit algorithm using preselected basic spectra from different regions of a cell (right). (C) Composite and pseudo-colored Raman image of a cell. Different cellular structures can be easily analyzed using spectral fingerprints. Inset shows a conventional brightfield image of the cell. Raman spectra were obtained using a WITec CRM 200 Confocal Raman Microscope using a x60 water-immersion objective and a frequency-doubled Nd:YAg laser (532 nm, 10 mW). Scan range represented region size of 100 × 100 pixels (10,000 spectra). Raman data shown are a courtesy of WITec GmbH (Ulm, Germany).
Recently, Raman spectroscopy employing a broad band (1,900 cm−1) spontaneous non-resonant scattering has been applied to investigate distribution of DNA, RNA, proteins and lipids in human HeLa cells undergoing apoptosis.80 Confocal Raman hyperspectral data sets allowed also a label-free visualization of mitochondrial distribution in living cells,81 as well as being used for detection of quantitative and conformational changes in proteins and nucleic acids in necroric cells.82 The ability of Raman micro-spectroscopy to non-invasively track cell cycle dynamics in single living cells has also been recently reported.83
The advent of surface enhanced Raman spectroscopy (SERS), which features signal enhancement of over 14 orders of magnitude over non-enhanced Raman scattering, opens up new avenues for real-time dynamic spectroscopy.84–86 New cellular SERS reporter probes based on silver and gold nanoparticles are already being developed.85–87 Nanostructures within a hybrid SERS probe provide sufficient enhancement in the electromagnetic field to analyze single biomolecular species while using only low laser powers and short data acquisition times.84,87 SERS has also been shown to provide valuable dynamic information on the local pH values with sub-endosomal resolution.87,88
Size and complexity of Raman spectroscopy data sets make detecting and classifying small biological changes difficult. Therefore, in recent studies of toxin- and heat shock-induced cell death Raman spectroscopy has been coupled with support vector machines (SVMs) learning algorithms.89 SVMs are artificial intelligence methods designed for automated learning from sets of known examples, usually well defined input:output pairs. They are perfectly suited for rapid searching and pattern recognition on established databases. The SVM algorithms proposed by Pyrgiotakis and colleagues allow rapid classification of the results from Raman micro-spectometry while ignoring random biological variations inherent to any cellular system. This elegant work is also first to apply sophisticated machine learning algorithms to non-invasively detect small biochemical differences in living cells using Raman spectroscopy in real-time.89
Multidimensional Atomic Force Microscopy (AFM)
The rich morphometric data can be also obtained using high resolution atomic force microscopy (AFM). AFM utilizes a micro-mechanical probe (cantilever) that moves (in contact mode) or oscillates (in tapping mode) up and down along the surface of the cell (Fig. 5A). Scanning probe technology allows resolution at ångström scale,90 which is theoretically limited only by the radius of the imaging probe. Collected AFM data reflect the relative size of structural features and give a topological visualization of the cell surface.91 In contrast to scanning electron microscopy (SEM), AFM provides an accurate 3D visualization of the cell surface that can be further enhanced by photo-realistic 3D volume software rendering (Fig. 5B). Importantly AFM does not require any sample processing steps or chemical dyes and can be performed at ambient pressures and temperatures.92 Moreover, during the AFM scans cells experience a very low mechanical stress, which provides a unique opportunity for the kinetic and real-time live cell scanning. Apart from cell surface, AFM scans can also reveal the distribution of subcellular structures localized in a close proximity to plasma membrane. This is possible as a result of the mechanical elasticity of the plasma membrane, and is somewhat analogical to palpation during medical examination.93 AFM technology has been also applied to study remodelling of cytoskeleton and plasma membrane during cell movement and division, tumor metastasis, viral infection as well as necrotic cell death.94–97 Recently AFM technology has begun to attract a growing interest in the real-time studies on programmed cell death. Kuznetsov and colleagues were first to investigate plasma membrane blebbing in osteosarcoma cells undergoing apoptosis.91 More recently, AFM was used in a multidimensional and kinetic study of volumetric changes during Staurosporine-induced casapase dependent apoptosis in human epidermoid carcinoma cells.90 Interestingly, the 3D virtualization of cell surface using AFM can be further enhanced by overlaying simultaneously acquired confocal microscopy images or even confocal Raman micro-spectroscopy data (Fig. 5C). Approaches that merge 3D rendering and volume measuring capabilities of AFM with chemical spectroscopy allow multidimensional imaging with many applications in real-time spectroscopy of programmed cell death.
