Abstract
The d-enantiomers of amino acids have been thought to have relatively minor functions in biological processes. While l-amino acids clearly predominate in nature, d-amino acids are sometimes found in proteins that are not synthesized by ribosomes, and d-Ala and d-Glu are routinely found in the peptidoglycan cell wall of bacteria. Here, we review recent findings showing that d-amino acids have previously unappreciated regulatory roles in the bacterial kingdom. Many diverse bacterial phyla synthesize and release d-amino acids, including d-Met and d-Leu, which were not previously known to be made. These noncanonical d-amino acids regulate cell wall remodeling in stationary phase and cause biofilm dispersal in aging bacterial communities. Elucidating the mechanisms by which d-amino acids govern cell wall remodeling and biofilm disassembly will undoubtedly reveal new paradigms for understanding how extracytoplasmic processes are regulated as well as lead to development of novel therapeutics.
Keywords: d-amino acid, Racemase, Stationary phase, Peptidoglycan, Biofilm, Regulation
Introduction
Objects are described as chiral if their reflected image in a mirror cannot be superimposed on the original. The term chiral is derived from the Greek word for hand, χειρ, as human hands provide a prime example of chirality. While very similar in almost all characteristics, the mirror image of a left hand—the right hand—cannot be superimposed on itself and interacts with other objects in a distinct manner [1]. Natural objects, such as vertebrate appendages, that exhibit chirality are readily apparent. Chirality is also widespread at the molecular level. Remarkably, in 1848 at the age of 25, before he carried out his pioneering studies in microbiology, the great French scientist Louis Pasteur (1822–1895) discovered molecular chirality in mirror-image tartaric acid crystals (revisited in [2]). Nine years later, while studying the fermentation of tartaric acid by microorganisms, Pasteur discovered that one mirror image of tartaric acid was consumed with greater preference over the other [2, 3]. In the ensuing 150 years, the great importance of molecular chirality in many biochemical processes has become clear.
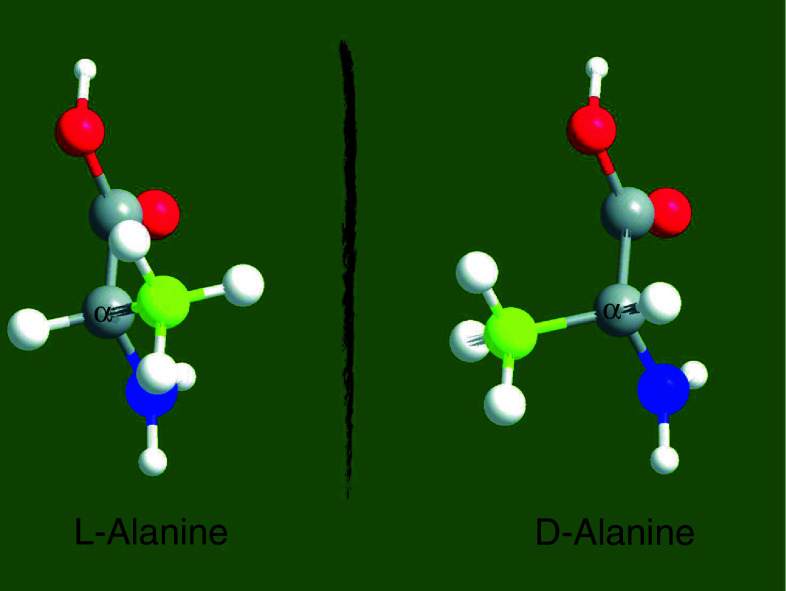
The two mirror images of a chiral molecule are referred to as optical isomers or enantiomers (from the Greek εχθρóς, “enemy”). Most physical properties of enantiomers are identical; however, they have distinct interactions with plane-polarized light. When plane-polarized light passes through a solution of chiral molecules, one enantiomer rotates the plane of polarization in a counterclockwise (left-hand) direction whereas the other enantiomer rotates the plane of polarization in a clockwise (right-hand) direction. In biological molecules, the most common basis of chirality is a carbon atom bonded to four different groups, which is termed a stereocenter (or chiral center). The three-dimensional arrangement of the atoms bound to a chiral center can be used to describe a molecule’s chirality. Different nomenclature systems have been developed to describe the chirality of molecules, but d- and l- prefixes are generally used for amino acids and sugars as described in [4]. The d/l nomenclature for amino acid configuration does not refer to the optical activity of the amino acid per se, but rather to the optical activity of the glyceraldehyde isomer that the amino acid can be overlayed upon. The structures of the l- and d-enantiomers of alanine are shown in Fig. 1.
Fig. 1.
Chirality of alanine. Ball-and-sticks representation of the enantiomeric forms of the amino acid alanine. Carboxyl group is colored in red, amino group in blue, and R-group in yellow. Chiral carbon is labeled as α. The molecules were designed with ChemDraw Ultra 12.0 and Chem3D Pro 12.0 software
While chemical synthesis routinely yields both enantiomers, most enzymes display marked substrate selectivity, and as a consequence many biochemical processes generally utilize and yield particular enantiomers. For example, l-amino acids are the predominant building blocks of proteins; d-amino acids cannot be incorporated into proteins via ribosomal synthesis. Similarly, nearly all naturally occurring monosaccharides exist as d-sugars. The origins of enantiospecificity in biological processes are unclear, but it has been proposed that enantiomerically enriched organic compounds were critical for the generation of proto-life forms [5–7].
Even though l-amino acids are the dominant substrates for ribosome-based protein synthesis, several roles for d-amino acids in other biological processes have been described. For example, d-aspartate is a major regulator of adult neurogenesis [8] and d-serine acts as a co-agonist of the N-methyl d-aspartate-type glutamate receptors in the brain, which are involved in learning, memory, and behavior in mammals [9–11]. d-serine is also the most abundant amino acid in human urine and alters gene expression of uropathogenic Escherichia coli (UPEC) [12].
It was noted more than 50 years ago that, in addition to free amino acids, some peptides contain d-amino acids [13]. d-configured residues in peptides provide resistance to proteases, which generally exhibit specificity for l-amino acid-containing peptides, and also contribute to their bioactivity (listed in Tables 1, 2). d-amino acids are incorporated into the peptides via two different mechanisms. The first mechanism is posttranslational conversion of l- to d-amino acids within peptides that were originally synthesized in ribosomes. The second mechanism requires the activity of nonribosomal peptide (NRP) synthetases, which, unlike ribosomal peptide synthesis, generates peptides independent of messenger RNA. While posttranslational modification occurs primarily in eukaryotes, NRP synthesis is more frequent in bacteria. For example, dermorphin is an analgesic 1,000 times more potent than morphine [14, 15], achatin-I is an excitatory neurotransmitter [16–18], and gramicidines and tyrocidines have antimicrobial activity [19–23]. Considering that Tables 1 and 2 summarize most of the known examples in the literature, d-amino acid-containing peptides seem relatively unusual. However, peptides containing d-amino acids have probably been overlooked. Furthermore, there have been increasing numbers of reports on this subject in the last few decades suggesting that there are likely to be many more yet-to-be-discovered examples of d-amino acids in nature.
Table 1.
d-amino acids in eukaryotic peptides
| Peptide | d-amino acid (position) | Source | Activity | Reference |
|---|---|---|---|---|
| Dermorphin | d-Ala (2nd) | Phyllomedusa sauvagei (skin secretions of Argentinian tree frog) | Binds to μ-type opiate receptors and acts as an analgesic 1,000 times more powerful than morphine | [14, 15] |
| Deltorphins | d-Met or d-Ala (2nd) | Phyllomedusa species (skin secretions) | Binds to δ-type opiate receptors | [107, 108] |
| Bombinins and bombinins H | d-allo-Ile (2nd) | Bombinatoridae (skin secretions of frogs) | Antimicrobial and hemolytic activity | [109–111] |
| Achatin-I | d-Phe (2nd) | Achatina fulica (ganglia and atrium of African snail) | Excitatory neurotransmitter controlling muscle contraction | [16–18] |
| Fucilin | d-Asn (2nd) | A. fulica (ganglia of African snail) | Excitatory neurotransmitter controlling penis contraction | [112, 113] |
| Contryphans | d-Trp (3rd or 4th) | Conus purpurascens and C. radiatus (venom of cone snail) | Causes tremor and mucous secretions when injected into fish | [114–116] |
| d-Leu (5th) | ||||
| FRF amide family | d-Leu (2nd) | Bivalves | Stimulates muscle contraction | [117] |
| Crustacean hyperglycemic hormone | d-Phe (3rd) | Decapod crustaceans | Neurohormone controlling hyperglycemia | [118, 119] |
| ω-Agatoxin-IVB | d-Ser (46th) | Agelenopsis aperta (venom of funnel-web spider) | Blocks voltage-sensitive calcium channels | [120, 121] |
| Paecilodepsipeptide A | Three d-amino acid residues including an unusual O-prenyl-d-Tyr | Insect pathogenic fungus Paecilomyces cinnamomeus BCC 9616 | Activity against the malarial parasite Plasmodium falciparum K1; cytotoxicity to cancer cell lines (KB and BC) | [122] |
Table 2.
d-amino acids in bacterial peptides
| Peptide | d-amino acid (position) | Source | Activity | Reference |
|---|---|---|---|---|
| Gramicidine S | d-Phe (4th, 9th) | Bacillus brevis (antimicrobial peptides) | Membrane disruption of lipid bilayer | [19, 20] |
| Gramicidine D | d-Leu (4th, 10th, 12th, 14th) | B. brevis (antimicrobial peptides) | Permeabilizes lipid membranes by forming ion channels that disrupt ion gradient | [20, 21] |
| d-Val (6th, 8th) | ||||
| Tyrocidines | d-Phe (1st) | B. brevis (antimicrobial peptides) | Permeabilizes lipid membranes | [22, 23] |
| d-Phe or d-Tyr (4th) | ||||
| Daptomycin (Cubicin) | d-Ala (8th) | Streptomycetes (S. roseosporus) | Bactericidal activity | [123–125] |
| Lipopeptidolactones | Arthrofactin (7 d-amino acids of the 11) | Pseudomonas strains | Biosurfactants | [126–132] |
| Syringomycin (2 d-amino acids of the 9) | ||||
| Syringopeptin (about 70% of the sequence is in d-configuration) | ||||
| DD-diketopiperazines | Only d-amino acids | Bacterial strains CF-20 (CECT5719) and C-148 (CECT5718), isolated from cultures of larvae of mollusks | Strong antibiotic activity against Vibrio anguillarum | [133] |
| Gassericin A | 1 d-Ala residue | Lactobacillus gasseri LA39 | Antimicrobial activity against Listeria monocytogenes, Bacillus cereus, and S. aureus | [134–136] |
Besides being occasionally incorporated in peptides, d-amino acids have been known to be utilized as nutrients to support bacterial growth [24–27], to regulate bacterial spore germination, and to be components of the bacterial cell wall. We briefly summarize the latter two previously recognized roles of d-amino acids below. Then, we discuss recent findings showing that d-amino acids have previously unappreciated regulatory roles in the bacterial kingdom.
d-amino acids and bacterial spore germination
Bacterial sporulation is an adaptive response to environmental stress, such as starvation, and involves a finely controlled developmental program. Initiation of sporulation leads to asymmetric cell division producing a mother cell and forespore with distinct cell fates. The forespore develops into the spore, whereas the mother cell nurtures the spore but ultimately lyses through programmed cell death [28].
