Abstract
In 1987, about 150 years after the discovery of Duchenne muscular dystrophy (DMD), its responsible gene, the dystrophin gene, was cloned by Kunkel. This was a new substance. During these 20 odd years after the cloning, our understanding on dystrophin as a component of the subsarcolemmal cytoskeleton networks and on the pathomechanisms of and experimental therapeutics for DMD has been greatly enhanced. During this paradigm change, I was fortunately able to work as an active researcher on its frontiers for 12 years. After we discovered that dystrophin is located on the cell membrane in 1988, we studied the architecture of dystrophin and dystrophin-associated proteins (DAPs) complex in order to investigate the function of dystrophin and pathomechanism of DMD. During the conduct of these studies, we came to consider that the dystrophin–DAP complex serves to transmembranously connect the subsarcolemmal cytoskeleton networks and basal lamina to protect the lipid bilayer. It then became our working hypothesis that injury of the lipid bilayer upon muscle contraction is the cause of DMD. During this process, we predicted that subunits of the sarcoglycan (SG) complex are responsible for respective types of DMD-like muscular dystrophy with autosomal recessive inheritance. Our prediction was confirmed to be true by many researchers including ourselves. In this review, I will try to explain what we observed and how we considered concerning the architecture and function of the dystrophin–DAP complex, and the pathomechanisms of DMD and related muscular dystrophies.
Keywords: dystrophin, Duchenne muscular dystrophy, dystrophin associated proteins, sarcoglycanopathy, dystrophin bolt, transverse fixation system
1. Background: Studies on muscular dystrophy in the pre-dystrophin era
1.1. Clinical and pathological aspects.
Muscular dystrophy is a group of genetic diseases encompassing 40–50 disorders, the symptoms of which are characterized by chronic generalized progressive atrophy of skeletal muscles. Among them, Duchenne muscular dystrophy (DMD), which is inherited in an X-linked recessive manner1) is the most prevalent worldwide. Disturbance of walking and running appears at the age of 3–5 years, loss of ambulation by the age of 15 years and being bed-bound thereafter.
Most symptomatologies1) and pathological changes2) of DMD were described in the 19th century. By the middle of 20th century, the mild type of DMD, namely, Becker muscular dystrophy (BMD), and autosomal recessive Duchenne-like muscular dystrophy (ARDMD, later renamed sarcoglycanopathy) which shows symptoms and course close to DMD were unambiguously described. In 20th century, especially in its later half, various congenital muscular dystrophies (CMD) were described.
1.2. Biological studies of muscular dystrophy in the pre-dystrophin era.
Presence of aldolase at higher levels in serum from patients with muscular dystrophy was described by Lehninger3) in 1949. Extending this finding, Ebashi4) together with Sugita discovered high serum creatine kinase (CK) level in patients in 1959. This has been used worldwide for biochemical examination of muscular dystrophy and other muscle diseases. At the same time, this finding triggered the classical biological research on these diseases. Serum CK has been considered to be derived mostly from the living muscle fibers leaking through the cell membrane. Later, many soluble cytoplasmic enzymes and proteins,5) such as myoglobin, were found to be released from DMD muscle fibers.
Mostly in 1970s, many important studies were described, some of which are still worthy of being seriously considered. (1) Movement, namely muscle contraction, is an important risk factor of DMD. In a short term experiment,6) the serum CK level of a DMD patient, whose baseline serum CK value at rest was already about one hundred times higher than that of normal boys, increased by about ten times from his baseline levels, when he visited the zoo. Myoglobin was similarly increased. (2) In DMD muscle, tiny tears on the lipid bilayer of the cell membrane were found by electron microscopy (EM).5) (3) Total calcium ions in DMD muscle fibers were found to be increased.5) (4) Calcium–calpain hypothesis was formulated7) (see Discussion (5)). However, further elucidation of these phenomena in causal relation to DMD was not feasible. Back then, DMD was only understood based on the complicated clinical and pathological findings, but the essence to connect such phenomena specifically to DMD was lacking. The classical DMD researches obviously came up against a wall and stayed there for a decade.
2. The beginning of the dystrophin era
In 1987, the wall was broken out by “laypersons of the medicine”. The responsible gene for DMD was discovered by young basic scientists, in spite of absence of knowledge on the responsible protein. This brought about a great revolution of the research not only of DMD but also of many genetic diseases. Soon, a number of new groups having various backgrounds of new biology joined in the research and grew to its mainstream. In this review, I summarize mainly our works and thoughts developed together with the progress of the new era. However, our works were related to those of others. To avoid confusion and to separately understand the works from each laboratory, I used the laboratory chiefs’ names to designate works cited. The names of the first authors of those works are found in the references.
2.1. The dystrophin and utrophin genes.
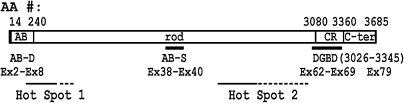
In 1983, Davies, Kunkel and Worton started to search for the responsible gene for DMD under the auspices of the Muscular Dystrophy Association of America. In 1987, Kunkel first cloned the gene,8,9) which was then named the dystrophin gene. Dystrophin was a new substance. Some characteristics of the gene and its products and other related matters are described here for better understanding of this review (Fig. 1 ).8,9) The dystrophin gene is localized at chromosome Xp21, its size is 3 megabases, occupying about 1/1,000 of the total genome size, and is composed of 79 exons. The size of the mRNA is 14 kb. The number of amino acid (AA) residues in dystrophin is 3,685, as deduced from the nucleotide sequence, and it has a molecular weight of 427 kDa. Dystrophin is a roughly slender protein. The primary sequence of the N-terminal portion is highly homologous to those of the N-terminal portion of α-actinin, and this portion was named the actin-binding domain (AB domain: AA #14–240, exons 2–8). Tandem domains of this region are the rod or triple helical segment (AA #253–3040, exons 8–61), the cysteine-rich domain (CR domain: AA #3080–3360, exons 62–69) and finally the C-terminal domain (AA #3361–3685, exons 69–79). The length of the rod was assumed to be 125 nm. Four potential small hinges were found in the rod. This molecular structure was constructed on the basis of the analogy of dystrophin and α-actinin, and dystrophin was initially considered to be present as an anti-parallel homodimer until 1997, except ourselves (see Discussion (2)).
Figure 1.
Dystrophin: Molecular domains and functional domains. AB-D: actin-binding domain that binds to γ-actin filament composing the subsarcolemmal cytoskelton network. Rod: also termed ‘triple helical segments.’ The rod contains AB-S (another binding site to γ-actin) encoded by exons (EX) 38–40. CR: cysteine rich domain. C-ter: C-terminal domain. DGBD:binding site on dystrophin for β-DG. Hot Spot 1 & 2: the sites that are deleted with high frequency in the dystrophin gene. The frequency of mutation in Hot Spot 2 is much higher than that in Hot Spot 1. AA #: Range of AA residues, spanning each domain.
Later in 1996, Ervasti10) reported the presence of a second binding site for actin filament in the middle of the rod of dystrophin. The binding site exists in AA #1416–1880 (exons 32–40) that corresponds to the triple repeat numbers 11–14 (Fig. 1). In this review, I tentatively call this site “the AB site” to discriminate this site from “the AB domain” at the N-terminus of dystrophin.
In 1989, Davies cloned the utrophin gene that is mapped to 6q24.11) Its identities to dystrophin are about 65% and 80% at the nucleotide and AA levels, respectively. Utrophin is expressed on the cell membrane of myotubes and is replaced by dystrophin as differentiation progresses.
In 1989, in the dystrophin gene of mdx mice, a nonsense mutation was discovered at nucleotide #3185.12) It is known that mdx mice lack dystrophin in muscle cells and show the dystrophic pathology in muscle. They have widely been used as an animal model of DMD.
2.2. Works of our project team: localizing dystrophin to sarcolemma.
Back in October 1983, I met Kunkel for the first time in a symposium held in Pennsylvania, where he gave a talk on his initial trials on the dystrophin gene cloning. After the meeting, I sometimes read papers reporting the course of cloning trials of this gene published from Davies, Kunkel and Worton groups. In the meanwhile, I mulled over the future tide of DMD research. “Sooner or later, the dystrophin gene must be cloned. After the gene is cloned, the next main ‘battlefield’ would no doubt be protein studies. In the biochemical studies, it is prerequisite to have a tool for detecting the protein, namely, to generate antibodies against the protein product of the dystrophin gene. To get antigen for dystrophin, we should synthesize polypeptides in accordance with the AA sequence deduced from the nucleotide sequence of the dystrophin gene that must be published in the near future. Once I have synthesized the polypeptides, their antibodies would easily be obtained.” In this context, I approached the Central Research Institute (CRI) of Ajinomoto Co. regarding the synthesis of the polypeptides, as a collaborative work. CRI has an excellent team for peptide synthesis headed by Eguchi. “The first work to be done should be the determination of the dystrophin locus in muscle fibers, which should provide the basis for dystrophin studies in various aspects.” Because I imagined that there must be many researchers worldwide who had the same idea, not wasting time was absolutely necessary for this study. Unfortunately, at that time our group did not have experience with immunohistochemical techniques. Thus, I decided to collaborate with Arahata who had good experience in the techniques in Sugita’s laboratory, just next door.