Figure 5.
Multidimensional atomic force microscopy (AFM). (A) Principles of the AFM technology. Piezoelectric actuator moves the microscope stage with ångström (Å) precision while very fine tip (probe) of the cantilever (100 nm) is moving (contact mode) or oscillates up and down (tapping mode) along the surface of the cell. The deflection of the tip is measured using laser beam reflected from the surface of the cantilever. Reflected light is subsequently projected onto the position sensitive photodiode array. From the output signals data processing electronics and visualization software creates 3D virtualization of the cell surface characteristics. Note that the tip of the cantilever (red) has subcellular dimensions [Inset photograph, courtesy of: Drs Alexandre Berquand (Veeco Instruments GmbH, Mannhaim, Germany), Ian Schroeder (Leica Microsystems) and Frank Lafont (Pasteur Institute, Lille, France)]. (B) Advanced visualization of AFM data showing improved recognition of unique morphometric features (indicated with white arrows). Standard 2D AFM height image (upper), pseudo (non-realistic) 3D AFM height image (middle) and photo-realistically rendered 3D height image (lower). Note that live realistic 3D volume rendering of AFM data allows even very small cell surface structures to be identified with great precision. (C) Hybrid images of a cell showing 3D representation of confocal fluorescence, height and deflection AFM modes (upper panel). Reconstruction of 3D cell surface visualized together with the fluorescence confocal data (green texture layer). Note volume measuring tool that provides unique morphometric capabilities (lower). Multiple surface visualization of atomic force microscopy and fluorescence data were performed using Surface3D PRO engine (ScienceGL Inc.,) (data used with permission from ScienceGL Inc., Attleboro, MA, USA).
Summary and Future Outlook
We have shown before that during the cell cycle cell populations become heterogeneous after cytokinesis which is asymmetrical and generates daughter cells unequal in size, RNA and protein content.98 Due to variation of their residence time in G1 allowing smaller cells to “catch up” with the larger ones the populations become more uniform at the time of entrance to S, to become again heterogenous after cytokinesis.98 We postulated that heterogeneity of cell populations generated this way along the cell cycle provides basis for cell variability in response to growth- and stress-factors and is one of the mechanisms of survival for cell populations in adverse conditions such as drug treatment.98 Recent approach of single cell encoding with quantum dots also shows that cell division is an asymmetric event.99 Moreover, the discoveries of new markers of apoptotic cell death go hand-in-hand with development of single-cell monitoring approaches, as in the case of detection of active acetylcholinesterase (AChE).100 While most standard analytical assays rely on averaging of cells populations, masking features of individual cells such as all-or-none effects or variable-time switch delays, the technological approaches presented in this review allow one to probe such variability. Speed and high-throughput, affordability, reliability and non-invasiveness are all being incorporated into the approaches of modern cell biology, such as microfluidic technologies (Lab-on-a-Chip) or cell microarrays. Advances in protein tagging, such as central dogma (SD) tagging approach based on retroviral introduction of a fluorescent tag into the chromosome to allow visualization of endogenous proteins that remain under their native regulation,101,102 opens new perspectives in kinetic studies of cell signaling. Moreover, computational modeling strategies probe phenomena not accessible with current experimental approaches.103–105 As we boldly go beyond the familiar territories of bioengineering, we are soon to witness massive technological advances in live-cell spectroscopy that can bring revolutionary discoveries in many fields of biology and medicine. It is our hope that by bridging the engineering, informatics, chemistry and biomedical arenas we can all contribute to a major “quantum leap” in drug discovery pipelines and personalized diagnostics, ultimately leading to a better and more affordable healthcare.
Acknowledgments
Authors thank the: Biotium Inc (Hayward, CA, USA) for DEVD-NucView 488 data; Evrogen JSC (Moscow, Russia) for Casper3-GR FRET sensor data; ScienceGL Inc, (Attleboro, MA, USA) for advanced visualization of AFM data using Surface3D PRO engine; WITec GmbH (Ulm, Germany) for Raman microspectroscopy data; and Drs Alexandre Berquand (Veeco Instruments GmbH, Mannhaim, Germany), Ian Schroeder (Leica Microsystems) and Frank Lafont (Pasteur Institute, Lille, France) for sharing confocal image with the AFM tip.