Bacterial spores are metabolically dormant and resistant to a number of harsh environments including heat, radiation, desiccation, pH extremes, and toxic chemicals [29]. This remarkable resistance is provided by a tough multilayered cell wall [30]. In the presence of specific germinants such as l-alanine or other nutrients, spores can reactivate metabolism and grow vegetatively. In 1949, it was discovered that d-alanine was a potent inhibitor of spore germination in many Bacillus species [31]. Subsequent work since this discovery has revealed that Bacillus species utilize d-alanine as an auto-inhibitor of spore germination at high spore density. This activity is mediated through expression of an alanine racemase in the spore exosporium that converts a spore germinant (l-Ala) to an anti-germinant (d-Ala) [32]. A nutrient receptor is responsible for recognition of l-Ala, and it has been suggested that d-Ala antagonizes l-Ala binding to this receptor [33]. Presumably, this mechanism of auto-inhibition is an evolutionary adaptation to prevent premature germination under low nutrient conditions and high population density, an environmental condition that would lead to rapid nutrient depletion and cell death. Recent work has also suggested that d-Ala alters the kinetics of germination in vivo to enhance the efficiency and timing of infection [34]. d-His has also been implicated as a germination inhibitor of Bacillus anthracis infection in murine macrophages [35], however its mechanism of action and whether it is physiologically produced by B. anthracis is unknown.
d-amino acids in peptidoglycan
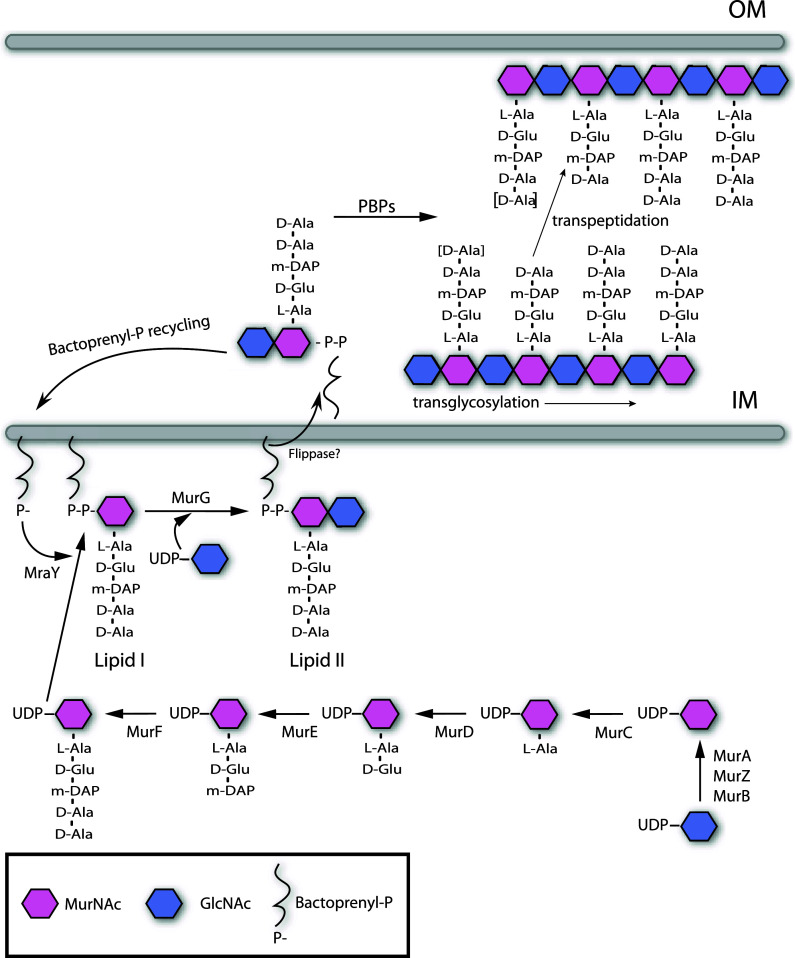
Bacteria have a formidable ability to withstand many physical, chemical, and biological insults. In large part, this is due to the peptidoglycan (PG) cell wall, which imparts to the cell its shape, strength, and resistance to osmotic pressure [36–38]. PG also serves as a scaffold for anchoring other cell envelope components [39, 40]. PG (also known as murein) is found on the outside of the cytoplasmic membrane of almost all bacteria [36, 41, 42]. It is a strong yet flexible net-like polymer composed of linear glycan strands made up of repeating disaccharide units of N-acetyl glucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) cross-linked by short peptides (Fig. 2). PG is essential for cell viability and therefore its synthesis and turnover must be tightly controlled; otherwise, the mechanical stability of the cell wall and cell integrity would be compromised. In gram-negative bacteria, a single layer of PG, which is found in the periplasmic space between the inner and outer cell membranes, is sufficient to maintain the cell’s mechanical stability [43]. In gram-positive bacteria, which lack an outer cell membrane, the cell wall is thicker, consisting of many layers of PG.
Fig. 2.
Biosynthesis of peptidoglycan (PG). General scheme of PG synthesis in gram negative bacteria. PG synthesis is initiated with the synthesis of the disaccharide pentapeptide precursors in the cytosol (GlcNAc-MurNAc-l-Ala-d-Glu-DAP-d-Ala-d-Ala) [42]. Then, PG precursors are translocated to the periplasmic space facilitated by the formation of lipidic complexes with bactoprenol [43, 44]. Once in the periplasm, PG monomers are incorporated into the murein polymer by transglycosylation and transpeptidation reactions carried out by the activity of the penicillin-binding proteins (PBPs) [45–48]. Also PBP activities (murein hydrolases) can affect the length of the stem peptides, depicted as d-Ala between brackets [43]. IM Inner membrane, OM outer membrane
The biosynthesis of PG is divided into three stages (Fig. 2). In the first step, the PG precursors nucleotide sugar-linked UDP-MurNAc-pentapeptide and UDP-GlcNAc are synthesized in the cytoplasm [44]. Second, lipid intermediates of these precursors are formed by transferring the phospho-MurNAc-pentapeptide moiety of UDP-MurNAc-pentapeptide to the membrane acceptor bactoprenyl-P, yielding lipid I. Subsequently, GlcNAc from UDP-GlcNAc is added to lipid I, yielding lipid II. The lipid II intermediate enables the cell to transport the hydrophilic precursors from the aqueous environment of the cytoplasm through the hydrophobic membrane, where they can act as substrates for incorporation into the PG polymer [45, 46]. PG polymerization, the third stage of PG biosynthesis, occurs outside of the cell membrane in the periplasmic space of gram-negative bacteria and outside the cell entirely in gram-positive organisms. Polymerization is carried out by penicillin-binding proteins (PBPs). These enzymes catalyze both the transglycosylation and transpeptidation reactions required to incorporate new muropeptides into the PG polymer [47–50].
One of the most striking features of PG composition is the presence of d-amino acids in the stem peptides (Fig. 2) [51]. d-amino acids contribute to the architecture of the murein and, more importantly, provide resistance to most known proteases. Thus, the presence of d-amino acids in PG likely constitutes a bacterial adaptation to protect a vital cellular structure. d-Ala and d-Glu are by far the most common d-amino acids present in the bacterial cell wall; however, the PG of some bacteria include other d-amino acids, such as d-Asp in Lactococcus [52] and Enterococcus [53], and d-Ser in vancomycin-resistant Staphylococcus aureus [54–56]. In some cases, such substitutions enable bacteria to tolerate bactericidal agents from the environment [57].
Production of d-amino acids
Since l-amino acids are the predominant amino acids found in living organisms, typically they act as the substrate for generation of d-amino acids. l- to d-conversion occurs by the action of racemases that change the stereochemistry of the chiral α-carbon atom in amino acids [58]. Amino acid racemases are classified into two groups, pyridoxal-5-phosphate (PLP)-dependent and PLP-independent enzymes, with distinct reaction mechanisms.