In the middle of June 1987, I proposed this collaborative project to Sugita. After I scrupulously explained this project to him, he at last agreed to my proposal. The muscle samples were to be provided by Nonaka. Thus, the project team of three laboratories of our institute and a laboratory of CRI started.
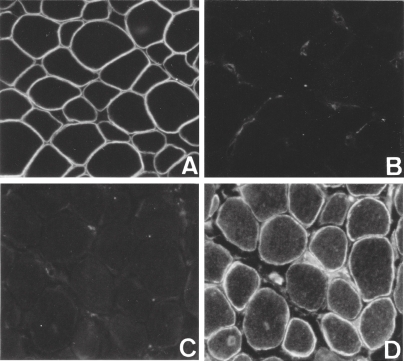
In the July 31 issue of Cell in 1987, I read Kunkel’s paper reporting the partial AA sequence of dystrophin.8) Immediately, I instructed Eguchi to start the polypeptide synthesis. Soon after, he produced and sent me two polypeptide fragments of dystrophin with 50 AA residues. Then, our team obtained two polyclonal antibodies against these polypeptides by conventional methods. Using these antibodies, Arahata stained the normal and DMD muscles. Our dystrophin antibodies clearly stained the sarcolemma in normal human but not in DMD in January 1988 (Fig. 2 A & C). Thus, absence of dystrophin on the sarcolemma was the cause of DMD. In addition to this, Arahata observed that dystrophin was distributed evenly throughout the sarcolemmas of human skeletal muscles without discriminating red and white muscle fibers. The first paper on localization of dystrophin was published in the February 1988 issue of this journal (PJA).13) Luckily enough, our next paper similar to the PJA paper was published in Nature on June 30, 1988.14) No similar paper had yet been published from other laboratories, before our papers.
Figure 2.
Reciprocal localization of dystrophin and utrophin on normal and DMD human sarcolemma. A & B, normal muscle; C & D, DMD muscle; A & C, anti-dystrophin antibody staining; and B & D, anti-utrophin antibody staining.
Function of dystrophin was at that time unknown. However, because our results were compatible with the hypothesis on the sarcolemmal injury as the cause of DMD that was widely believed in the pre-dystrophin era, our results were soon accepted. This method has been widely used both for DMD diagnostics and basic research on dystrophin including the gene therapy study of DMD. In any case, our results indicated the direction of my following studies, i.e., how dystrophin is associated with the cell membrane, and what is its function, which when defective must cause DMD.
2.3. Sarcolemma.
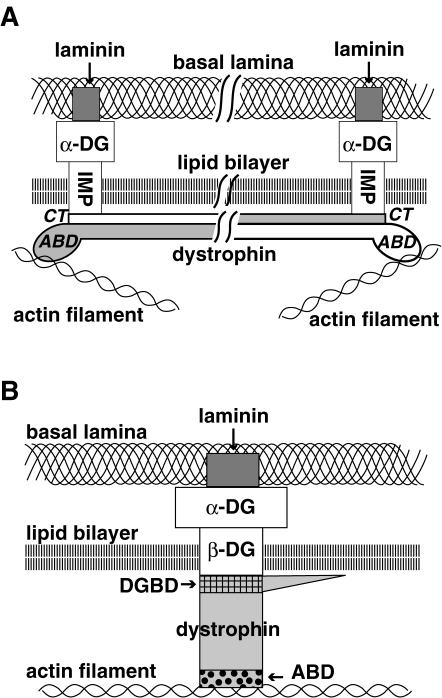
In the following description, the term sarcolemma is used to designate the basal lamina, lipid bilayer and subsarcolemmal cytoskeleton networks together (Fig. 3B ). This usage is rational, because this term was first used by Bowman in 1840 to designate the amorphous structure present on the surface of the muscle fiber.
Figure 3.
Dystrophin models and dystrophin bolt. A. Antiparallel dimer model. C-terminal (CT) domain and AB domain (ABD) are present at both ends of antiparallel dimer oriented dystrophin fixed by respective IMP. Although this figure is drawn as if two dystrophin molecules were composed of solid metal that were attaching throughout, how these long molecules were attached to each other and to IMP was not well considered. Note that in this model, dystrophin is fixed in parallel to the lipid bilayer and perpendicular to IMP. This imaginary figure was drawn based on the models of Kunkel19) and Campbell29) and with binding of α-DG to laminin taken into consideration. For details see the text. B. Monomer model. Three strata of the sarcolemma, namely, an actin filament in the subsarcolemmal cytoskeleton networks, lipid bilayer and basal lamina, are stitched and fixed by dystrophin:β-DG:α-DG.50,51) These three proteins, dystrophin:β-DG:α-DG, altogether are considered as a functional unit and termed the dystrophin bolt. Thus, an actin filament and laminin serve as scaffolds of dystrophin bolt. When dystrophin is replaced by utrophin, the utrophin bolt composed of utrophin:β-DG:α-DG is formed. Because the function of the dystrophin bolt and that of the utrophin bolt are qualitatively the same, they together are termed fixation bolts in this review.
The lipid bilayer is the sole layer among the sarcolemma that serves as the barrier to maintain the intracellular environment constant, because ionized substances are not permeable. Thus, its intactness is prerequisite for keeping life for the cell. However, the lipid bilayer that is penetrated with a lot of proteins is mechanically weak. Thus, the muscle fiber upon contraction threatens its own life by generating strong tension.
The basal lamina is a thick and mechanically tough sheet that extracellularly attaches to the lipid bilayer, but is permeable to various molecules including proteins.
The subsarcolemmal cytoskeleton networks present beneath the lipid bilayer of muscle fibers, especially at costamere, contain a number of γ-actin filaments.15) The γ-actin filaments are connected to desmin by plectin to form the main components of the networks that form the lining of the lipid bilayer. It was known from the early days of new era that the γ-actin filament serves as a scaffold for the N-terminus of dystrophin. Unfortunately, only a few EM studies of the subsarcolemmal cytoskeleton networks in relation to dystrophin have been made.16,17)
Shortly, the basal lamina and the subsarcolemmal cytoskeleton networks mechanically protect the lipid bilayer.
2.4. The Kunkel model of dystrophin attachment to the cell membrane in 1988.
Kunkel’s paper18) reporting the presence of dystrophin on the cytoplasmic surface of the sarcolemma was published in the same issue of Nature as our paper above-mentioned.14) At the end of 1988, Kunkel19) published a hypothetical model of how dystrophin is placed on the sarcolemma, based on the following: (1) that dystrophin does not have a membrane spanning domain; (2) an assumption, that dystrophin is a long molecule and thus must be fixed at both ends: (2a) the N-terminus must bind to an actin filament and (2b) the C-terminus must bind to an imaginary “integrated membrane protein” (IMP); and (3) an assumption that dystrophin is present as an antiparallel dimer as described above. The assumption (3) obligatorily results in that dystrophin has binding sites to IMP at its both ends and is attached parallel to the cell membrane. This Kunkel model was composed only of IMP present in the lipid bilayer, dystrophin dimer and actin filaments (cf. Fig. 3A; The basal lamina, including laminin, and α-DG drawn in this figure, were not included in the Kunkel model).
When Kunkel published this model, no experimental data to show the existence of the IMP was available. However, this model greatly influenced on the formation of the 1991-models of Campbell and later models of ourselves as explained below.
2.5. Our main works (1): Immunohistochemistry.
Having introduced the immunohistochemical techniques into my laboratory, we carried out various studies, including developmental expression of dystrophin20) but I will cite here only those closely relevant to this review.
In 198921) and 1991,22) we discovered that utrophin is expressed on the sarcolemma of DMD human and mdx mouse muscles in place of dystrophin (Fig. 2). In 199323) and 1994,24) we found that β-dystroglycan (β-DG; the dystrophin binding protein as explained below) expression is reduced in the DMD and mdx muscles but is clearly present on the sarcolemma. Because utrophin is anchored to β-DG in DMD muscle, utrophin must be distributed less densely on DMD sarcolemma than dystrophin present on normal sarcolemma.