Footnotes
Authors declare no conflicting financial interest.
References
- 1.Legewie S, Blüthgen N, Herzel H. Mathematical modeling identifies inhibitors of apoptosis as mediators of positive feedback and bistability. PLoS Comput Biol. 2006;2:120. doi: 10.1371/journal.pcbi.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK. Modeling a Snap-Action, Variable-Delay Switch Controlling Extrinsic Cell Death. PLoS Biol. 2008;6:2831–52. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–32. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 5.Khosravi-Far R, Esposti MD. Death receptor signals to mitochondria. Cancer Biol Ther. 2004;3:1051–7. doi: 10.4161/cbt.3.11.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy ME, Leu JI, George DL. p53 moves to mitochondria: a turn on the path to apoptosis. Cell Cycle. 2004;3:836–9. [PubMed] [Google Scholar]
- 7.Muñoz-Pinedo C, Guío-Carrión A, Goldstein JC, Fitzgerald P, Newmeyer DD, Green DR. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc Natl Acad Sci USA. 2006;103:11573–8. doi: 10.1073/pnas.0603007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohr A, Zwacka RM, Debatin KM, Stahnke K. A novel method for the combined flow cytometric analysis of cell cycle and cytochrome c release. Cell Death Differ. 2004;11:1153–4. doi: 10.1038/sj.cdd.4401480. [DOI] [PubMed] [Google Scholar]
- 9.Valentijn AJ, Upton JP, Gilmore AP. Analysis of endogenous Bax complexes during apoptosis using blue native PAGE: implications for Bax activation and oligomerization. Biochem J. 2008;412:347–57. doi: 10.1042/BJ20071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh I, Tomimori Y, Ikawa Y, Kurata S. Dimerization and processing of procaspase-9 by redox stress in mitochondria. J Biol Chem. 2004;279:15515–23. doi: 10.1074/jbc.M311819200. [DOI] [PubMed] [Google Scholar]
- 11.Brooks C, Dong Z. Regulation of mitochondrial morphological dynamics during apoptosis by Bcl-2 family proteins: a key in Bak? Cell Cycle. 2007;6:3043–7. doi: 10.4161/cc.6.24.5115. [DOI] [PubMed] [Google Scholar]
- 12.Luetjens CM, Kögel D, Reimertz C, Düssmann H, Renz A, Schulze-Osthoff K, et al. Multiple kinetics of mitochondrial cytochrome c release in drug-induced apoptosis. Mol Pharmacol. 2001;60:1008–19. doi: 10.1124/mol.60.5.1008. [DOI] [PubMed] [Google Scholar]
- 13.Bhola PD, Mattheyses AL, Simon SM. Spatial and temporal dynamics of mitochondrial membrane permeability waves during apoptosis. Biophys J. 2009;97:2222–31. doi: 10.1016/j.bpj.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000;2:156–62. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 15.Heiskanen KM, Bhat MB, Wang HW, Ma J, Nieminen AL. Mitochondrial depolarization accompanies cyto-chrome c release during apoptosis in PC6 cells. J Biol Chem. 1999;274:5654–8. doi: 10.1074/jbc.274.9.5654. [DOI] [PubMed] [Google Scholar]
- 16.Rehm M, Düssmann H, Prehn JH. Real-time single cell analysis of Smac/DIABLO release during apoptosis. J Cell Biol. 2003;162:1031–43. doi: 10.1083/jcb.200303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–92. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Chang DC. Dynamics and structure of the Bax-Bak complex responsible for releasing mitochondrial proteins during apoptosis. J Cell Sci. 2008;121:2186–96. doi: 10.1242/jcs.024703. [DOI] [PubMed] [Google Scholar]
- 19.Pollok BA, Heim R. Using GFP in FRET-based applications. Trends Cell Biol. 1999;9:57–60. doi: 10.1016/s0962-8924(98)01434-2. [DOI] [PubMed] [Google Scholar]
- 20.Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–8. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 21.Harpur AG, Wouters FS, Bastiaens PI. Imaging FRET between spectrally similar GFP molecules in single cells. Nat Biotechnol. 2001;19:167–9. doi: 10.1038/84443. [DOI] [PubMed] [Google Scholar]