PLP-dependent amino acid racemases
Alanine racemases
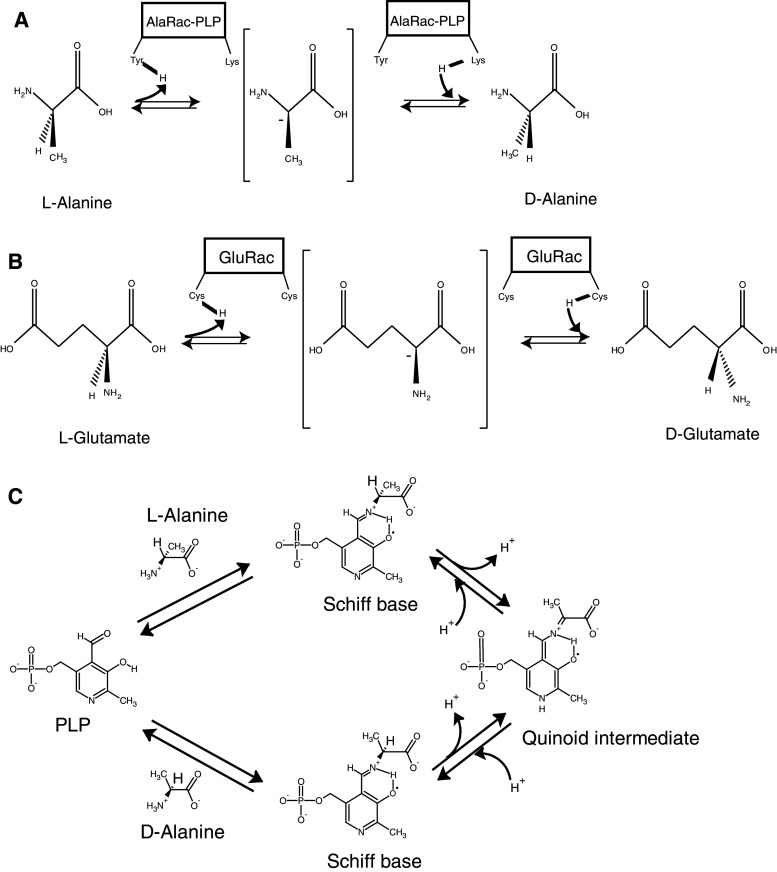
Alanine racemases use a PLP-dependent mechanism to deprotonate the α-carbon of alanine. Reprotonation of the α-carbon on the opposite side generates the antipodal amino acid (Fig. 3a). In the racemase molecule, PLP is bound to a Lys residue to form an internal Schiff base. The substrate alanine molecule reacts with PLP by transaldimination to form an external Schiff base. Abstraction of the α-hydrogen of the substrate amino acid moiety of the Schiff base generates an anionic form that is stabilized as a quinoid intermediate with the PLP moiety (Fig. 3c). This intermediate is subsequently protonated and releases the antipodal form of alanine through a second transaldimination reaction with the same Lys residue involved in the initial reaction, thus regenerating the enzyme (Fig. 3c) [59]. One- and two-base reaction mechanisms have been proposed for the abstraction/addition of the α-hydrogen. In the former the α-hydrogen of both isomers is abstracted and added with a single catalytic residue (Lys). In the latter the α-hydrogen of either d-alanine or l-alanine is abstracted and added by a different catalytic residue (Tyr and Lys, respectively). Although the two-base mechanism is more probable, further experimentation is under way to show this conclusively [60].
Fig. 3.
Mechanism of amino acid racemase reactions. a PLP-dependent alanine racemase (AlaRac) and b PLP-independent glutamate racemase (GluRac) reactions are depicted showing the amino acidic residues of the enzymes that participate in the stereochemical transformation of alanine and glutamic acid, respectively. The achiral anionic quinoid intermediate is shown in brackets. c Schematic of the catalytic mechanism of the PLP-dependent alanine racemase reaction. The molecules were designed with ChemDraw Ultra 12.0 software
Most bacteria encode two different alanine racemases, DadB and Alr. While both racemases use the same reaction mechanism, they are components of distinct molecular pathways. DadB participates in l-Ala catabolism, producing d-Ala to be used as a substrate to form pyruvate via a d-Ala dehydrogenase. In contrast, Alr generates the d-Ala that is utilized in formation of muropeptide precursors for PG synthesis [61, 62].
Serine racemase
Bacterial serine racemases are homologues of alanine racemases and play an important role in resistance to vancomycin [54, 56], a glycopeptide antibiotic that inhibits PG synthesis by binding to the d-Ala-d-Ala moiety in muropeptide precursors. In enterococci, vancomycin resistance arises through modification of the vancomycin target site from d-Ala-d-Ala to either d-Ala-d-lactate or d-Ala-d-Ser [63]. d-Ser is generated from l-Ser by VanT, a serine racemase, which, like alanine racemase, is also PLP-dependent and probably uses a two-base catalyzed reaction. The similarity of VanT to alanine racemase suggests that the two enzymes share a common evolutionary history [64].
PLP-independent amino acid racemases
Glutamate racemase
Like d-Ala, d-Glu is a component of the PG cell wall in bacteria, but this d-amino acid is produced by a PLP-independent racemase. Similar to alanine racemases, glutamate racemases are thought to follow a two-base mechanism, however, two cysteines have been suggested to be involved in the catalysis of the latter [65–67] (Fig. 3b).
In some organisms, such as E. coli, expression of glutamate racemase is activated 100-fold in the presence of UDP-MurNAc-L-Ala. This intermediate is ligated to d-Glu in generating PG precursor units, lipid I and lipid II. Thus, expression of the glutamate racemase is regulated by the factors that require its presence [68, 69].
Bacillus subtilis encodes two glutamate racemases, Glr and YrpC. Though both proteins have similar biochemical properties, Glr is mainly involved in d-glutamate synthesis for poly-γ-d-glutamate, a structural component of the spore capsule, whereas YrpC is the racemase that creates d-Glu for the cell wall PG [70, 71].
Aspartate and proline racemases
d-Asp occurs in the PG layer of some bacterial cell walls and is produced from l-Asp by an aspartate racemase. The enzyme is present in various gram-positive bacteria, including Lactococcus lactis [52], Enterococcus faecium [53], Lactobacillus fermenti [72], and Streptococcus faecalis [73], as well as some archea [74]. Like glutamate racemase, aspartate racemase requires no cofactors, contains an essential cysteine residue, and has been proposed to act via a two-base mechanism to remove and return the α-proton of the substrate [75].
Proline racemase is also a member of the PLP-independent enzyme family. Proline racemases have been identified in Clostridium difficile and Clostridium sticklandii. Currently, it is not known if d-Pro is a component of PG in these spore-forming organisms, but it has been hypothesized that d-Pro may enable these pathogens to evade the host immune response [76].
Newly appreciated regulatory roles for d-amino acids in bacteria
As outlined above, the roles of d-amino acids in bacteria were thought to be fairly limited. However, recently it has become clear that d-amino acids are synthesized and released by bacteria from diverse phyla, at up to millimolar concentrations [77]. Furthermore, released d-amino acids function to regulate cell wall chemistry and architecture as well as biofilm development in bacteria [78]. These findings and their implications are discussed below.
d-amino acids govern cell wall remodeling in bacteria
Although PG functions as an exoskeleton, bacterial growth and survival depends on PG plasticity [41, 42, 79]. When bacteria grow, covalent bonds in the PG polymer must be cleaved by murein hydrolases and new bonds formed to insert new subunits into the sacculus. Furthermore, new PG synthesis is crucial for the formation of the septum during cell division (with subsequent generation of the new poles in the daughter cells) [80] and for sporulation/germination PG remodeling [29, 81, 82]. Finally, in times of stress, PG also can be remodeled. For example, many rod-shaped bacteria reorganize their PG in stationary phase, becoming smaller and more coccoid in shape [83]. Currently, there is relatively little understanding of the factors that govern PG remodeling during exponential growth; however, d-amino acids were recently shown to control changes in PG composition and architecture as cells enter stationary phase (see below).
More than six decades ago, high concentrations of exogenous d-amino acids were found to inhibit bacterial growth [84–86]. This effect, which was attributed to alterations in PG metabolism in the treated cells [87–90], required addition of seemingly nonphysiological levels of amino acids, and hence was not thought to be biologically meaningful; however, it did inspire the use of exogenously added components to investigate murein segregation. De Pedro and colleagues demonstrated that low concentrations of d-Cys were relatively innocuous to cells but that several bacterial species incorporated this sulfhydryl-bearing amino acid into murein [89, 90]. Immunodetection of the -SH groups in purified sacculi enabled the distribution of d-Cys in PG to be tracked [91], and such studies have provided valuable insights into cell growth, polarity, and PG synthesis and maintenance [92, 93].
Discovery of production and release of d-amino acids by bacteria
Our group recently made the unexpected discovery that many diverse phyla of bacteria produce types of d-amino acids that were not previously known to be synthesized and release them into the environment [77]. We made this surprising finding while investigating the genetic basis for the curved rod-shape of Vibrio cholerae, the gram-negative bacterium that causes cholera. A genetic screen for V. cholerae with altered cell shape yielded a mutant that exhibited a growth phase-dependent cell morphology defect. We observed that nearly all mrcA mutant cells in stationary phase cultures were spherical, although their morphology did not differ from that of wild-type cells during exponential growth. The mrcA gene encodes PBP1A, an inner membrane-anchored enzyme that elongates the glycan chains and establishes the cross-links between the peptides of the PG [37]. Thus, PBP1A-mediated PG synthesis appears to be required for the maintenance of V. cholerae rod shape in stationary phase. In contrast, deletion of mrcB, which encodes PBP1B, had no effect on V. cholerae morphology suggesting that these two homologous enzymes have clearly separable functions [77].
We hypothesized that the rod-to-sphere transition of the mrcA cells might be stimulated by an extracellular factor present in stationary phase supernatants, since the morphology of mrcA cells changed as cultures became saturated and entered stationary phase. In fact, exponentially growing rod-shaped mrcA cells rapidly became spherical when incubated in a cell-free supernatant from stationary phase cultures. We purified the sphere-inducing component and identified that the active factors were four amino acids: Met, Leu, Val, and Ile. The d- rather than l-forms of these particular amino acids were the active agents in V. cholerae supernatants that influenced mrcA’s morphology. Remarkably, stationary phase V. cholerae supernatants contained a ~1 mM total concentration of these four d-amino acids, an amount sufficient to account for the sphere-inducing activity in the supernatants. Except for d-Pro and d-Gln, other d-amino acids also had some capacity to stimulate the mrcA mutant’s shape transition from rod to sphere, suggesting that sphere-inducing activity is promoted by the chirality rather than the side chain of the amino acid [77].