3. Dystrophin and the dystrophin-associated protein (DAP) complex and its architecture
3.1. Our main works (2): Biochemical purification of dystrophin and DAPs. Our modification of Campbell’s method.
We started to develop purification methods for dystrophin in 1988, soon after our studies localized it on the sarcolemma. However, Campbell’s first paper25) on the preparation of dystrophin and many other proteins that were copurified with dystrophin was published in Nature on March 16, 1989.
Campbell at first prepared the sarcolemmal fraction from rabbit muscle. Then, its lipid bilayer was dissolved with a detergent, digitonin. The resulting membrane proteins were fractionated using a Wheat-germ agglutinin (WGA) column and anion exchange column, followed by anti-dihydropyridine-receptor (DR) antibody column. His final sample showed about 20 bands upon SDS polyacryl amide gel electrophoresis (SDS PAGE) in addition to the dystrophin band. Some of them were glycoproteins that must be membrane-integrated or extracellular proteins.
Soon, we adopted his method with some modifications.26) After the WGA and anion exchange column separation procedures, we used a gel filtration column instead of the anti-DR antibody column. Our final sample was assumed to contain about 10 proteins (Table 1 ), judging from the bands that were separated by SDS PAGE in addition to the band corresponding to dystrophin. We confirmed that some of them were glycoproteins.
Table 1.
Dystrophin-associated proteins
| Current terms | Abbreviations | Previous terms | Molecular weight (kDa) |
|
|---|---|---|---|---|
| Campbell | Ozawa | |||
| α-dystroglycan | α-DG | 156DAG | — | 156 |
| β-dystroglycan | β-DG | 43DAG | A3a | 43 |
| α-sarcoglycan | α-SG | 50DAG | A2 | 50 |
| β-sarcoglycan | β-SG | — | A3b | 43 |
| γ-sarcoglycan | γ-SG | 35DAG | A4 | 35 |
| δ-sarcoglycan | δ-SG | — | — | 35 |
| sarcospan | SPN | 25DAP | A5 | 25 |
| dystrobrevin-1 | DB-1 | — | A0 | 94 |
| α-syntrophin | α-SYN | 59DAP | A1 (acidic) |
59 |
| β-syntrophin | β-SYN | A1 (basic) |
62 | |
Dystrobrevin-1 is the largest isoform of this protein. α-Syntrophin is an acidic protein, present abundantly on the protoplasmic surface of the lipid bylayer. β-Syntrophin is a basic protein, present almost restricted to the neuromuscular junction. The prefixes α and β are often omitted, when discrimination of α- and β-syntrophin is not necessary.
We then found that on treatment with a protein cross-linker, bis-(sulfosuccinimidyl) suberate, all of these proteins excluding A2 formed a huge cross-linked product that moved only a short distance from the original level by SDS PAGE. This suggested that these proteins were directly or indirectly associated with dystrophin excluding A2. However, we later showed that A2 indirectly bound to dystrophin.
In 1990, after we submitted our manuscript, Campbell published his second paper27) on a revised protein preparation method. By this revised method, he also obtained about 10 protein bands in addition to dystrophin band. He showed the association of dystrophin with these proteins in a way different from ours.
Although Campbell also obtained about 10 bands similar to us, it was soon revealed that individual proteins discovered by both groups were not completely the same (Table 1). In our first biochemical analysis, we did not find Campbell’s 156DAG (α-dystroglycan, α-DG). In Campbell’s list, our A0 (dystrobrevin-1) was not included. Furthermore, although Campbell reported in 1990 that 43 kDa proteins migrated as a doublet, he reported in 199228) that these two proteins were immunologically indistinguishable, concluding that the doublet was derived from a single protein. In contrast, we tenaciously persisted with our claim since 1990 that there are two types of 43 kDa glycoprotein, namely, A3a (β-dystroglycan, β-DG) and A3b (β-sarcoglycan: β-SG) on the basis of isoelectric focusing analysis.
3.2. The Campbell 1991-model of the dystrophin–DAP complex and cloning of dystroglycan gene in 1992.
In the following description, I will use the current terminology of dystrophin-associated proteins (DAPs), although each DAP term was coined later through several steps as our understanding of the characteristics of the DAP progressed.
Thereafter, Campbell published several papers on dystrophin and DAPs. In Cell published in 1991,29) he classified dystrophin and DAPs into three categories, namely, extracellular (α-DG), membrane-integrated (α-SG, β-DG, γ-SG and sarcospan) and intracellular (dystrophin and dystrobrevin-1) proteins, based on their experiments on membrane fractions treated with alkaline buffers. Extracellular and the membrane-integrated proteins except sarcospan were glycoproteins and could be stained with lectins. All the glycoproteins except α-DG were stained by 3-trifluoromethyl-3-iodophenyl diazirine (TID), a marker of the hydrophobic AA sequence that spans the lipid bilayer. Sarcospan was unique among the DAPs in that it was not glycosylated but strongly binds to TID. Furthermore, he showed that α-DG has both O- and N-sugar chains.
Then, he constructed his 1991-model, modifying the Kunkel model on the basis of above findings.29) Campbell replaced IMP with the above-described four membrane-integrated proteins and putting α-DG on these proteins outside the sarcolemma. Thus, his model was essentially similar to Fig. 3A, except that the basal lamina including laminin was not included. In addition, syntrophin was supposed to bind with the intracellular domains of these membrane-integrated glycoproteins. The binding partner of dystrophin was not determined among DAPs.
The impact of his model, albeit immature, was so strong that on seeing the model, many readers took it as if he had actually determined all individual protein–protein bindings depicted in the figure, although he did not describe evidence to show that he had determined those bindings. Contrary to this tendency, Davies in her review30) provided a well-reasoned critique discriminating between the findings given by Campbell, by us and by others in 1994. This review is important as an explanation and criticism of the model by a third person.
In 1992, Campbell28) reported that α-DG and β-DG are encoded by a single gene, of which protein product must be post-translationally processed, although a cleavage site on the protein was not known. He named the precursor protein dystroglycan (DG). The processed proteins were called α-DG (156DAG) and β-DG (43DAG). Then, he showed that native glycosylated α-DG binds to laminin, a major component of the basal lamina, in the presence of calcium ions, whereas recombinant non-glycosylated DG does not, suggesting that the extracellular protein α-DG connects to the basal lamina via the sugar chains. This showed the connection of the dystophin–DAP complex with the basal lamina. Thus, he discussed as follows: “the function of dystrophin is to link the subsarcolemmal cytoskeleton through a transmembrane complex to an extracellular glycoprotein which binds laminin.” Then, his model was redrawn similar to Fig. 3A where IMP was composed of four proteins as described above, namely, β-DG, α-SG and γ-SG and sarcospan. But still, which DAG corresponds to IMP and what portion of dystrophin binds to IMP was not yet determined.
Initially, studies constructing the architecture composed of the “native” dystrophin–DAP complex were carried out by Campbell’s group. However, after his 1991-model and DG cloning was published, with some overlapped period, the study was mostly succeeded by our group. It seemed to me that he had a stronger interest in determination of the characteristics of “individual” DAPs including their gene cloning and in analysis of human clinical and animal model materials with his antibodies rather than in finding out new interactions between the protein molecules of the complex. Thereafter, he looked almost ceasing from exploring new protein–protein interactions, until he only returned to this study in 1997 (see below).
3.3. Our model of the dystrophin–DAP complex in 2000.
For a few years after establishment of the purification methods, we prepared for promoting our study on the architecture of the dystrophin-DAP complex.31) We considered that it is inevitable to determine the binding partner of each protein to construct the architecture. Because DAPs are composed of about 10 kinds of protein, it is difficult to determine each binding partners using the whole dystrophin-DAP complex or to search for each pair with in vitro methods. Accordingly, we decided to biochemically extract subcomplexes composed of a few proteins from the whole complex, because binding partners can be easily determined from such small subcomplexes. In addition, the bindings thus elucidated must represent those working in vivo. Furthermore, once the binding partners are determined using native sample, details of each binding can easily be analyzed by in vitro techniques.
By means of this strategy, we got several subcomplexes (Fig. 4 A, B and C) and determined the binding partners during 8 years since 1992. Each time when we got new data, we constructed small models for the subcomplexes. Based on them, we further constructed and published our model taking both the Kunkel and Campbell models into consideration. Our model was constructed and repeatedly revised step by step as our understanding progressed. It is troublesome to show each of our models at different stages. Here, I show only our final model published in 2000,32) which was revised later by addition of actin filament associated to the AB site as described by Ervasti10) (Fig. 4D).