- 22.Onuki R, Nagasaki A, Kawasaki H, Baba T, Uyeda TQ, Taira K. Confirmation by FRET in individual living cells of the absence of significant amyloid beta-mediated caspase 8 activation. Proc Natl Acad Sci USA. 2002;99:14716–21. doi: 10.1073/pnas.232177599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao X, Deng X, May WS. Cleavage of Bax to p18 Bax accelerates stress-induced apoptosis, and a cathepsin-like protease may rapidly degrade p18 Bax. Blood. 2003;102:2605–14. doi: 10.1182/blood-2003-01-0211. [DOI] [PubMed] [Google Scholar]
- 24.Gräf R, Rietdorf J, Zimmermann T. Live cell spinning disk microscopy. Adv Biochem Eng Biotechnol. 2005;95:57–75. doi: 10.1007/b102210. [DOI] [PubMed] [Google Scholar]
- 25.Zhivotovsky B. Apoptosis, necrosis and between. Cell Cycle. 2004;3:64–6. [PubMed] [Google Scholar]
- 26.Lee BW, Johnson GL, Hed SA, Darzynkiewicz Z, Talhouk JW, Mehrotra S. DEVDase detection in intact apoptotic cells using the cell permeant fluorogenic substrate, (z-DEVD)2-cresyl violet. Biotechniques. 2003;35:1080–5. doi: 10.2144/03355pf01. [DOI] [PubMed] [Google Scholar]
- 27.Packard BZ, Toptygin DD, Komoriya A, Brand L. Profluorescent protease substrates: intramolecular dimers described by the exciton model. Proc Natl Acad Sci USA. 1996;93:11640–5. doi: 10.1073/pnas.93.21.11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komoriya A, Packard BZ, Brown MJ, Wu ML, Henkart PA. Assessment of caspase activities in intact apoptotic thymocytes using cell-permeable fluorogenic caspase substrates. J Exp Med. 2000;191:1819–28. doi: 10.1084/jem.191.11.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Packard BZ, Komoriya A, Brotz TM, Henkart PA. Caspase 8 activity in membrane blebs after anti-Fas ligation. J Immunol. 2001;167:5061–6. doi: 10.4049/jimmunol.167.9.5061. [DOI] [PubMed] [Google Scholar]
- 30.Packard BZ, Komoriya A. Intracellular protease activation in apoptosis and cell-mediated cytotoxicity characterized by cell-permeable fluorogenic protease substrates. Cell Res. 2008;18:238–47. doi: 10.1038/cr.2008.17. [DOI] [PubMed] [Google Scholar]
- 31.Telford WG, Komoriya A, Packard BZ. Detection of localized caspase activity in early apoptotic cells by laser scanning cytometry. Cytometry. 2002;47:81–8. doi: 10.1002/cyto.10052. [DOI] [PubMed] [Google Scholar]
- 32.Cen H, Mao F, Aronchik I, Fuentes RJ, Firestone GL. DEVD-NucView488: a novel class of enzyme substrates for real-time detection of caspase-3 activity in live cells. FASEB J. 2008;22:2243–52. doi: 10.1096/fj.07-099234. [DOI] [PubMed] [Google Scholar]
- 33.Antczak C, Takagi T, Ramirez CN, Radu C, Djaballah H. Live-cell imaging of caspase activation for high-content screening. J Biomol Screen. 2009;14:956–69. doi: 10.1177/1087057109343207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JH. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J Biol Chem. 2002;277:24506–14. doi: 10.1074/jbc.M110789200. [DOI] [PubMed] [Google Scholar]
- 35.Rehm M, Huber HJ, Dussmann H, Prehn JH. Systems analysis of effector caspase activation and its control by X-linked inhibitor of apoptosis protein. EMBO J. 2006;25:4338–49. doi: 10.1038/sj.emboj.7601295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takemoto K, Nagai T, Miyawaki A, Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J Cell Biol. 2003;160:235–43. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyas L, Brophy VA, Pope A, Rivett AJ, Tavaré JM. Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer. EMBO Rep. 2000;1:266–70. doi: 10.1093/embo-reports/kvd050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones J, Heim R, Hare E, Stack J, Pollok BA. Development and application of a GFP-FRET intrac-ellular caspase assay for drug screening. J Biomol Screen. 2000;5:307–18. doi: 10.1177/108705710000500502. [DOI] [PubMed] [Google Scholar]