BsrV racemase
Genomic analysis allowed us to identify a novel V. cholerae racemase that proved to be necessary and sufficient for the synthesis of the unusual d-amino acids in V. cholerae supernatants. Besides the two genes that encode the V. cholerae Glu and Ala racemases, the V. cholerae genome contains an additional gene (vc1312) that encodes a putative PLP-dependent amino acid racemase. A strain with a deletion of this gene was highly defective in the production of d-Met, d-Leu, d-Val, and d-Ile, suggesting that it encodes the principal racemase for the generation of these four d-amino acids. This enzyme, which was named broad spectrum racemase in Vibrio (BsrV) also exhibits in vitro racemase activity for these d-amino acids confirming BsrV as a broad spectrum racemase [77]. Since extracellular d-amino acids were not detected in supernatants from exponential phase cultures of wild-type cells, BsrV is likely only active during stationary phase. In addition, unlike nearly all other bacterial amino acid racemases, which are cytoplasmic, the BsrV racemase was found exclusively in the periplasm, suggesting that d-amino acids produced by BsrV primarily act on periplasmic targets [77].
Notably, we found that the release of d-amino acids into the media is not limited to V. cholerae and other vibrios; instead, stationary phase supernatants from many species representing diverse phyla including B. subtilis, S. aureus, Pseudomonas aeruginosa, and Deinococcus radiodurans contained different d-amino acids. Consistent with this observation, most bacterial genomes appear to encode one or more amino acid racemases in addition to the enzymes required to generate d-Ala and d-Glu for PG synthesis [77]. This suggests that production of broad spectrum racemases may be a conserved trait amongst diverse bacteria.
d-amino acids regulate peptidoglycan composition, amount, and strength
Even though the morphology of wild-type cells was not altered by d-amino acids, we hypothesized that the production of d-amino acids by wild-type V. cholerae has an important physiological role since their generation represents a significant metabolic expenditure. Several observations supported the idea that release of noncanonical d-amino acids could regulate cell wall remodeling. First, d-amino acids (principally d-Ala and d-Glu) are known to be cell wall components, and thus it seemed reasonable that additional d-amino acids could influence PG metabolism. Second, we observed that wild-type V. cholerae cells became spherical if d-amino acids were supplemented with β-lactam antibiotics. Since PG is the principal determinant of bacterial cell shape, these observations suggested that d-amino acids in combination with either genetic (e.g., deletion of mrcA) or chemical inactivation of PBPs lead to weakened PG that is unable to maintain V. cholerae’s rod shape. Finally, as mentioned above, previous work by de Pedro and colleagues revealed that exogenous d-Cys or d-Met could (at high concentrations) be incorporated into PG [89–91, 94].
We compared the chemical composition, structure, and amount of PG isolated from stationary phase wild-type and bsrV V. cholerae to ascertain the effects of the physiologic production of d-amino acids on PG metabolism. Remarkably, the bsrV mutant, which fails to produce and release significant amounts of d-Met, d-Leu, d-Val, and d-Ile, but still produces d-Ala and d-Glu, contained twice the amount of PG found in wild-type cells in stationary phase. Furthermore, exogenous addition of d-amino acids reduced the amount of PG in V. cholerae. Thus, d-amino acids negatively regulate the amount of PG produced by V. cholerae in stationary phase. Moreover, because the accumulation of d-amino acids coincides with the transition into stationary phase and appears to downregulate PG synthesis, d-amino acids may enable coordination of metabolic slowing in cell wall and cytoplasmic compartments when resources become scarce [77].
The noncanonical d-amino acids are incorporated into the PG polymer. HPLC-based analyses of muropeptides from wild-type cells demonstrated that d-Met and d-Leu were present in PG in stationary phase. d-Met and d-Leu replaced d-Ala in the fourth position of the peptide bridge in 3–4% of the stationary phase muropeptides but were not detected in PG from the bsrV mutant. As previously suggested for the incorporation of exogenous d-amino acid into the PG [94], the incorporation of physiologically released d-Met and d-Leu may occur via the action of a periplasmic, β-lactam-insensitive PG-modifying enzyme.
BsrV-generated d-amino acids also influenced the structure of stationary phase PG. In comparison to wild-type PG, the glycan chains from the bsrV mutant were ~20% shorter, and there was an approximately 50% reduction in the amount of full-length pentapeptides and an increase in the amount of trimer muropeptides. Importantly, the changes in PG structure and abundance in the bsrV mutant appear to reduce PG strength. Wild-type cells were far more resistant to an osmotic challenge, suggesting that the PG in wild-type cells is stronger than that in the bsrV mutant. The osmotic hypersensitivity of bsrV mutant cells was unexpected since the bsrV mutant cells contain twice as much PG as wild-type cells. The “weakness” of the PG in bsrV cells presumably is a consequence of the alterations in cell wall structure that result from the absence of d-amino acids in these cells. In this regard, variations in the proportions of pentapeptides and tetrapeptides should have little, if any, effect on the physical strength of the sacculus (as these peptides do not participate in bridging). However, the decrease in the length of the glycan strands in the bsrV mutant could weaken the PG because the number of covalent bonds closing the net-like peptidoglycan molecule is reduced. Regardless of the chemical changes that weaken the PG in the bsrV mutant, general weakening of the entire sacculus is not required to bestow osmotic sensitivity, only localized weakening of the PG would be sufficient. Together, these observations suggest that d-amino acid production by BsrV as cells enter stationary phase provides an autocrine-like signal for V. cholerae to remodel its PG and decrease PG synthesis in adaptation to stationary phase conditions [77].
Regulation of PG by d-amino acids is not limited to V. cholerae. We found that B. subtilis produces different d-amino acids than does V. cholerae (mainly d-Phe and d-Tyr) in stationary phase, but these d-amino acids appear to influence PG synthesis and chemistry in similar ways as d-Met and d-Leu in V. cholerae. Thus, evolutionarily distant bacteria have a common strategy to modulate PG synthesis in stationary phase through production and release of d-amino acids. Remarkably, B. subtilis growth was inhibited when cultured in the presence of exogenous physiologic concentrations of d-amino acids produced by B. subtilis in stationary phase. This suggests that d-amino acids may be a mechanism to simultaneously slow down growth and PG synthesis as population density becomes saturating.
Finally, we also found that exogenous physiologic concentrations of d-Met that are produced by V. cholerae were incorporated into E. coli PG at the same position in the peptide bridge. Thus, E. coli is capable of incorporating noncanonical d-amino acid into its murein sacculus even though this bacterium does not produce or release noncanonical d-amino acids. Therefore, d-amino acids can also act as paracrine-like effectors to influence PG physiology in species other than those that produce them.
Mechanisms of d-amino acid cell wall regulation
Exactly how d-amino acid-dependent cell wall remodeling occurs remains to be determined. Incorporation of unusual d-amino acids (such as d-Met or d-Leu) into the PG polymer could modulate the strength and flexibility of this polymer (Fig. 4). Furthermore, PBPs and other enzymes that modify PG may have altered affinity for and activity on d-amino acid-modified muropeptides. However, it is unlikely that all of the differences in PG composition, structure, and amount observed between V. cholerae wild-type and bsrV mutant strains can be solely attributed to the incorporation of d-amino acids into PG. Supporting this idea, we found that 2.0 mM d-Ala stimulated the conversion of rod-shaped mrcA cells to spheres, even though d-Ala is already present at the site where d-Met or d-Leu is incorporated. Thus, d-Ala, and presumably other d-amino acids, have effects that are not merely consequences of their incorporation into PG. In this regard, d-amino acids likely regulate the periplasmic enzymes that synthesize and modify the PG polymer. We found that d- but not l-Met blocks the binding of a fluorescent derivative of penicillin G to several V. cholerae PBPs [77]. This result suggests that free amino acids accumulated in the periplasm might compete with muropeptide moieties for PBP active sites, thereby serving as regulators of PBP activity. While the exact identity of these PBPs remains to be determined, this observation suggests that d-amino acids may be direct modulators of PBP activity under stationary phase conditions. Further genetic and biochemical analyses of d-amino acid targets are necessary to define the mechanism(s) of d-amino acid action on cell wall PG.
Fig. 4.
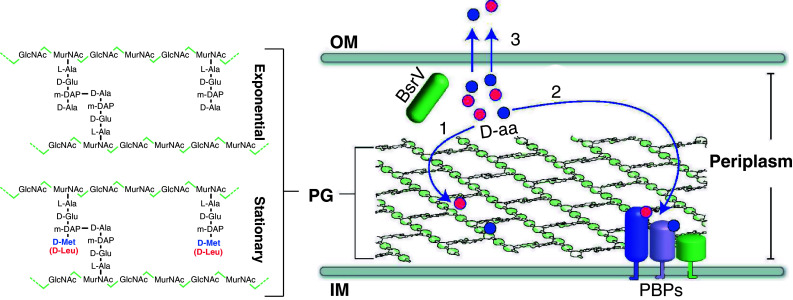
Model of PG remodeling governed by d-amino acid release in stationary phase. PG in V. cholerae is composed of linear glycan strands made up of repeating disaccharide units of GlcNAc and MurNAc cross-linked by short peptides that consist of l-Ala, d-Glu, meso-diaminopimelic acid (m-DAP), and d-Ala. In stationary phase, d-Met (blue circles) and d-Leu (red circles) are produced by BsrV, a periplasmic racemase. These d-amino acids (1) are incorporated at the 4th position of the PG-peptide bridge where d-Ala is usually found, (2) regulate the activity of periplasmic enzymes including penicillin-binding proteins (PBPs), which synthesize and modify PG, and (3) are released into the extracellular milieu where d-amino acids regulate the PG of other bacteria. OM Outer membrane, IM inner membrane
Consistent with reports in other bacterial species [95, 96], we observed additive effects of exogenous d-amino acids in combination with β-lactam antibiotics on wild-type V. cholerae shape and growth. For example, the combination of d-Met with the cephalosporin cefmetazole or the monobactam aztreonam caused wild-type V. cholerae to become spherical (similar to stationary V. cholerae mrcA), whereas when added independently, only minor morphological changes were induced. These observations support the idea that d-amino acids exert regulatory effects on PG-modifying enzymes.