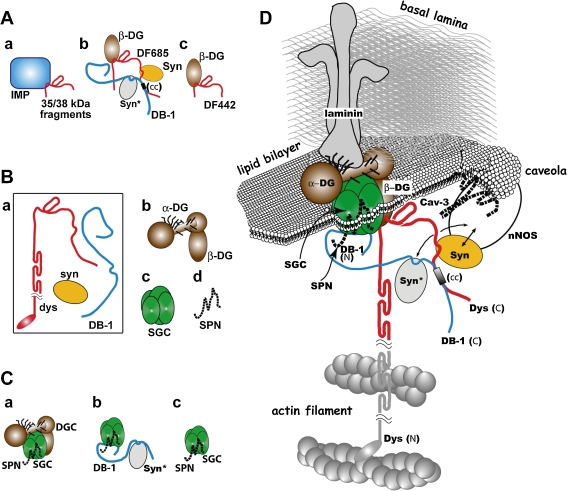
Figure 4.
Our model of the dystrophin–DAP architecture in 2000. IMP: integrated membrane protein. For other abbreviations, see Table 1. For further details, see text. A: Models of the subcomplex architecture published in 1992–5. a: Subcomplex obtained by proteinase treatment composed of a glycoprotein complex (blue box) including IMP and dystrophin fragments (35/38 kDa fragments spanning mainly CR domain expressed in red line).40) b: DF685 binds to β-DG, syntrophin (shown in yellow oval) and dystrobrevin in overlay binding assay.42,57) For Syn* (gray oval) and (cc) (small box), see text. c: DF442 binds to β-DG (full binding) in overlay binding assay.42) B: Subcomplexes derived from the dystrophin–DAP complex treated by octyl glucoside in 1994.52) a: The void fraction of gel filtrate that contained dystrophin, dystrobrevin-1 and syntrophin. b: the DG complex; c: the SG complex; and d: sarcospan. C: Subcomplexes published in 2000.32) Complex a, b and c were constructed on the basis of the data obtained from the experiment described in Our Main Works (6). The complex c was previously discovered by Campbell in 1997.33) D: The colored part of this model was constructed in 2000 by superimposing our submodels, mainly those shown in A, B and C.
With a few exceptions, the colored part of this model composed of the DAPs was constructed by superimposing small models deduced from our subcomplex preparations, as explained below in Our Main Works (3), (4) and (6) (Fig. 4A, B and C). The exceptions were the binding of dystrobrevin-1 with syntrophin (syn*, gray oval) and that of sarcospan with the SG complex (buried under the colored part) that were discovered by Froehner33) and by Campbell,34) respectively in 1997. On the other hand, the dark parts of Fig. 4D were drawn in accordance with the findings of other laboratories. All the associations of DAPs to its adjacent structural proteins were determined in other laboratories, namely, the associations of dystrophin:actin, α-DG:laminin and syntrophin:nNOS were experimentally determined by Perry,35) Campbell,28) and Bredt,36) respectively.
Our initial descriptions on the protein binding were not necessarily detailed, although we later studied details of some of the binding partners. With the lapse of time, many of the bindings that we determined were analyzed in detail by other researchers, on which I reviewed elsewhere.37) In constructing Fig. 4A and D, we included the detailed binding site of dystrophin and dystrobrevin-1 reported by Kunkel in 1997.38)
3.4. Our main works (3): Determination of the binding domain of dystrophin to β-DG, the most important interaction for understanding the function of DMD.
Back in 1992, while we39) were examining the proteinase-sensitive loci of dystrophin, we40) collected glycoproteins present in the enzyme-treated dystrophin–DAP complex sample using a lectin column. We obtained a few types of small fragment of dystrophin that bound to these glycoproteins. We considered that the portion common among these fragments must include the domain by which dystrophin bound to IMP. We examined these fragments using various anti-dystrophin antibodies and partially by AA analysis,40) and found that these dystrophin fragments had molecular weights ranging from 35 to 38 kDa. We further determined that the common portion spans a tiny portion of rod, the CR domain and the initial portion of the C-terminal domain. Therefore, the IMP binding potion was not the C-terminus of dystrophin and different from the anticipations of Kunkel19) and Campbell29) (Fig. 4A-a). It was located at more proximal site of dystrophin.
A few years before our findings, Kunkel41) pointed out based on their dystrophin gene mutation analysis that the nucleotide sequence corresponding to the CR domain was lacking in most DMD cases. On this basis, we interpreted our own findings as follows: the absence of this dystrophin binding site for IMP results in the complete loss of dystrophin from the sarcolemma leading to DMD.40) In other words, the absence of binding between dystrophin and IMP makes dystrophin unable to be fixed at the subsarcolemmal cytoskeleton networks and loss of the connection between actin filament and IMP causes the disease.
In 1994, to determine IMP, we42) constructed various recombinant peptide fragments of dystrophin that were considered to encompass the domain for binding IMP (Fig. 4A-b and A-c). Using an overlay binding assay method with these fragments, we found that IMP is a single glycoprotein, β-DG. Dystrophin must bind to the intracellular domain of β-DG. Indeed, Campbell43) later showed that the intracellular C-terminal 15 AA residues of β-DG binds to the above described dystrophin fragment.
We42) further found that a fragment AA #3026–3264 (DF264) bound to β-DG, but the binding was weak (reduced binding), whereas a longer fragment AA #3026–3442 (DF442) strongly bound to β-DG (full binding). Later, we confined the full binding domain to AA #3026–3345.44) All of these fragments included a small portion of the C-terminal portion of the rod as later pointed out by others to be a part of the WW domain, a protein–protein binding consensus domain.
By specifying the AA residues of β-DG binding domain (DGBD) and identifying the IMP as β-DG, we came to the conclusion that dystrophin is fixed to the actin filament in the subsarcolemmal cytoskeleton networks at its N-terminus, as found by others, and to a membrane-spanning protein β-DG at DGBD. These two binding sites are the basis of our understanding on dystrophin function and DMD pathomechanism. This concept was used by Takeda45) in 1998 for designing the micro-dystrophin used for experimental gene therapy of DMD as explained in Discussion (6). Thereafter, several papers from other laboratories elucidated the details of the association between DGBD and β-DG using various methods, including X-ray diffraction (for review37)).
In 2004, we44) specified that physiological DGBD has AA #3026–3345, namely, DGBD ranges from the last 54 AA residues of the rod to the CR domain excluding the last 15 AA residues. The end of this physiological DGBD was determined by comparing our data to the in vivo experiment of Chamberlain.46) We further suggested that to convert from the reduced binding to full binding, a certain intra-molecular structure of this domain was required. As for utrophin, which was shown also to bind to β-DG in a few laboratories, we44) showed that the binding affinity of utrophin to β-DG is about one half that of dystrophin.
3.5. Discussion (1): Kunkel’s reading frame rule from the viewpoint of protein–protein interaction.
The relationship between the translational reading frame and the AA sequence was already known in 1960. What was new and important in Kunkel’s work in 198847) was that he applied this formula to the prediction of the relationship between reading frame changes and the clinical severity of patients with dystrophin gene mutations. His hypothesis predicted that when the change in the number of nucleotides in a mutation is 3N + 1 or 3N + 2 (N: integer), namely, out-of-frame mutation, the down stream AA sequence changes and a stop codon is formed. Thus, a truncated dystrophin missing the C-terminal side of the protein may be generated. When the number changes by 3N (the N of AA residues at the mutation site is deleted or inserted) the downstream AA sequence does not change except at the mutation site. Today, this beautiful formula is often called the reading frame “rule”. According to a recent report from a French project team,48) among 2,084 independent mutational events, the reading frame rule holds true for 96% of DMD patients and 93% of BMD patients. However, soon after Kunkel published this rule, Worton49) reported exceptions to the rule in 1988 when a mutation occurs at or close to the AB domain.
Since 1995,50) we repeatedly claimed that we have to consider the reading frame rule on the basis of changes in dystrophin at the protein level. As stated above, when any of the structures that mediate dystrophin binding to actin filaments or of DGBD are defective, dystrophin cannot be fixed to the sarcolemma, resulting in the development of DMD. I considered that the DMD cases with in-frame mutations among Worton’s exceptions49) to the rule must be due to the loss of function of dystrophin for binding to an actin filament. However, this discussion is not enough and for further details, see my review published elsewhere.51)
In an in-frame mutation, the AA sequence is not disturbed in the DGBD. Although the size of the rod is decreased (or increased) in this case, dystrophin can bind to the actin filament and β-DG and is functional.