- 39.Tawa P, Tam J, Cassady R, Nicholson DW, Xanthoudakis S. Quantitative analysis of fluorescent caspase substrate cleavage in intact cells and identification of novel inhibitors of apoptosis. Cell Death Differ. 2001;8:30–7. doi: 10.1038/sj.cdd.4400769. [DOI] [PubMed] [Google Scholar]
- 40.Luo KQ, Yu VC, Pu Y, Chang DC. Measuring dynamics of caspase-8 activation in a single living HeLa cell during TNFalpha-induced apoptosis. Biochem Biophys Res Commun. 2003;304:217–22. doi: 10.1016/s0006-291x(03)00559-x. [DOI] [PubMed] [Google Scholar]
- 41.Xu X, Gerard AL, Huang BC, Anderson DC, Payan DG, Luo Y. Detection of programmed cell death using fluorescence energy transfer. Nucleic Acids Res. 1998;26:2034–5. doi: 10.1093/nar/26.8.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Xing D, Chen WR. Single cell FRET imaging for determination of pathway of tumor cell apoptosis induced by photofrin-PDT. Cell Cycle. 2006;5:729–34. doi: 10.4161/cc.5.7.2630. [DOI] [PubMed] [Google Scholar]
- 43.Shcherbo D, Souslova EA, Goedhart J, Chepurnykh TV, Gaintzeva A, Shemiakina II, et al. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol. 2009;9:24. doi: 10.1186/1472-6750-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–60. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Simone J, Hewgill D, Siegel R, Lipsky PE, He L. Measurement of two caspase activities simultaneously in living cells by a novel dual FRET fluorescent indicator probe. Cytometry A. 2006;69:477–86. doi: 10.1002/cyto.a.20300. [DOI] [PubMed] [Google Scholar]
- 46.Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidyl-serine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148:2207–16. [PubMed] [Google Scholar]
- 47.Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84:1415–20. [PubMed] [Google Scholar]
- 48.van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31:1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 49.Kietselaer BL, Hofstra L, Dumont EA, Reutelingsperger CP, Heidendal GA. The role of labeled Annexin A5 in imaging of programmed cell death. From animal to clinical imaging. Q J Nucl Med. 2003;47:349–61. [PubMed] [Google Scholar]
- 50.De Saint-Hubert M, Prinsen K, Mortelmans L, Verbruggen A, Mottaghy FM. Molecular imaging of cell death. Methods. 2009;48:178–87. doi: 10.1016/j.ymeth.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 51.Belhocine TZ, Blankenberg FG. The imaging of apoptosis with the radiolabelled annexin A5: a new tool in translational research. Curr Clin Pharmacol. 2006;1:129–37. doi: 10.2174/157488406776872541. [DOI] [PubMed] [Google Scholar]
- 52.Faley SL, Copland M, Wlodkowic D, Kolch W, Seale KT, Wikswo JP, et al. Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab Chip. 2009;9:2659–64. doi: 10.1039/b902083g. [DOI] [PubMed] [Google Scholar]
- 53.Dicker DT, Kim SH, Jin Z, El-Deiry WS. Heterogeneity in non-invasive detection of apoptosis among human tumor cell lines using annexin-V tagged with EGFP or Qdot-705. Cancer Biol Ther. 2005;4:1014–7. doi: 10.4161/cbt.4.9.2150. [DOI] [PubMed] [Google Scholar]
- 54.Le Gac S, Vermes I, van den Berg A. Quantum dots based probes conjugated to annexin V for photostable apoptosis detection and imaging. Nano Lett. 2006;6:1863–9. doi: 10.1021/nl060694v. [DOI] [PubMed] [Google Scholar]
- 55.Kim YE, Chen J, Chan JR, Langen R. Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration. Nat Methods. 2010;7:67–73. doi: 10.1038/nmeth.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaiswal JK, Simon SM. Potentials and pitfalls of fluorescent quantum dots for biological imaging. Trends Cell Biol. 2004;14:497–504. doi: 10.1016/j.tcb.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 57.Koeppel F, Jaiswal JK, Simon SM. Quantum dot-based sensor for improved detection of apoptotic cells. Nanomed. 2007;2:71–8. doi: 10.2217/17435889.2.1.71. [DOI] [PubMed] [Google Scholar]