The challenge of regulating PG composition, amount, and structure
Regulation of PG amount, chemistry, and architecture constitutes a particularly challenging issue for the bacterial cell. This complex polymer as well as the enzymes that assemble and modify it are located beyond the cytoplasmic membrane and thus outside of the carefully controlled milieu of the cytoplasm. d-amino acid production by BsrV in the periplasmic space enables the cell to produce d-amino acids in the cell compartment (the periplasm) where they will act. d-amino acid incorporation into the pre-existing PG polymer in the periplasm can be thought of as cell wall “editing.” Such post-synthetic regulation of PG is analogous to posttranslational modification of proteins by glycosylation and to methylation of DNA, processes that alter the properties/activities of other biopolymers. It is likely that incorporation of noncanonical d-amino acids into PG alters the polymers’ physical properties as well as influences the ability of d-amino acid-modified PG to serve as a substrate for periplasmic PG-modifying enzymes. Controlling the concentration of d-amino acids in the periplasm could constitute a reversible mechanism for inhibiting PBPs, thereby enabling cells to rapidly transition between states of active and inactive PG metabolism under changing environmental conditions.
d-amino acids coordinate cell wall metabolism in bacterial populations
Our work has revealed that noncanonical d-amino acids regulate cell wall metabolism. Because the accumulation of d-amino acids coincides with the transition into stationary phase and appears to downregulate PG synthesis, d-amino acids may couple metabolic slowing in cell wall and cytoplasmic compartments under conditions of stress. Additionally, rapid diffusion of small molecule regulators like d-amino acids in aqueous environments enables a quick and synchronized response from the whole bacterial population. In times of scarce nutrients (and potentially additional undefined cellular stresses) the release of extracellular d-amino acids can signal to the whole population to regulate PG amount, composition, and strength. This mechanism may be crucial in nutrient-depleted environments where secretion of secondary metabolites such as organic acids, antibiotics, or small lipids can put the integrity of the cell at risk. Finally, since bacteria are more likely to grow in polymicrobial communities than in monoculture, d-amino acid control may mediate interspecies regulation among bacteria or other organisms that occupy the same niche. It remains to be seen whether d-amino acids mediate both mutualistic and competitive behaviors among different bacteria that share a niche.
d-amino acids control biofilm dispersal
Many bacteria are able to switch between two different “lifestyles.” They can exist as either single (planktonic) cells or in communities known as biofilms. A biofilm is defined as a sessile microbial community that adheres to a solid surface and is surrounded by a bacterially produced extracellular matrix. This matrix is typically composed of polysaccharides, but it can also include proteins and/or DNA. The transition between planktonic and biofilm growth is regulated by a variety of environmental and physiological cues, including bacterial cell density, nutrient availability, and cellular stress [97, 98].
Biofilms are prevalent both in natural environments and in industrial and hospital settings [99–105]. Bacterial infections associated with biofilms adherent to medical devices, such as intravascular catheters, are extremely difficult to eradicate, as the extracellular matrices protect biofilm-associated bacteria from antimicrobials and the host immune system. Thus, discovery of new agents to prevent biofilm formation and/or disrupt established biofilms is of considerable interest.
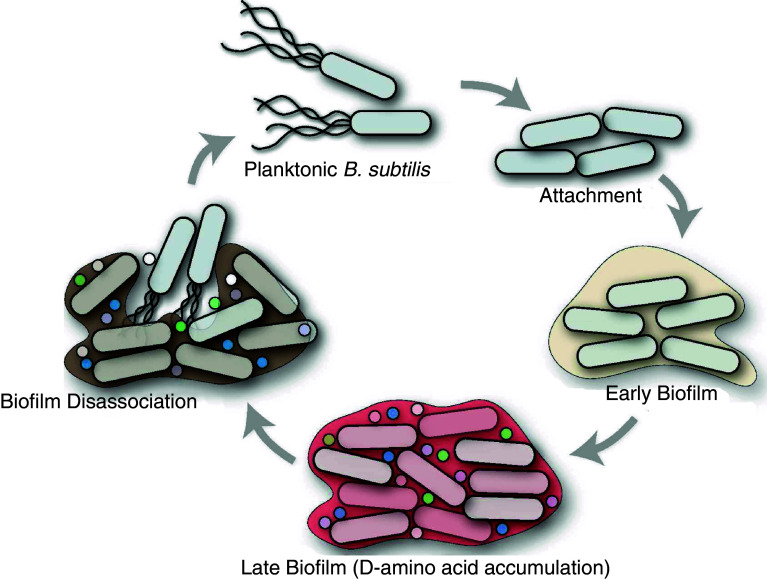
B. subtilis forms biofilms that can be visualized in the laboratory as pellicles at the liquid-to-surface interface of standing cultures or on semi-solid agar plates. Biofilms on standing cultures begin to disassemble after ~6–8 days, and Losick and colleagues recently reported that dissolution is induced by a mixture of d-amino acids (d-Leu, d-Met, d-Trp, and d-Tyr) produced by B. subtilis in these biofilms [78]. In these experiments, d-amino acid accumulation was found to be restricted to mature biofilms (6–8 days) and to be produced at least in part by the racemases YlmE and RacX. However, exogenous addition of d-amino acids to standing cultures prevented B. subtilis biofilm formation altogether. Individually, d-Tyr showed the highest potency, but the mixture of all four amino acids was more potent and had a minimum inhibitory concentration of ~10 nM. In contrast, a mixture of the corresponding l-amino acids neither inhibited biofilm formation nor disrupted existing biofilms [78]. Koldkin-Gal et al. hypothesized that d-amino acid production signals for biofilm disassembly by B. subtilis under conditions when nutrients have become limiting and metabolic waste products have accumulated, so escape into a planktonic lifestyle is therefore beneficial (Fig. 5).
Fig. 5.
Model of biofilm life cycle. B. subtilis cells associated in biofilm communities produce d-amino acids (d-Tyr, d-Met, d-Trp, and d-Leu represented with colored circles), which accumulate and trigger biofilm dispersal
The mechanisms of d-amino acid-regulated biofilm dispersal are being dissected. Koldkin-Gal et al. [78] showed that d-amino acids can induce the disassembly of matrix-associated amyloid fibers that link B. subtilis cells within the biofilm. Maintenance of these fibers seems likely to contribute to biofilm durability, since mutants that could form biofilms in the presence of d-amino acids contained alterations within yqxM, whose gene product is required for attachment of amyloid fibers to the cell. However, it is likely that disruption of amyloid networks is not the only means by which d-amino acids promote biofilm disassembly, since Koldkin-Gal et al. found that d-amino acids led to biofilm dispersal in additional bacterial species, such as S. aureus and P. aeruginosa, that are not known to produce biofilm matrix-associated amyloid fibers. Thus, a universal mechanism of biofilm dispersal is unlikely. Regardless of the mechanism(s) by which d-amino acids promote biofilm dispersal, use of d-amino acids to combat biofilm-associated infections holds considerable promise, especially since d-amino acids are likely to have favorable pharmacokinetic properties and to lack significant toxicity [106].
Conclusions and perspectives for future studies
Recent studies have revealed two exciting and unexpected findings regarding d-amino acids in the bacterial kingdom. First, highly diverse bacteria release various d-amino acids into the environment. And second, the released d-amino acids have heretofore unappreciated roles in regulating key processes, including controlling stationary phase cell wall remodeling and biofilm disassembly in aging bacterial communities. Thus, d-amino acids are signaling molecules that control processes that occur at high cell densities, probably when nutrients become limited. However, the conditions that stimulate d-amino acid production have not been thoroughly explored; additional research is needed to define the stimuli that promote the expression and activity of the racemases that generate d-amino acids.
d-amino acid signaling can be regarded as both autocrine and paracrine in nature since released d-amino acids act on both the cells that release them as well as neighboring cells. In many cases, neighboring cells are of the identical bacterial species as the cells that produced them, but d-amino acids can also act on nearby cells that are of different species. For example, we found that d-Met or d-Leu could be incorporated into the PG of E. coli [77]. In addition, in nature, biofilms are thought to often consist of more than one species [106]. Therefore, released d-amino acids in mixed biofilms may be a paracrine signal, which, along with other chemical regulators, control the architecture of the bacterial community. d-amino acids may be considered as a “chemical language” akin to quorum-sensing molecules, such as acyl homoserine-lactones, that mediate interspecies communication. It remains to be seen whether different d-amino acids mediate distinct types of signals in mixed communities. For example, d-Tyr may primarily function as a signal for biofilm disassembly, whereas d-Met may primarily promote cell wall remodeling. Furthermore, d-amino acids are likely mediators of more traditional types of signaling that regulate gene expression, such as the altered gene expression of UPEC by d-serine in human urine [12].
Many intriguing questions/issues regarding the regulatory roles of d-amino acids in bacterial biology remain to be addressed. Elucidating the mechanisms by which d-amino acids govern cell wall remodeling and biofilm disassembly will undoubtedly reveal new paradigms for understanding how processes outside of the cell membrane are controlled. Finally, application of the regulatory roles of d-amino acids to solve environmental and clinical problems has considerable promise.