It is also important to consider that premature stop codon mutations in dystrophin lead to nonsense mediated decay (NMD) of the mRNA. If NMD functions efficiently, the reading frame rule might be explained by this mechanism in most cases, as little mRNA of mutated dystrophin will become available for translation by the ribosome. However, it is also known that NMD is often not completely efficient and a surviving mRNA with a nonsense mutation must be translated resulting in a defective dystrophin protein lacking the DGBD. The function of such a defective dystrophin must be considered as described above.
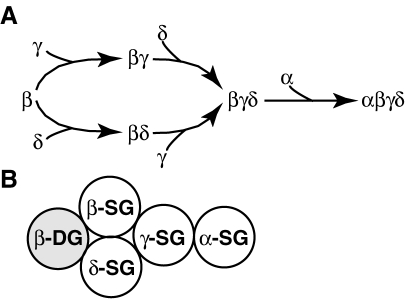
3.6. Our main works (4): Discovery of the DG and SG complexes.
In 1994, we found the DG and SG complexes,52) and a non-glycosylated (intracellular) protein complex (Fig. 4B). These complexes were obtained by treatment of the purified dystrophin–DAP complex with a detergent, n-octyl β-d-glucoside, followed by fractionation using a gel filtration column.52) We coined the term sarcoglycan (SG) at this time. The classification of glycoproteins into the DG and SG complexes was widely accepted thereafter, and used as the standard classification of dystrophin-associated glycoproteins.
[1] The DG complex composed of α-DG and β-DG (Fig. 4B-b): As shown by Campbell28) in 1992, they are encoded by a single gene and posttranslationally processed. Here, we showed that these two proteins are present forming a single protein complex after processing, which we named ‘the DG complex’ after Campbell’s term DG. Campbell’s discovery is concerning the genotype and synthesis of DG, whereas ours is concerning the DG subcomplex in the architecture of dystrophin–DAP complex at the phenotype level.
[2] The SG complex (Fig. 4B-c): This complex is now known to be composed of 4 subunits. We initially considered that the SG complex was composed of 3 subunits, α-SG (A2 or 50DAG), β-SG (A3b) and γ-SG (A4 or 35DAG).52) We termed it the sarcoglycan complex. Details of the characteristics of these subunits were described elsewhere.53)
In 1996, Nigro54) cloned a new gene highly homologous to γ-SG and termed it δ-SG (see below). In collaboration with him, we55) showed that δ-SG (glycoprotein) is also contained in the purified SG complex preparation. Therefore, the SG complex is composed of four subunits. The molecular architecture of the SG complex is described in Our Main Works (6). The number of subunits forming the SG complex does not change even after other two subunits ε-SG and ζ-SG were discovered (for review56)). These are not major components of the SG complex in skeletal muscle.
Sarcospan was separated by octyl glucoside treatment from the SG complex.
Each subunit of the SG complex and O-sugar chains of α-DG is responsible for 4 kinds of ARDMDs (sarcoglycanopathy) and 6 kinds of CMDs, respectively (see below). Sarcospan is not involved in the cause of muscular dystrophy.
[3] The dystrophin–dystrobrevin–syntrophin complex: In 1994, we52) detected dystrophin, dystrobrevin-1 and syntrophin in the void fraction of gel filtration after treatment of the dystrophin–DAP complex with n-octyl β-glucoside (Fig. 4B-a). It was possible that these three proteins form a subcomplex, because we had previously found that they were included in the dystrophin–DAP complex.26) In 1994 in a separate work, we42) examined this question with the overlay binding assay of these proteins using two kinds of recombinant dystrophin fragments spanning DF442 and AA #3026–3685 (DF685: dystrophin fragment spanning from DGBD to the terminus of dystrophin molecule) (Fig. 4A-b and A-c). In addition to the binding to β-DG, DF685 bound to dystrobrevin-1 and syntrophin, whereas DF442 did not. Therefore, dystrobrevin-1 and syntrophin bind to the C-terminal domain of dystrophin. In 1995, we57) found that syntrophin bound to the dystrophin fragment AA #3443–3494. Simultaneously, Kunkel58) also reported the same syntrophin binding site of dystrophin. Furthermore, we57) found that dystrobrevin-1 bound to the dystrophin fragment AA #3495–3544 (Fig. 4A-b). In 1997, Kunkel38) specified the binding sites to be the coiled-coil (cc) domains present on both proteins, and Froehner33) found binding of syntrophin (Syn*) to dystrobrevin-1 by the overlay binding assay.
3.7. Discussion (2): The dystrophin bolt: A complex of dystrophin and dystroglycans that connect actin and laminin.
The following items are generally accepted: (1) Muscle fibers contract generating force and heat. Upon contraction, chemical energy contained in ATP is changed to the mechanical energy that cannot be analyzed by biochemical methods but by physiological methods.
(2) It is certain that muscle fibers in health are toughly constructed so as not to be injured by the force generated during their contraction.
(3) It was becoming likely with time that dystrophin and the glycoproteins do not have biochemical activities, such as enzymatic activity.
(4) DMD ensues, when dystrophin and its associated glycoproteins (DAG) are decreased or lost. Dystrophin and DAG must be related to defects of the tough construction of the muscle fibers.
(5) Muscle contraction is a strong risk factor of DMD. In other words, muscle fibers are prone to injury by contraction. The injury causes leakage of soluble cytoplasmic proteins such as CK.5)
Taking these together, I consider that DMD is primarily caused by impairment of some constituents of a large architecture on the sarcolemma including dystrophin, which must play essential roles in preventing the cell body from injury by contractile force. Thus, I conceived of a structure composed of 3 proteins, dystrophin, β-DG and α-DG from the architecture of the dystrophin–DAP complex (Fig. 4D) and collectively named this triad the dystrophin axis50) (this was later renamed the dystrophin bolt59)). The dystrophin bolt transmembraneously connects subsarcolemmal cytoskeleton networks with the basal lamina (Fig. 3B). An actin filament and laminin serve as the scaffolds of the dystrophin bolt. By stitching the networks and basal lamina, the lipid bilayer that is inserted between them can be mechanically and firmly protected. The dystrophin bolt is distributed with a high density all over the sarcolemma.13,14) In DMD muscle, dystrophin is replaced by utrophin with a low density,22) whose binding ability to β-DG is weaker than that of dystrophin.44) As shown by Davies,60) forced expression of utrophin in mdx mice rescued them from muscle atrophy. Thus, the utrophin bolt, composed of utrophin and α-DG and β-DG, is functionally of the same quality as the dystrophin bolt. Together they both are called “fixation bolts”.
In human DMD muscles, the dystrophin bolt is lost but the utrophin bolt is weakly expressed.22) Thus, the pathogenesis of DMD can be considered primarily as functional insufficiency of the fixation bolt.
When dystrophin was discovered, it was claimed that dystrophin assembles as an antiparallel dimer.9) Both the Kunkel and Campbell models were constructed on the basis of this claim (see Fig. 3A). In contrast, we considered that dystrophin is a monomer on the basis of the EM observations on the sarcolemma by Cullen17) in 1991. He showed that gold-labeled anti-dystrophin antibody against the C-terminus stained the site over or outside of the lipid bilayer, whereas that against the rod (AA #1183–1388. Roughly the middle of rod.) stained the deeper position at 15–20 nm (some bound as deep as at 75 nm) from the lipid bilayer. In contrast to Cullen’s discussion, we interpreted his observations that (1) its C-terminus is located at close to the lipid bilayer, (2) its N-terminal should present deeper apart from the lipid bilayer and thus (3) dystrophin is present in vivo as a monomer (see Fig. 3B). Dystrophin spans between β-DG and an actin filament with some angle to the lipid bilayer or with some bending of dystrophin molecules probably at their hinges upon relaxation, but may be turned and stretched perpendicularly toward the sarcolemma by cytoplasmic flow upon contraction (see Discussion (3)).
While we used the monomer model in our publications since 1994,50,61) Kunkel62) and Ervasti63) independently showed in 1997 that dystrophin can be present as a monomer. Then, Campbell also adopted the monomer model, which was soon accepted worldwide.
To determine whether dystrophin is an antiparallel dimer or a monomer is not merely of biochemical interest; there is a crucial difference between them in relation to the function of dystrophin. (1) In the antiparallel dimer model (Fig. 3A), the AB domain and C-terminus (Originally, the C-terminus was assumed to be the IMP-binding site.) must be present at both ends. Both ends of the dimer must be fixed by a transmembrane IMP, and must be closely localized to the lipid bilayer. Thus, dystrophin must be fixed parallel to the lipid bilayer and roughly perpendicular to IMP. In the dimer model, force directed to dystrophin would not be effectively conducted to IMP. (2) In the monomer model (Fig. 3B), dystrophin DGBD binds to IMP (β-DG) and the AB domain anchors to an actin filament located within the length of dystrophin. Thus, dystrophin can be fixed roughly perpendicular to the lipid bilayer, at least upon contraction. In other words, dystrophin is fixed extending strait from IMP to the subsarcolemmal network. In the monomer model, force directed to dystrophin must be effectively conducted to IMP.