- 58.van Tilborg GA, Mulder WJ, Chin PT, Storm G, Reutelingsperger CP, Nicolay K, et al. Annexin A5-conjugated quantum dots with a paramagnetic lipidic coating for the multimodal detection of apoptotic cells. Bioconjug Chem. 2006;17:865–8. doi: 10.1021/bc0600463. [DOI] [PubMed] [Google Scholar]
- 59.Zhao L, Cheng P, Li J, Zhang Y, Gu M, Liu J, et al. Analysis of Nonadherent Apoptotic Cells by a Quantum Dots Probe in a Microfluidic Device for Drug Screening. Anal Chem. 2009 doi: 10.1021/ac901121f. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 60.Zhao H, Traganos F, Dobrucki J, Wlodkowic D, Darzynkiewicz Z. Induction of DNA damage response by the supravital probes of nucleic acids. Cytometry A. 2009;75:510–9. doi: 10.1002/cyto.a.20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wlodkowic D, Skommer J, Darzynkiewicz Z. Flow cytometry-based apoptosis detection. Methods Mol Biol. 2009;559:19–32. doi: 10.1007/978-1-60327-017-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wlodkowic D, Skommer J, McGuinness D, Faley S, Kolch W, Darzynkiewicz Z, et al. Chip-Based Dynamic Real-Time Quantification of Drug-Induced Cytotoxicity in Human Tumor Cells. Anal Chem. 2009;81:6952–9. doi: 10.1021/ac9010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wlodkowic D, Faley S, Zagnoni M, Wikswo JP, Cooper JM. Microfluidic single-cell array cytometry for the analysis of tumor apoptosis. Anal Chem. 2009;81:5517–23. doi: 10.1021/ac9008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenfeldt MT, Ryan KM. The role of autophagy in tumour development and cancer therapy. Expert Rev Mol Med. 2009;11:36. doi: 10.1017/S1462399409001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu L, Lenardo MJ, Baehrecke EH. Autophagy and caspases: a new cell death program. Cell Cycle. 2004;3:1124–6. [PubMed] [Google Scholar]
- 66.Tasdemir E, Galluzzi L, Maiuri MC, Criollo A, Vitale I, Hangen E, et al. Methods for assessing autophagy and autophagic cell death. Methods Mol Biol. 2008;445:29–76. doi: 10.1007/978-1-59745-157-4_3. [DOI] [PubMed] [Google Scholar]
- 67.O’Prey J, Skommer J, Wilkinson S, Ryan KM. Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy. Cell Cycle. 2009;8:2260–5. doi: 10.4161/cc.8.14.9050. [DOI] [PubMed] [Google Scholar]
- 68.Farkas T, Høyer-Hansen M, Jäättelä M. Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy. 2009;5:1018–25. doi: 10.4161/auto.5.7.9443. [DOI] [PubMed] [Google Scholar]
- 69.Jun YW, Sheikholeslami S, Hostetter DR, Tajon C, Craik CS, Alivisatos AP. Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level. Proc Natl Acad Sci USA. 2009;106:17735–40. doi: 10.1073/pnas.0907367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lasne D, Blab GA, Berciaud S, Heine M, Groc L, Choquet D, et al. Single nanoparticle photothermal tracking (SNaPT) of 5-nm gold beads in live cells. Biophys J. 2006;91:4598–604. doi: 10.1529/biophysj.106.089771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Giorgi F, Lartigue L, Bauer MK, Schubert A, Grimm S, Hanson GT, et al. The permeability transition pore signals apoptosis by directing Bax translocation and multimerization. FASEB J. 2002;16:607–9. doi: 10.1096/fj.01-0269fje. [DOI] [PubMed] [Google Scholar]
- 72.Minor LK. Label-free cell-based functional assays. Comb Chem High Throughput Screen. 2008;11:573–80. doi: 10.2174/138620708785204072. [DOI] [PubMed] [Google Scholar]