Acknowledgments
This work was supported by Howard Hughes Medical Institute (HHMI), NIH AI-R37-42347 (M.K.W.), MEC Fellowship (F.C.), and Jane Coffin Childs Fellowship (H.L.). We thank Brigid Davis and Simon Ringgaard for helpful comments on this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- NRP
Nonribosomal peptide
- PG
Peptidoglycan
- GlcNAc
N-acetyl glucosamine
- MurNAc
N-acetylmuramic acid
- PBPs
Penicillin-binding proteins
- PLP
Pyridoxal-5-phosphate
- BsrV
Broad spectrum racemase in Vibrio
References
- 1.Bentley R. Chiral: a confusing etymology. Chirality. 2010;22:1–2. doi: 10.1002/chir.20699. [DOI] [PubMed] [Google Scholar]
- 2.Flack HD. Louis Pasteur’s discovery of molecular chirality and spontaneous resolution in 1848, together with a complete review of his crystallographic and chemical work. Acta Crystallogr A. 2009;65:371–389. doi: 10.1107/S0108767309024088. [DOI] [PubMed] [Google Scholar]
- 3.Gal J. The discovery of biological enantioselectivity: Louis Pasteur and the fermentation of tartaric acid, 1857—a review and analysis 150 yr later. Chirality. 2008;20:5–19. doi: 10.1002/chir.20494. [DOI] [PubMed] [Google Scholar]
- 4.Meierhenrich U. Stereochemistry for the study of the origin of life. Heidelberg: Springer; 2008. [Google Scholar]
- 5.Kondepudi DK, Kaufman RJ, Singh N. Chiral symmetry breaking in sodium chlorate crystallization. Science. 1990;250:975–976. doi: 10.1126/science.250.4983.975. [DOI] [PubMed] [Google Scholar]
- 6.Podlech J. Origin of organic molecules and biomolecular homochirality. Cell Mol Life Sci. 2001;58:44–60. doi: 10.1007/PL00000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wachtershauser G. Biomolecules: the origin of their optical activity. Med Hypotheses. 1991;36:307–311. doi: 10.1016/0306-9877(91)90001-F. [DOI] [PubMed] [Google Scholar]
- 8.Kim PM, Duan X, Huang AS, Liu CY, Ming GL, Song H, Snyder SH. Aspartate racemase, generating neuronal d-aspartate, regulates adult neurogenesis. Proc Natl Acad Sci USA. 2010;107:3175–3179. doi: 10.1073/pnas.0914706107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleckner NW, Dingledine R. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes . Science. 1988;241:835–837. doi: 10.1126/science.2841759. [DOI] [PubMed] [Google Scholar]
- 10.Wolosker H. NMDA receptor regulation by d-serine: new findings and perspectives. Mol Neurobiol. 2007;36:152–164. doi: 10.1007/s12035-007-0038-6. [DOI] [PubMed] [Google Scholar]
- 11.Wolosker H, Panizzutti R, De Miranda J. Neurobiology through the looking-glass: d-serine as a new glial-derived transmitter. Neurochem Int. 2002;41:327–332. doi: 10.1016/S0197-0186(02)00055-4. [DOI] [PubMed] [Google Scholar]
- 12.Anfora AT, Halladin DK, Haugen BJ, Welch RA. Uropathogenic Escherichia coli CFT073 is adapted to acetatogenic growth but does not require acetate during murine urinary tract infection. Infect Immun. 2008;76:5760–5767. doi: 10.1128/IAI.00618-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodanszky M, Perlman D. Peptide antibiotics. Science. 1969;163:352–358. doi: 10.1126/science.163.3865.352. [DOI] [PubMed] [Google Scholar]
- 14.Glaser T, Hubner K, de Castiglione R, Hamprecht B. Dermorphins, opioid peptides from amphibian skin, act on opioid receptors of mouse neuroblastoma x rat glioma hybrid cells. J Neurochem. 1981;37:1613–1617. doi: 10.1111/j.1471-4159.1981.tb06336.x. [DOI] [PubMed] [Google Scholar]
- 15.Broccardo M, Erspamer V, Falconieri Erspamer G, Improta G, Linari G, Melchiorri P, Montecucchi PC. Pharmacological data on dermorphins, a new class of potent opioid peptides from amphibian skin. Br J Pharmacol. 1981;73:625–631. doi: 10.1111/j.1476-5381.1981.tb16797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto K, Kubota I, Yasuda-Kamatani Y, Minakata H, Nomoto K, Yoshida M, Harada A, Muneoka Y, Kobayashi M. Purification of achatin-I from the atria of the African giant snail, Achatina fulica, and its possible function. Biochem Biophys Res Commun. 1991;177:847–853. doi: 10.1016/0006-291X(91)91867-C. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi H, Kim KH, Liu GJ, Yasuda-Kamatani Y, Minakata H, Nomoto K. Achatin-I, an excitatory neurotransmitter having a d-phenylalanine residue of Achatina giant neurones. Acta Biol Hung. 1992;43:147–158. [PubMed] [Google Scholar]
- 18.Santos DE, Liu GJ, Takeuchi H. Blockers for excitatory effects of achatin-I, a tetrapeptide having a d-phenylalanine residue, on a snail neurone. Eur J Pharmacol. 1995;272:231–239. doi: 10.1016/0014-2999(94)00659-U. [DOI] [PubMed] [Google Scholar]
- 19.Prenner EJ, Lewis RN, McElhaney RN. The interaction of the antimicrobial peptide gramicidin S with lipid bilayer model and biological membranes. Biochim Biophys Acta. 1999;1462:201–221. doi: 10.1016/S0005-2736(99)00207-2. [DOI] [PubMed] [Google Scholar]
- 20.Marahiel MA, Nakano MM, Zuber P. Regulation of peptide antibiotic production in Bacillus . Mol Microbiol. 1993;7:631–636. doi: 10.1111/j.1365-2958.1993.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 21.Burkhart BM, Gassman RM, Langs DA, Pangborn WA, Duax WL, Pletnev V. Gramicidin D conformation, dynamics and membrane ion transport. Biopolymers. 1999;51:129–144. doi: 10.1002/(SICI)1097-0282(1999)51:2<129::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Kleinkauf H, von Dohren H. Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochem. 1990;192:1–15. doi: 10.1111/j.1432-1033.1990.tb19188.x. [DOI] [PubMed] [Google Scholar]
- 23.Mootz HD, Marahiel MA. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang YF, Adams E. d-lysine catabolic pathway in Pseudomonas putida: interrelations with l-lysine catabolism. J Bacteriol. 1974;117:753–764. doi: 10.1128/jb.117.2.753-764.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conrad RS, Massey LK, Sokatch JR. d- and l-isoleucine metabolism and regulation of their pathways in Pseudomonas putida. J Bacteriol. 1974;118:103–111. doi: 10.1128/jb.118.1.103-111.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pioli D, Venables WA, Franklin FC. d-Alanine dehydrogenase. Its role in the utilisation of alanine isomers as growth substrates by Pseudomonas aeruginosa PA01. Arch Microbiol. 1976;110:287–293. doi: 10.1007/BF00690240. [DOI] [PubMed] [Google Scholar]
- 27.Roesch PL, Redford P, Batchelet S, Moritz RL, Pellett S, Haugen BJ, Blattner FR, Welch RA. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol Microbiol. 2003;49:55–67. doi: 10.1046/j.1365-2958.2003.03543.x. [DOI] [PubMed] [Google Scholar]
- 28.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis . Annu Rev Genet. 1996;30:241–297. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 29.Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hills GM. Chemical factors in the germination of spore-bearing aerobes; the effect of yeast extract on the germination of Bacillus anthracis and its replacement by adenosine. Biochem J. 1949;45:353–362. doi: 10.1042/bj0450353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halvorson HO, Spiegelman S. The inhibition of enzyme formation by amino acid analogues. J Bacteriol. 1952;64:207–221. doi: 10.1002/path.1700640121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atluri S, Ragkousi K, Cortezzo DE, Setlow P. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J Bacteriol. 2006;188:28–36. doi: 10.1128/JB.188.1.28-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKevitt MT, Bryant KM, Shakir SM, Larabee JL, Blanke SR, Lovchik J, Lyons CR, Ballard JD. Effects of endogenous d-alanine synthesis and autoinhibition of Bacillus anthracis germination on in vitro and in vivo infections. Infect Immun. 2007;75:5726–5734. doi: 10.1128/IAI.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu H, Emerson J, Aronson AI. Factors involved in the germination and inactivation of Bacillus anthracis spores in murine primary macrophages. FEMS Microbiol Lett. 2007;272:245–250. doi: 10.1111/j.1574-6968.2007.00766.x. [DOI] [PubMed] [Google Scholar]
- 36.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli . Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 38.Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dramsi S, Magnet S, Davison S, Arthur M. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol Rev. 2008;32:307–320. doi: 10.1111/j.1574-6976.2008.00102.x. [DOI] [PubMed] [Google Scholar]
- 40.Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nanninga N. Morphogenesis of Escherichia coli . Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JT. The convergence of murein recycling research with beta-lactamase research. Microb Drug Resist. 1996;2:105–112. doi: 10.1089/mdr.1996.2.105. [DOI] [PubMed] [Google Scholar]
- 43.Holtje JV. From growth to autolysis: the murein hydrolases in Escherichia coli . Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 44.Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 45.van Heijenoort J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev. 2007;71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 47.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Donk WA. Lighting up the nascent cell wall. ACS Chem Biol. 2006;1:425–428. doi: 10.1021/cb600308w. [DOI] [PubMed] [Google Scholar]
- 49.Buynak JD. Cutting and stitching: the cross-linking of peptidoglycan in the assembly of the bacterial cell wall. ACS Chem Biol. 2007;2:602–605. doi: 10.1021/cb700182u. [DOI] [PubMed] [Google Scholar]
- 50.Young KD. Approaching the physiological functions of penicillin-binding proteins in Escherichia coli . Biochimie. 2001;83:99–102. doi: 10.1016/S0300-9084(00)01205-0. [DOI] [PubMed] [Google Scholar]