Needless to say, biomaterials are flexible and not the same as solid metals. However, I would like to emphasize that a vertical component against the lipid bilayer of the force given to dystrophin is effectively conducted to IMP, when the monomer model is adopted. In this way, stitching of the three layers by the dystrophin bolt can work well. Details of this concept will be described in Discussion (3) and in Discussion (4).
4. The second stage of the dystrophin era
In August of 1994, the 8th Neuromuscular Congress, which is held once every four years, was held in Kyoto. Almost all of the top researchers from various fields of muscular dystrophy research, especially those of genetics, gathered together and intensely discussed their recent progresses. New leaders from new generations gave lectures on their newly emerging fields of the science. This meeting was an unprecedented success both in quality and quantity.
Despite the success of the meeting, many participants must have become aware that the tide of muscular dystrophy research was clearly changing from dystrophin genetics to many new directions. Mainly, one was change in focus from gene to protein, and the other was that from genetics of DMD to those of other neurological disorders as well as other muscular dystrophies. Trials on DMD gene therapy were about to thrive, as the fundamentals for such trials became available.
4.1. Our main works (5): Sarcoglycanopathy (former ARDMD; SG-pathy): proposal and verification of a hypothesis.
In 1992, Campbell64) together with Fardeau showed that 50DAG was absent from the cell membrane of ARDMD muscle fibers and Vance65) reported that the Tunisian ARDMD locus mapped to chromosome 13. In the next year, Zatz66) claimed that ARDMD is genetically heterogenous, because some Brazilian ARDMD gene loci did not map to 13q. Getting a hint from these works, we examined whether ARDMD muscles could be immunostained using 3 types of anti-SG subunit antibodies. As expected, we found that all three subunits were absent in the sarcolemma of ARDMD muscle fibers. Thus, it was likely that the SG complex is responsible for ARDMD.
In accordance with this finding, we61) predicted in 1994 immediately after the Kyoto meeting that if any of the SG genes is mutated, its protein product is missing, which then results in loss of the whole SG complex, resulting in ARDMD. This may explain the genetic heterogeneity.
In this statement, we had implicitly claimed that the number of varieties of ARDMD must be the same as that of subunits of the SG complex. All the patients with varieties of ARDMD must be classified according to the respective SG gene that is mutant. This hypothesis further implicitly claimed that in the case of ARDMD, we can clone each responsible gene by a conventional method on the basis of the partial AA sequences of each SG subunit, and the complicated positional cloning method is not necessarily required.
In 1994, the Campbell–Fardeau team67) cloned an ARDMD responsible gene that mapped to chromosome 17q21 and called this molecule “adhalin”. In 1995, the Kunkel–Ozawa team68) (We provided partial AA sequences of A3b to Kunkel who cloned the gene on the basis of the sequences) and the Campbell–Beckmann team69) simultaneously cloned the A3b gene. This gene was mapped to chromosome 4q12 and mutations in the A3b gene were found in the corresponding ARDMD patients. The SG complex was also absent in the muscle fibers of these patients.
Based on the suggestion of the editor of Nature Genetics to which both teams had submitted their papers, Kunkel proposed a name for this molecule to the relevant researchers including Campbell: Let us name the present molecule β-SG. The names of the molecules of this family should be called γ- and δ-SG, etc. In this context, the term “adhalin” given to 50DAG is abandoned and 50DAG should be renamed α-SG. His proposal was agreed on by all the researchers involved in the cloning of these genes.
At that time of the proposal, the γ-SG gene had already been cloned in our laboratory,70) and other necessary data were being collected in Kunkel’s and Vance’s laboratories using materials from Hentati and Middleton. It was mapped at 13q12 corresponding to the Tunisean type of ARDMD.65)
In 1996, the gene for the fourth ARDMD was mapped to chromosome 5q33 by Zatz.71) Independently, as described above, the δ-SG gene was cloned and was also mapped to chromosome 5q33 by Nigro.54) They72) showed that the 5q33 mapping patients had mutations on the δ-SG gene. We, in collaboration with Nigro,55) showed that its protein product is actually present in a purified SG complex. After a while, Zatz and Kunkel73) reported that in all the cases originating from mutations in any of four SG genes, the patient muscles lack the entire SG complex by immunohistochemistry, although low levels of some subunits are sometimes observed. These studies confirmed that our hypothesis61) was true. Afterwards, I summarized these processes in detail in a review published elsewhere.53)
Another claim, that the number of the types of ARDMD that can be genetically discriminated is the same as that of the SG subunits, was also considered to be valid. It is because no new SG-pathy other than these 4 types has been reported.
4.2. Our main works (6): Finalizing the model of the architecture of the dystrophin–DAP complex and the formation of the architecture in development.
[1] Finalizing our model of the architecture of the dystrophin–DAP complex: We obtained 3 subcomplexes from a purified dystrophin–DAP sample by the following treatment32): (Fig. 4C) (1) The dystrophin–DAP sample was incubated in 0.1 M NaCl at pH 7.5 at 37 ℃ for 6 hours and separated by gel filtration chromatography. One of the peaks was refractionated by gel filtration chromatography and finally the fraction for the DG–SG–sarcospan complex (Fig. 4C-a) was obtained. This was the first direct evidence for the association of the DG and SG complexes, although their association had long been accepted without doubt on the basis of some other indirect evidence. (2) The dystrophin–DAP sample was incubated in 1 M NaCl at pH 7.5 for 6 hours and separated by gel filtration chromatography. One of the fractions was immunoprecipitated with anti-α-SG antibody and the fraction for the syntrophin–dystrobrevin–SGs–sarcospan complex (Fig. 4C-b) was obtained. Binding of dystrobrevin-1 to the SG complex must be noted, because many of the current models ignored this binding. Yoshida later gave definitive evidence on the binding of syntrophin to dystrobrevin-1 (personal communication, unpublished data). Although their in vitro binding was previously described,33) the evidence for the binding from the native complex was first shown here. (3) The fraction of (2) before immunoprecipitation was further incubated with 1 M NaCl for an additional 6 hours and separated by gel filtration chromatography to obtain the SG-sarcospan complex (Fig. 4C-c): this final complex was previously reported by Campbell.34)
In order to construct a model for the overall architecture of dystrophin and DAPs, all the subunit architectures illustrated in Fig. 4A, B & C were superimposed on the actin filaments, the lipid bilayer and the basal lamina. A larger protein architecture model obtained on the basis of our data is shown as the colored parts of the model in Fig. 4D, with a few exceptions described above. Ten years have elapsed since the publication of this model, and no modification of this colored model has been made.
[2] Developmental biology of the SG-complex: At the early stage of myogenesis in vitro, when myoblasts are predominant in a muscle cell culture, we74) found that α-, β-, γ- and δ-SG are detected at both the mRNA and protein levels, although their amounts are small. When myotubes are formed, the expression level of all of the SG proteins increases with those of other muscle-specific proteins. However, their differential patterns at the level of SG mRNAs were not the same. The expressed level of α- and γ-SG mRNAs increased, whereas those of β- and δ-SG mRNAs did not, suggesting that the transcription of α- and γ-SG, and that of β- and δ-SG are differentially controlled.
To clarify the problem of how α- and γ-SG mRNAs are regulated, we75) studied the γ-SG gene. We observed that the γ-SG mRNA promoter has an E-box that is a consensus site for myogenic gene b-HLH type protein, such as myoD, as well as an A/T-rich element that binds to MEF2. These observations suggest that expression of γ-SG is regulated by classic myogenic differentiation under the control of myogenic regulatory genes.
We76) found that at a specific stage of early myogenic differentiation, it was possible to detect the incompletely formed SG complex composed of a small number of subunits binding to β-DG. We analyzed these nascent SG complexes and determined the order of binding of the SG subunits to β-DG, as shown in Fig. 5A. This was roughly compatible with the report of Zatz and Kunkel73) who assumed the order of SG complex formation from the residual of SG subunit in SG-pathy muscle biopsy samples from the patients. On the bases of our findings together with their and Sakamoto’s reports,77) the SG complex directly bound to β-DG with β- and δ-SGs and then γ-SG and finally α-SG (Fig. 5B). In the cases when ζ-and ε-SGs are expressed, they may replace γ- and α-SG, respectively, forming the different SG complexes with four subunits.