- 73.Sohn LL, Saleh OA, Facer GR, Beavis AJ, Allan RS, Notterman DA. Capacitance cytometry: measuring biological cells one by one. Proc Natl Acad Sci USA. 2000;97:10687–90. doi: 10.1073/pnas.200361297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tong C, Shi B, Xiao X, Liao H, Zheng Y, Shen G, et al. An Annexin V-based biosensor for quantitatively detecting early apoptotic cells. Biosens Bioelectron. 2009;24:1777–82. doi: 10.1016/j.bios.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 75.Chan LL, Gosangari SL, Watkin KL, Cunningham BT. A label-free photonic crystal biosensor imaging method for detection of cancer cell cytotoxicity and proliferation. Apoptosis. 2007;12:1061–8. doi: 10.1007/s10495-006-0031-y. [DOI] [PubMed] [Google Scholar]
- 76.Tamaki E, Sato K, Tokeshi M, Sato K, Aihara M, Kitamori T. Single-cell analysis by a scanning thermal lens microscope with a microchip: direct monitoring of cytochrome c distribution during apoptosis process. Anal Chem. 2002;74:1560–4. doi: 10.1021/ac011092t. [DOI] [PubMed] [Google Scholar]
- 77.Matthäus C, Bird B, Miljkovi_ M, Chernenko T, Romeo M, Diem M. Chapter 10: Infrared and Raman microscopy in cell biology. Methods Cell Biol. 2008;89:275–308. doi: 10.1016/S0091-679X(08)00610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Toms SA, Konrad PE, Lin WC, Weil RJ. Neurooncological applications of optical spectroscopy. Technol Cancer Res Treat. 2006;5:231–8. doi: 10.1177/153303460600500306. [DOI] [PubMed] [Google Scholar]
- 79.Notingher I, Hench LL. Raman microspectroscopy: a noninvasive tool for studies of individual living cells in vitro. Expert Rev Med Devices. 2006;3:215–34. doi: 10.1586/17434440.3.2.215. [DOI] [PubMed] [Google Scholar]
- 80.Uzunbajakava N, Lenferink A, Kraan Y, Volokhina E, Vrensen G, Greve J, et al. Nonresonant confocal Raman imaging of DNA and protein distribution in apoptotic cells. Biophys J. 2003;84:3968–81. doi: 10.1016/S0006-3495(03)75124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matthäus C, Chernenko T, Newmark JA, Warner CM, Diem M. Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy. Biophys J. 2007;93:668–73. doi: 10.1529/biophysj.106.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kunapareddy N, Freyer JP, Mourant JR. Raman spectroscopic characterization of necrotic cell death. J Biomed Opt. 2008;13:54002. doi: 10.1117/1.2978061. [DOI] [PubMed] [Google Scholar]
- 83.Swain RJ, Jell G, Stevens MM. Non-invasive analysis of cell cycle dynamics in single living cells with Raman micro-spectroscopy. J Cell Biochem. 2008;104:1427–38. doi: 10.1002/jcb.21720. [DOI] [PubMed] [Google Scholar]
- 84.Kneipp J, Kneipp H, Rice WL, Kneipp K. Optical probes for biological applications based on surface-enhanced Raman scattering from indocyanine green on gold nanoparticles. Anal Chem. 2005;77:2381–5. doi: 10.1021/ac050109v. [DOI] [PubMed] [Google Scholar]
- 85.Kneipp J, Kneipp H, Kneipp K. SERS—a single-molecule and nanoscale tool for bioanalytics. Chem Soc Rev. 2008;37:1052–60. doi: 10.1039/b708459p. [DOI] [PubMed] [Google Scholar]
- 86.Jett JH. Raman spectroscopy comes to flow cytometry. Cytometry A. 2008;73:109–10. doi: 10.1002/cyto.a.20526. [DOI] [PubMed] [Google Scholar]
- 87.Kneipp J, Kneipp H, Wittig B, Kneipp K. Novel optical nanosensors for probing and imaging live cells. Nanomedicine. 2009 doi: 10.1016/j.nano.2009.07.009. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 88.Kneipp J, Kneipp H, Wittig B, Kneipp K. One- and two-photon excited optical ph probing for cells using surface-enhanced Raman and hyper-Raman nanosensors. Nano Lett. 2007;7:2819–23. doi: 10.1021/nl071418z. [DOI] [PubMed] [Google Scholar]
- 89.Pyrgiotakis G, Kundakcioglu OE, Finton K, Pardalos PM, Powers K, Moudgil BM. Cell death discrimination with Raman spectroscopy and support vector machines. Ann Biomed Eng. 2009;37:1464–73. doi: 10.1007/s10439-009-9688-z. [DOI] [PubMed] [Google Scholar]