- 51.Nagata Y, Fujiwara T, Kawaguchi-Nagata K, Fukumori Y, Yamanaka T. Occurrence of peptidyl d-amino acids in soluble fractions of several eubacteria, archaea and eukaryotes. Biochim Biophys Acta. 1998;1379:76–82. doi: 10.1016/s0304-4165(97)00084-6. [DOI] [PubMed] [Google Scholar]
- 52.Veiga P, Piquet S, Maisons A, Furlan S, Courtin P, Chapot-Chartier MP, Kulakauskas S. Identification of an essential gene responsible for d-Asp incorporation in the Lactococcus lactis peptidoglycan crossbridge. Mol Microbiol. 2006;62:1713–1724. doi: 10.1111/j.1365-2958.2006.05474.x. [DOI] [PubMed] [Google Scholar]
- 53.Bellais S, Arthur M, Dubost L, Hugonnet JE, Gutmann L, van Heijenoort J, Legrand R, Brouard JP, Rice L, Mainardi JL. Aslfm, the d-aspartate ligase responsible for the addition of d-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium . J Biol Chem. 2006;281:11586–11594. doi: 10.1074/jbc.M600114200. [DOI] [PubMed] [Google Scholar]
- 54.Reynolds PE, Courvalin P. Vancomycin resistance in enterococci due to synthesis of precursors terminating in d-alanyl-d-serine. Antimicrob Agents Chemother. 2005;49:21–25. doi: 10.1128/AAC.49.1.21-25.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sieradzki K, Tomasz A. A highly vancomycin-resistant laboratory mutant of Staphylococcus aureus . FEMS Microbiol Lett. 1996;142:161–166. doi: 10.1111/j.1574-6968.1996.tb08424.x. [DOI] [PubMed] [Google Scholar]
- 56.De Jonge BL, Gage D, Xu N. The carboxyl terminus of peptidoglycan stem peptides is a determinant for methicillin resistance in Staphylococcus aureus . Antimicrob Agents Chemother. 2002;46:3151–3155. doi: 10.1128/AAC.46.10.3151-3155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol. 2007;10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanner ME. Understanding nature’s strategies for enzyme-catalyzed racemization and epimerization. Acc Chem Res. 2002;35:237–246. doi: 10.1021/ar000056y. [DOI] [PubMed] [Google Scholar]
- 59.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 60.Yoshimura T, Goto M. D-amino acids in the brain: structure and function of pyridoxal phosphate-dependent amino acid racemases. FEBS J. 2008;275:3527–3537. doi: 10.1111/j.1742-4658.2008.06516.x. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe A, Yoshimura T, Mikami B, Hayashi H, Kagamiyama H, Esaki N. Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5′-phosphopyridoxyl)alanine. J Biol Chem. 2002;277:19166–19172. doi: 10.1074/jbc.M201615200. [DOI] [PubMed] [Google Scholar]
- 62.Walsh CT. Enzymes in the d-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem. 1989;264:2393–2396. [PubMed] [Google Scholar]
- 63.Arthur M, Reynolds PE, Depardieu F, Evers S, Dutka-Malen S, Quintiliani R, Jr, Courvalin P. Mechanisms of glycopeptide resistance in enterococci. J Infect. 1996;32:11–16. doi: 10.1016/S0163-4453(96)80003-X. [DOI] [PubMed] [Google Scholar]
- 64.Arias CA, Martin-Martinez M, Blundell TL, Arthur M, Courvalin P, Reynolds PE. Characterization and modelling of VanT: a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarum BM4174. Mol Microbiol. 1999;31:1653–1664. doi: 10.1046/j.1365-2958.1999.01294.x. [DOI] [PubMed] [Google Scholar]
- 65.Choi SY, Esaki N, Yoshimura T, Soda K. Reaction mechanism of glutamate racemase, a pyridoxal phosphate-independent amino acid racemase. J Biochem. 1992;112:139–142. doi: 10.1093/oxfordjournals.jbchem.a123853. [DOI] [PubMed] [Google Scholar]
- 66.Tanner ME, Gallo KA, Knowles JR. Isotope effects and the identification of catalytic residues in the reaction catalyzed by glutamate racemase. Biochemistry. 1993;32:3998–4006. doi: 10.1021/bi00066a021. [DOI] [PubMed] [Google Scholar]
- 67.Hwang KY, Cho CS, Kim SS, Sung HC, Yu YG, Cho Y. Structure and mechanism of glutamate racemase from Aquifex pyrophilus . Nat Struct Biol. 1999;6:422–426. doi: 10.1038/8223. [DOI] [PubMed] [Google Scholar]
- 68.Doublet P, van Heijenoort J, Bohin JP, Mengin-Lecreulx D. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J Bacteriol. 1993;175:2970–2979. doi: 10.1128/jb.175.10.2970-2979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ho HT, Falk PJ, Ervin KM, Krishnan BS, Discotto LF, Dougherty TJ, Pucci MJ. UDP-N-acetylmuramyl-l-alanine functions as an activator in the regulation of the Escherichia coli glutamate racemase activity. Biochemistry. 1995;34:2464–2470. doi: 10.1021/bi00008a009. [DOI] [PubMed] [Google Scholar]
- 70.Kada S, Nanamiya H, Kawamura F, Horinouchi S. Glr, a glutamate racemase, supplies d-glutamate to both peptidoglycan synthesis and poly-gamma-glutamate production in gamma-PGA-producing Bacillus subtilis . FEMS Microbiol Lett. 2004;236:13–20. doi: 10.1016/j.femsle.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 71.Ashiuchi M, Soda K, Misono H. Characterization of yrpC gene product of Bacillus subtilis IFO 3336 as glutamate racemase isozyme. Biosci Biotechnol Biochem. 1999;63:792–798. doi: 10.1271/bbb.63.792. [DOI] [PubMed] [Google Scholar]
- 72.Wallinder IB, Neujahr HY. Cell wall and peptidoglycan from Lactobacillus fermenti . J Bacteriol. 1971;105:918–926. doi: 10.1128/jb.105.3.918-926.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staudenbauer W, Willoughby E, Strominger JL. Further studies of the d-aspartic acid-activating enzyme of Streptococcus faecalis and its attachment to the membrane. J Biol Chem. 1972;247:5289–5296. [PubMed] [Google Scholar]
- 74.Matsumoto M, Homma H, Long Z, Imai K, Iida T, Maruyama T, Aikawa Y, Endo I, Yohda M. Occurrence of free d-amino acids and aspartate racemases in hyperthermophilic archaea. J Bacteriol. 1999;181:6560–6563. doi: 10.1128/jb.181.20.6560-6563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamauchi T, Choi SY, Okada H, Yohda M, Kumagai H, Esaki N, Soda K. Properties of aspartate racemase, a pyridoxal 5′-phosphate-independent amino acid racemase. J Biol Chem. 1992;267:18361–18364. [PubMed] [Google Scholar]
- 76.Goytia M, Chamond N, Cosson A, Coatnoan N, Hermant D, Berneman A, Minoprio P. Molecular and structural discrimination of proline racemase and hydroxyproline-2-epimerase from nosocomial and bacterial pathogens. PLoS One. 2007;2:e885. doi: 10.1371/journal.pone.0000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. d-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mengin-Lecreulx D, Lemaitre B. Structure and metabolism of peptidoglycan and molecular requirements allowing its detection by the Drosophila innate immune system. J Endotoxin Res. 2005;11:105–111. doi: 10.1179/096805105X35233. [DOI] [PubMed] [Google Scholar]
- 80.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Makino S, Moriyama R. Hydrolysis of cortex peptidoglycan during bacterial spore germination. Med Sci Monit. 2002;8:RA119–RA127. [PubMed] [Google Scholar]
- 82.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 83.Nystrom T. Stationary-phase physiology. Annu Rev Microbiol. 2004;58:161–181. doi: 10.1146/annurev.micro.58.030603.123818. [DOI] [PubMed] [Google Scholar]
- 84.Graham CE, Hier SW, Waitkoff HK, Saper SM, Bibler WG, Pentz EI. Studies on natural and racemic amino acids with rats. J Biol Chem. 1950;185:97–102. [PubMed] [Google Scholar]
- 85.Teeri A, Josselyn D. Effect of excess amino acids on growth of certain Lactobacilli . J Bacteriol. 1953;66:72–73. doi: 10.1128/jb.66.1.72-73.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bopp M. Inhibition of Agrobacterium tumefaciens by d-amino acids. Z Naturforsch B. 1965;20:899–905. [PubMed] [Google Scholar]
- 87.Izaki K, Matsuhashi M, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. 8. Peptidoglycan transpeptidase and d-alanine carboxypeptidase: penicillin-sensitive enzymatic reaction in strains of Escherichia coli . J Biol Chem. 1968;243:3180–3192. [PubMed] [Google Scholar]
- 88.Hammes WP. The LD-carboxypeptidase activity in Gaffkya homari. The target of the action of d-amino acids or glycine on the formation of wall-bound peptidoglycan. Eur J Biochem. 1978;91:501–507. doi: 10.1111/j.1432-1033.1978.tb12703.x. [DOI] [PubMed] [Google Scholar]
- 89.Caparros M, Torrecuadrada JL, de Pedro MA. Effect of d-amino acids on Escherichia coli strains with impaired penicillin-binding proteins. Res Microbiol. 1991;142:345–350. doi: 10.1016/0923-2508(91)90050-K. [DOI] [PubMed] [Google Scholar]
- 90.Caparros M, Pisabarro AG, de Pedro MA. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli . J Bacteriol. 1992;174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Pedro MA, Quintela JC, Holtje JV, Schwarz H. Murein segregation in Escherichia coli . J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus . Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 93.de Pedro MA, Young KD, Holtje JV, Schwarz H. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J Bacteriol. 2003;185:1147–1152. doi: 10.1128/JB.185.4.1147-1152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caparros M, Aran V, de Pedro MA. Incorporation of S-[3H]methyl-d-cysteine into the peptidoglycan of ether-treated cells of Escherichia coli . FEMS Microbiol Lett. 1992;72:139–146. doi: 10.1016/0378-1097(92)90519-T. [DOI] [PubMed] [Google Scholar]
- 95.Lark C, Lark KG. The effects of d-amino acids on Alcaligenes fecalis . Can J Microbiol. 1959;5:369–379. doi: 10.1139/m59-046. [DOI] [PubMed] [Google Scholar]