Figure 5.
Formation of the SG complex. Sequence of binding of the SG subunits. A. Sequence of formation of the SG complex. B. Architecture of the SG complex that binds to β-DG. This model was constructed on the basis of reports from several laboratories including ours. Note that non-specific β- and δ-SGs occupy essential loci for the binding to β-DG, but muscle-specific γ- and α-SGs are placed outside.
Generally, a membrane-integrated protein is translated in the endoplasmic reticulum (ER) and then transported to the Golgi apparatus. The apparatus, in turn, is partially torn off to form transport vesicles that are transported to the sarcolemma. We76) analyzed the locus and the process of formation of the complexes using brefeldin A, which destroys the Golgi apparatus, and found that DG and the SG subunits are all translated in the ER. Both complexes are separately formed in the ER as soon as their subunits are synthesized. In the Golgi apparatus, the SG complex binds to both DG and sarcospan to form a larger transmembrane complex, which then becomes progressively glycosylated.
For the subsequent fate of this complex, based on evidence observed by us76) as well as on the general rule of protein synthesis, we can assume the following. Dystrophin is synthesized in free ribosomes and becomes associated through its DGBD to the protoplasmic domain of β-DG that is already bound to the SG complex in Golgi complex. After binding dystrophin, the vesicles are transported to and fused with the lipid bilayer, extruding dystrophin into the protoplasma. Since the AB domain remains free, it would bind to the subsarcolemmal actin filament available for the length of dystrophin. This may be the mechanism by which the dystrophin–DAP complex is fixed on the sarcolemma.
[3] Production of knock-out (KO) mice: During this period, we78–80) generated mice carrying KO gene for the β-SG, γ-SG or caveolin-3 gene. The β- and γ-SG KO mice showed muscular dystrophy phenotypes, as described above. Caveolin-3 KO mice showed only a limited muscular dystrophy phenotype.80)
5. My trials to elucidate the function of dystrophin and the pathomechanisms of DMD
When I retired at the end of March 2000, 12 years had already elapsed since the discovery of dystrophin. The main trend in DMD research in the third stage was shifting to the experimental gene therapy. Before and more frequently after my retirement, I fortunately had chances to write comprehensive reviews,53,56,59,81) and chapters of large books,5,37,51,82) on the dystrophin–DAP complex and muscular dystrophies, and learned more certainly that the dystrophin–DAP complex is a biochemically mute cytoskeleton component.
Accordingly, my question was why absence of dystrophin causes progressive muscle atrophy, in spite that dystrophin does not show any specified function in vitro. It is current trend to search for a biochemically active substance(s) as a responsible factor(s) of DMD, which must secondarily be disturbed due to absence of dystrophin. However, these trials are not successful and almost no answer to my question is available. Therefore, I must answer to my own question on the basis of our works and thoughts.
To answer this question, I tried to unite evidence from various laboratories concerning (1) experimental studies on dystrophin and DAPs, especially of molecular associations, (2) biology of muscle contraction that is a powerful risk factor of muscular dystrophy, and (3) clinical observations and related experimental studies including those derived in the pre-dystrophin era. Although available evidence was insufficient and my explanations deduced from these studies have gaps, I consider that it is worthy to raise questions for future investigators.
5.1. Discussion (3): Further understanding of function of the dystrophin bolt in relation to muscle contraction in normal muscles.
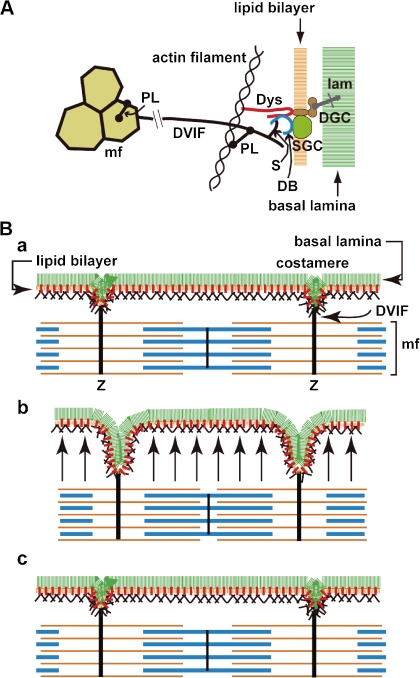
[1] The dystrophin bolt functions in two ways: First, the dystrophin bolt stitching the sarcolemma is diffusely distributed throughout the sarcolemma, as explained in Discussion (2) (Fig. 2 & 3B). Second, densely located at the costameres, it also serves as a part of larger structure that links the Z-bands of myofibrils and the basal lamina (Fig. 6A ).51,83) Desmin–vimentin intermediate filaments (DVIF)84) wind around individual myofibrils at the level of each Z-band fixing all myofibrils together to form a myofibril bundle. These DVIF further radiate away from the myofibrils to the costamere, a condensed portion of the subsarcolemmal cytoskeleton networks, where DVIF indirectly connect to the dystrophin bolt. This system was named the transverse fixation system (TFS),37) details of which were described elsewhere.51) Thus, I understood that the dystrophin bolt was a missing link of TFS that transmembranously connects the DVIF and the basal lamina.
Figure 6.
Transverse fixation system (TFS). A. Molecular architecture of TFS: DVIF that surrounds and fixes many myofibrils (mf) at the Z-band level and radiates outward to the sarcolemma. Roughly, desmin in the DIVF connects to actin filament in the subsarcolemmal cytoskeleton networks via plectin, then to the dystrophin bolt which is finally fixed to the basal lamina. It also connected to dystrobrevin via syncoilin and/or β-synemin that bind to the C-terminal domain of dystrophin, which is finally fixed to the basal lamina via dystroglycan. PL, plectin; S, syncoilin and/or β-synemin; lam, laminin. For other abbreviations see Table 1. B. Function of TFS: Festooning of the sarcolemma upon contraction observed in longitudinal section (B-b). a, relaxation; b, contraction; and c, relaxation. Red bar on the lipid bilayer indicates the dystrophin bolt. Arrow in b: flux of cytoplasm toward sarcolemma upon contraction. Note that the sarcolemma balloons upon contraction owing to the flux of the cytoplasm from the core of the muscle fiber. However, at costameres, which are moored to the myofibrils by TFS, the sarcolemma retracts. The ballooning of the sarcolemma becomes flat upon relaxation. For further details see the text.
[2] Balooning of the sarcolemma on muscle contraction: When we estimate a muscle fiber to be a cylinder whose radius is r (r being 25–100 μm), the radius increases with contraction. When a sarcomere (∼2.5 μm) shortens by 1/5, the radius increases to 1.12r, because the volume of the sarcomere remains constant at least at the initial stage of contraction. If we adopt the square pole model for a muscle fiber, the result is similar. We must note that the increase of the radius occurs within 10–20 milliseconds upon contraction.
Upon contraction, the cylinder does not thicken evenly along its length longitudinally (Fig. 6B), because the radius does not increase at costameres due to fixation by the TFS. Instead, the radius increases inbetween the costameres in a balloon-like manner. Indeed, in longitudinal section of contracted muscle fibers display a repeating pattern of retractions and balloonings when observed by EM, a pattern termed festooning.85) Note that because of this retraction at the costameres, the total surface area of the lipid bilayer is not decreased but is instead stretched upon contraction.
[3] Myofibrils, which are composed of a series of sarcomeres, are the only machine that generates a force that is exclusively directed along its longitudinal axis. The force upon contraction is not primarily directed toward the sarcolemma. Upon contraction, all sarcomeres shorten similarly, the pressure inside the adjacent sarcomeres simultaneously increases to the same level. Cytoplasm in a given sarcomere cannot longitudinally flow to the adjacent sarcomere over the Z-band. Thus, the cytoplasm must be forced to move in a transverse direction to push the sarcolemma outward. This movement gives rise to festooning (Fig. 6B).
Almost no study is available on the cytoplasmic movement upon contraction. It was known by X-ray diffraction study of living skeletal muscle fibers at tetanus when shortening reached at the plateau, the diameter of the cross section of sarcomeres increased reversely proportional to the size of contraction (effect of constant volume of the sarcomere),86) whereas in a saponin-treated (skinned) muscle fibers, whose lipid bilayer became permeable to cytoplasma,87) the diameter remained almost similar to that of the resting muscle.88) In my context, this difference in diameter can be interpreted as follows: In the skinned muscle fibers in tetanus, the cytoplasm squeezed from the shortened sarcomere must leak extracellularly through the permeable sarcolemma leaving the sarcomere diameter unchanged, whereas in the intact muscle, the cytoplasm once squeezed out of the shortened sarcomere would be forced back by the elasticity of the sarcolemma making the diameter larger. If this interpretation is the case, the approximate volume of the cytoplasm can be assumed that shuttles between inside and outside spaces of sarcomere during the contraction-relaxation cycle. Thus, the mean value of hydrodynamic pressure due to movement of cytoplasm may be obtained by studies on this line.