- 90.Hessler JA, Budor A, Putchakayala K, Mecke A, Rieger D, Banaszak Holl MM, et al. Atomic force microscopy study of early morphological changes during apoptosis. Langmuir. 2005;21:9280–6. doi: 10.1021/la051837g. [DOI] [PubMed] [Google Scholar]
- 91.Kuznetsov YG, Malkin AJ, McPherson A. Atomic force microscopy studies of living cells: visualization of motility, division, aggregation, transformation and apoptosis. J Struct Biol. 1997;120:180–91. doi: 10.1006/jsbi.1997.3936. [DOI] [PubMed] [Google Scholar]
- 92.Jena BP. Atomic force microscopy: Unraveling the fundamental principles governing secretion and membrane fusion in cells. Ultramicroscopy. 2009;109:1094–104. doi: 10.1016/j.ultramic.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Frederix PL, Bosshart PD, Engel A. Atomic force microscopy of biological membranes. Biophys J. 2009;96:329–38. doi: 10.1016/j.bpj.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuznetsov YG, Malkin AJ, McPherson A. Atomic force microscopy studies of living cells: visualization of motility, division, aggregation, transformation and apoptosis. J Struct Biol. 1997;120:180–91. doi: 10.1006/jsbi.1997.3936. [DOI] [PubMed] [Google Scholar]
- 95.Kumar S, Weaver VM. Mechanics, malignancy and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–27. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dufrêne YF. Towards nanomicrobiology using atomic force microscopy. Nat Rev Microbiol. 2008;6:674–80. doi: 10.1038/nrmicro1948. [DOI] [PubMed] [Google Scholar]
- 97.Parot P, Dufrêne YF, Hinterdorfer P, Le Grimellec C, Navajas D, Pellequer JL, et al. Past, present and future of atomic force microscopy in life sciences and medicine. J Mol Recognit. 2007;20:418–31. doi: 10.1002/jmr.857. [DOI] [PubMed] [Google Scholar]
- 98.Darzynkiewicz Z, Crissman HA, Traganos F, Steinkamp J. Cell heterogeneity during the cell cycle. J Cell Physiol. 1982;112:465–74. doi: 10.1002/jcp.1041130316. [DOI] [PubMed] [Google Scholar]
- 99.Errington RJ, Brown MR, Silvestre OF, Njoh KL, Chappell SC, Khan IA, et al. Single cell nanoparticle tracking to model cell cycle dynamics and compartmental inheritance. Cell Cycle. 2010;9:121–30. doi: 10.4161/cc.9.1.10246. [DOI] [PubMed] [Google Scholar]
- 100.Huang X, Lee B, Johnson G, Naleway J, Guzikowski A, Dai W, Darzynkiewicz Z. Novel assay utilizing fluorochrome-tagged physostigmine (Ph-F) to in situ detect active acetylcholinesterase (AChE) induced during apoptosis. Cell Cycle. 2005;4:140–7. doi: 10.4161/cc.4.1.1322. [DOI] [PubMed] [Google Scholar]
- 101.Sigal A, Danon T, Cohen A, Milo R, Geva-Zatorsky N, Lustig G, et al. Generation of a fluorescently labeled endogenous protein library in living human cells. Nat Protoc. 2007;2:1515–27. doi: 10.1038/nprot.2007.197. [DOI] [PubMed] [Google Scholar]
- 102.Cohen-Saidon C, Cohen AA, Sigal A, Liron Y, Alon U. Dynamics and variability of ERK2 response to EGF in individual living cells. Mol Cell. 2009;36:885–93. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 103.Aguda BD, Algar CK. A structural analysis of the qualitative networks regulating the cell cycle and apoptosis. Cell Cycle. 2003;2:538–44. doi: 10.4161/cc.2.6.550. [DOI] [PubMed] [Google Scholar]
- 104.Raychaudhuri S, Skommer J, Henty K, Birch N, Brittain T. Neuroglobin protects nerve cells from apoptosis by inhibiting the intrinsic pathway of cell death. Apoptosis. 2009;15:401–11. doi: 10.1007/s10495-009-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Raychaudhuri S, Willgohs E, Nguyen TN, Khan EM, Goldkorn T. Monte Carlo simulation of cell death signaling predicts large cell-to-cell stochastic fluctuations through the type 2 pathway of apoptosis. Biophys J. 2008;95:3559–62. doi: 10.1529/biophysj.108.135483. [DOI] [PMC free article] [PubMed] [Google Scholar]