- 96.Tuttle AL, Gest H. Induction of morphological aberrations in Rhodospirillum rubrum by d-amino acids. J Bacteriol. 1960;79:213–216. doi: 10.1128/jb.79.2.213-216.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spormann AM. Physiology of microbes in biofilms. Curr Top Microbiol Immunol. 2008;322:17–36. doi: 10.1007/978-3-540-75418-3_2. [DOI] [PubMed] [Google Scholar]
- 98.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 99.Liu YC, Post JC. Biofilms in pediatric respiratory and related infections. Curr Allergy Asthma Rep. 2009;9:449–455. doi: 10.1007/s11882-009-0066-6. [DOI] [PubMed] [Google Scholar]
- 100.Kassar R, Hachem R, Jiang Y, Chaftari AM, Raad I. Management of Bacillus bacteremia: the need for catheter removal. Medicine (Baltimore) 2009;88:279–283. doi: 10.1097/MD.0b013e3181b7c64a. [DOI] [PubMed] [Google Scholar]
- 101.Homoe P, Bjarnsholt T, Wessman M, Sorensen HC, Johansen HK. Morphological evidence of biofilm formation in Greenlanders with chronic suppurative otitis media. Eur Arch Otorhinolaryngol. 2009;266:1533–1538. doi: 10.1007/s00405-009-0940-9. [DOI] [PubMed] [Google Scholar]
- 102.Reslinski A, Mikucka A, Szmytkowski J, Gospodarek E, Dabrowiecki S. In vivo biofilm on the surface of a surgical mesh implant. Pol J Microbiol. 2009;58:367–369. [PubMed] [Google Scholar]
- 103.Yankah AC, Pasic M, Klose H, Siniawski H, Weng Y, Hetzer R. Homograft reconstruction of the aortic root for endocarditis with peri-annular abscess: a 17-year study. Eur J Cardiothorac Surg. 2005;28:69–75. doi: 10.1016/j.ejcts.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 104.Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis . Clin Microbiol Rev. 2008;21:26–59. doi: 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kobayashi H. Airway biofilms: implications for pathogenesis and therapy of respiratory tract infections. Treat Respir Med. 2005;4:241–253. doi: 10.2165/00151829-200504040-00003. [DOI] [PubMed] [Google Scholar]
- 106.Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng. 2008;10:145–167. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- 107.Auvynet C, Seddiki N, Dunia I, Nicolas P, Amiche M, Lacombe C. Post-translational amino acid racemization in the frog skin peptide deltorphin I in the secretion granules of cutaneous serous glands. Eur J Cell Biol. 2006;85:25–34. doi: 10.1016/j.ejcb.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 108.Lacombe C, Cifuentes-Diaz C, Dunia I, Auber-Thomay M, Nicolas P, Amiche M. Peptide secretion in the cutaneous glands of South American tree frog Phyllomedusa bicolor: an ultrastructural study. Eur J Cell Biol. 2000;79:631–641. doi: 10.1078/0171-9335-00085. [DOI] [PubMed] [Google Scholar]
- 109.Simmaco M, Kreil G, Barra D. Bombinins, antimicrobial peptides from Bombina species. Biochim Biophys Acta. 2009;1788:1551–1555. doi: 10.1016/j.bbamem.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 110.Mangoni ML, Papo N, Saugar JM, Barra D, Shai Y, Simmaco M, Rivas L. Effect of natural l- to d-amino acid conversion on the organization, membrane binding, and biological function of the antimicrobial peptides bombinins H. Biochemistry. 2006;45:4266–4276. doi: 10.1021/bi052150y. [DOI] [PubMed] [Google Scholar]
- 111.Mangoni ML, Grovale N, Giorgi A, Mignogna G, Simmaco M, Barra D. Structure-function relationships in bombinins H, antimicrobial peptides from Bombina skin secretions. Peptides. 2000;21:1673–1679. doi: 10.1016/S0196-9781(00)00316-8. [DOI] [PubMed] [Google Scholar]
- 112.Fujisawa Y, Masuda K, Minakata H. Fulicin regulates the female reproductive organs of the snail, Achatina fulica . Peptides. 2000;21:1203–1208. doi: 10.1016/S0196-9781(00)00260-6. [DOI] [PubMed] [Google Scholar]
- 113.Ohta N, Kubota I, Takao T, Shimonishi Y, Yasuda-Kamatani Y, Minakata H, Nomoto K, Muneoka Y, Kobayashi M. Fulicin, a novel neuropeptide containing a d-amino acid residue isolated from the ganglia of Achatina fulica . Biochem Biophys Res Commun. 1991;178:486–493. doi: 10.1016/0006-291X(91)90133-R. [DOI] [PubMed] [Google Scholar]
- 114.Jimenez EC, Olivera BM, Gray WR, Cruz LJ. Contryphan is a d-tryptophan-containing conus peptide. J Biol Chem. 1996;271:28002–28005. doi: 10.1074/jbc.271.45.28002. [DOI] [PubMed] [Google Scholar]
- 115.Jacobsen R, Jimenez EC, Grilley M, Watkins M, Hillyard D, Cruz LJ, Olivera BM. The contryphans, a d-tryptophan-containing family of conus peptides: interconversion between conformers. J Pept Res. 1998;51:173–179. doi: 10.1111/j.1399-3011.1998.tb01213.x. [DOI] [PubMed] [Google Scholar]
- 116.Jacobsen RB, Jimenez EC, De la Cruz RG, Gray WR, Cruz LJ, Olivera BM. A novel d-leucine-containing conus peptide: diverse conformational dynamics in the contryphan family. J Pept Res. 1999;54:93–99. doi: 10.1034/j.1399-3011.1999.00093.x. [DOI] [PubMed] [Google Scholar]
- 117.Cropper EC, Brezina V, Vilim FS, Harish O, Price DA, Rosen S, Kupfermann I, Weiss KR. FRF peptides in the ARC neuromuscular system of Aplysia: purification and physiological actions. J Neurophysiol. 1994;72:2181–2195. doi: 10.1152/jn.1994.72.5.2181. [DOI] [PubMed] [Google Scholar]
- 118.Mykles DL, Adams ME, Gade G, Lange AB, Marco HG, Orchard I. Neuropeptide action in insects and crustaceans. Physiol Biochem Zool. 2010;83:836–846. doi: 10.1086/648470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Soyez D, Toullec JY, Ollivaux C, Geraud G. l- to d-amino acid isomerization in a peptide hormone is a late post-translational event occurring in specialized neurosecretory cells. J Biol Chem. 2000;275:37870–37875. doi: 10.1074/jbc.M007302200. [DOI] [PubMed] [Google Scholar]
- 120.Turner TJ, Adams ME, Dunlap K. Calcium channels coupled to glutamate release identified by omega-Aga-IVA. Science. 1992;258:310–313. doi: 10.1126/science.1357749. [DOI] [PubMed] [Google Scholar]
- 121.Lundy PM, Hamilton MG, Frew R. Pharmacological identification of a novel Ca2+ channel in chicken brain synaptosomes. Brain Res. 1994;643:204–210. doi: 10.1016/0006-8993(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 122.Isaka M, Palasarn S, Lapanun S, Sriklung K. Paecilodepsipeptide A, an antimalarial and antitumor cyclohexadepsipeptide from the insect pathogenic fungus Paecilomyces cinnamomeus BCC 9616. J Nat Prod. 2007;70:675–678. doi: 10.1021/np060602h. [DOI] [PubMed] [Google Scholar]
- 123.Miao V, Coeffet-Legal MF, Brian P, Brost R, Penn J, Whiting A, Martin S, Ford R, Parr I, Bouchard M, Silva CJ, Wrigley SK, Baltz RH. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology. 2005;151:1507–1523. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- 124.Baltz RH. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr Opin Chem Biol. 2009;13:144–151. doi: 10.1016/j.cbpa.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 125.Eisenstein BI, Oleson FB Jr, Baltz RH (2010) Daptomycin: from the mountain to the clinic, with essential help from Francis Tally, MD. Clin Infect Dis 50 Suppl 1:S10–S15 [DOI] [PubMed]
- 126.Morikawa M, Daido H, Takao T, Murata S, Shimonishi Y, Imanaka T. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J Bacteriol. 1993;175:6459–6466. doi: 10.1128/jb.175.20.6459-6466.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lim SP, Roongsawang N, Washio K, Morikawa M. Flexible exportation mechanisms of arthrofactin in Pseudomonas sp. MIS38. J Appl Microbiol. 2009;107:157–166. doi: 10.1111/j.1365-2672.2009.04189.x. [DOI] [PubMed] [Google Scholar]
- 128.Segre A, Bachmann RC, Ballio A, Bossa F, Grgurina I, Iacobellis NS, Marino G, Pucci P, Simmaco M, Takemoto JY. The structure of syringomycins A1, E and G. FEBS Lett. 1989;255:27–31. doi: 10.1016/0014-5793(89)81054-3. [DOI] [PubMed] [Google Scholar]
- 129.Ballio A, Barra D, Bossa F, Collina A, Grgurina I, Marino G, Moneti G, Paci M, Pucci P, Segre A, Simmaco M. Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae. FEBS Lett. 1991;291:109–112. doi: 10.1016/0014-5793(91)81115-O. [DOI] [PubMed] [Google Scholar]
- 130.Balibar CJ, Vaillancourt FH, Walsh CT. Generation of d amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem Biol. 2005;12:1189–1200. doi: 10.1016/j.chembiol.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 131.Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hutchison ML, Gross DC. Lipopeptide phytotoxins produced by Pseudomonas syringae pv. syringae: comparison of the biosurfactant and ion channel-forming activities of syringopeptin and syringomycin. Mol Plant Microbe Interact. 1997;10:347–354. doi: 10.1094/MPMI.1997.10.3.347. [DOI] [PubMed] [Google Scholar]
- 133.Fdhila F, Vazquez V, Sanchez JL, Riguera R. dd-diketopiperazines: antibiotics active against Vibrio anguillarum isolated from marine bacteria associated with cultures of Pecten maximus . J Nat Prod. 2003;66:1299–1301. doi: 10.1021/np030233e. [DOI] [PubMed] [Google Scholar]
- 134.Kawai Y, Saito T, Toba T, Samant SK, Itoh T. Isolation and characterization of a highly hydrophobic new bacteriocin (gassericin A) from Lactobacillus gasseri LA39. Biosci Biotechnol Biochem. 1994;58:1218–1221. doi: 10.1271/bbb.58.1218. [DOI] [PubMed] [Google Scholar]
- 135.Kawai Y, Kemperman R, Kok J, Saito T. The circular bacteriocins gassericin A and circularin A. Curr Protein Pept Sci. 2004;5:393–398. doi: 10.2174/1389203043379549. [DOI] [PubMed] [Google Scholar]
- 136.Kawai Y, Ishii Y, Arakawa K, Uemura K, Saitoh B, Nishimura J, Kitazawa H, Yamazaki Y, Tateno Y, Itoh T, Saito T. Structural and functional differences in two cyclic bacteriocins with the same sequences produced by Lactobacilli . Appl Environ Microbiol. 2004;70:2906–2911. doi: 10.1128/AEM.70.5.2906-2911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]