If the cytoplasm flow pushed the lipid bilayer very slowly in a quasi-static manner upon contraction, the pressure change would be slight and no harmful damage would occur on the lipid bilayer. However, because muscle contracts quickly and cytoplasm must be rapidly forced out through narrow clefts between the thin and thick fibers of myofibrils, the hydrodynamic force applied to the lipid bilayer by viscous cytoplasmic fluid may become far larger than the hydrostatic force.
[4] The muscle lipid bilayer has to bear the hydrodynamic force upon contraction. To prevent it from the destructive hydrodynamic force, it has a robust protective system, as described above. In a healthy muscle, this protective system must be sufficient to resist mechanical stress of the cytoplasmic flow. Thus, all the muscles of the body could endure severe movements, such as athletics.
However, the protective system has some limitations or thresholds for the CK leakage even in healthy muscles. It is known that after severe exercise, for example running 90 km,89) serum CK levels markedly increases to the levels found in DMD patients, although there are some individual differences. Serum CK levels return to the normal baseline level within 2 days in normal subjects. This suggests that some damage to the sarcolemma occurs by severe exercise even in healthy persons.
[5] It has recently been discovered that the sarcolemma is equipped with sarcolemmal membrane repair machinery.90) This suggests that microtears of the lipid bilayer occurring even in healthy muscle fibers, if they happen, are repaired quickly by this machinery.
All the phenomena described above including Discussion (2) are compatible with our working hypothesis that dystrophin bolts that stitch the sarcolemma mechanically protect the lipid bilayer from injury that might occur upon contraction of the muscle.
5.2. Discussion (4): Hypothesis on the role of protection systems for the lipid bilayer in the pathomechanisms of DMD and some other muscular dystrophies.
Difference between normal and DMD sarcolemma resides primarily in the difference in strength of the fixation bolts distributed on the sarcolemma.
[1] Strenuous exercise for a DMD patient results in a severe increase of serum CK and myoglobin release from DMD muscle,6) even if it is not strenuous for normal persons. Many soluble enzymes leak from DMD muscles.5) These intracellular soluble proteins are too large and lack hydrophobicity to pass through the lipid bilayer, as long as the lipid bilayer is intact. Increase of the release of soluble proteins describes that the lipid bilayer in DMD muscle must be much more easily injured than that in normal muscle, forming microtears upon contraction allowing leakage of the soluble cytoplasmic proteins. However, such microtears are so tiny and must be resealed very soon and may not necessarily be revealed with EM, so long as their sizes do not exceed the ability of the repairing machinery. Such damages might better be called “nanotears” rather than microters.
Experimentally, when the sarcolemma is subjected to negative pressure by sucking with an ultrathin glass tube, in other words, to an artificial force directed outwards on the sarcolemma, the mdx sarcolemma which lacks strong protective systems is more easily damaged than the wild-type sarcolemma, showing its mechanical fragility.91)
Shortly, microtear formation must be the first step of muscle damage leading to atrophy in DMD.
[2] The mutations that result in the dysfunction of the dystrophin bolt cause other types of muscular dystrophy with very high serum CK level.
[2A] DMD group: (1) A significant decrease in fixation bolt causes DMD and BMD. The difference in the symptomatology between DMD and BMD may be due to the difference in the amount of the fixation bolt present in their muscles. (2) A decrease in fixation between dystrophin and β-DG in the absence of the SG complex92) may cause 4 kinds of SG-pathy.53)
[2B] CMD group: Mutation of the DG gene may be embryonic lethal. The connection between the O-sugar chain of α-DG and laminin28) is prerequisite for the function of the dystrophin bolt. Failure in the connection causes CMD.
(1) The sugar sequence of the O-sugar chain of α-DG, composed of 4 sugar molecules, was determined by Endo in 1997.93) Six CMDs have been associated with the disturbance in sugar chain synthesis. This results in weak connection between the α-DG and laminin. These CMDs are muscle–eye–brain disease, Fukuyama-type CMD, Walker–Warburg syndromes 1 & 2, Fukutin-related protein deficiency and LARGE (for review94)).
(2) The absence of laminin α2 (merosin), the α-dystroglycan scaffold protein, causes CMD.95)
Roughly, these 7 types of CMDs and 4 types of SG-pathy can be discussed in a similar manner to those of DMD on the basis of the above-mentioned discussion for DMD, because the dystrophin bolt is impaired due to a weak or absent binding to laminin and thus cannot function sufficiently to prevent damage of the lipid bilayer.
One question is why the muscle damage appears in the intra-uterine stage in these CMDs and in the childhood in DMD, in spite that muscle fibers of both diseases have insufficiency of the fixation bolt. This difference can be ascribed to the difference in the defective portion of the fixation bolt.37,51) In DMD and SG-pathies, intracellularly the connection between the subsarcolemmal cytoskeleton networks and β-DG is defective but weakly present, whereas in these CMDs, extracellularly that between α-DG and laminin is defective (cf. Fig. 3B).
In in utero normal development, the utrophin bolt at first appears in myotube stage and is replaced by the dystrophin bolt before birth. Thus, the utrophin bolt with lower density is switched to the dystrophin bolt with higher density to bear the increase in the strength of movements with development of the body.
In these CMDs, the connection between α-DG and laminin is defective both in utero and after birth, regardless of switch from the utrophin bolt to the dystrophin bolt. The basal lamina with defective fixation easily exfoliates from the lipid bilayer, which becomes exposed to risk of injury even by weak movements in utero. Thus, the myotubes and muscle fibers of the CMD fetus can be easily injured. Stunted development of the muscles may occur. Thus, the symptoms are noticed at or soon after birth of the patients. Indeed, in merosin-deficient CMD cases, partial loss of the basal lamina is sometimes observed,59) and the degree of muscle atrophy was mild, when the basal lamina was well preserved.96)
In DMD, the utrophin bolts, even though they are present only at lower density, work to fix the sarcolemma to some extent (Fig. 2). Immediately after birth, while the patient moves only weakly, the symptoms do not become apparent. But when the patient starts to walk, run and jump, the sarcolemma fixed by the utrophin bolt becomes not strong enough to bear his violent movements. Thus, the lipid bilayer may be injured and atrophy and weakness of the muscles gradually appear.
Thus, DMD, SG-pathy and these CMDs can be summarized as “fixation bolt insufficiencies” that eventually induces injury of the lipid bilayer.
[3] In 2006, a mouse was reported with a mutation in the choline kinase gene that results in a decrease in phosphatidyl choline levels and disruption of the lipid bilayer in muscle fibers as observed in DMD muscle fibers.97) This mouse showed a muscular dystrophic muscle phenotype and elevated serum CK levels, even though dystrophin was present at the sarcolemma. Although many questions remain to be answered, the existence of muscular dystrophy due to abnormality of a component of the lipid bilayer strongly supports my central idea that the essential cause of muscular dystrophy is repeated disruptions of the lipid bilayer.
5.3. Discussion (5): The next step after microtear formation that leads to muscle atrophy.
When microtear formation and repair of the lipid bilayer occurs repeatedly, how do muscle fibers atrophy? Atrophy may be caused by loss of cytoplasm, or by disturbance of either synthesis or degradation of proteins or both.51)
[1] It is possible that leakage of soluble cytoplasmic enzymes and other proteins, if the amount lost is large, may have some influence on the decrease in the cell volume, their catalytic processes and other functions. In addition, because AA derived from degraded proteins is used for synthesis of new proteins, recycling of AA in protein turnover may be disturbed, if the protein leaks chronically and large amounts of proteins are lost. But even if this occurs at a high level, this might not be the sole cause of muscle atrophy.
[2] For increased digestion of proteins, the calcium–calpain hypothesis was proposed in the 1970’s.7) This hypothesis states that through minute pores of lipid bilayer, Ca++ ions from the extracellular fluid enter into the cytoplasm. Then, Ca++ ions activate calpain, a calcium-activated neutral protease. Activated calpain, in turn, digests muscle proteins leading to muscle atrophy. This idea on the mechanisms underlying DMD has long been believed worldwide. This may be one of the probable hypotheses. However, it seems to me that this hypothesis is too preliminary to be accepted as the cause of DMD. Although calpain enzymology has largely progressed, it has been scarcely clarified how calpain in living DMD muscle fibers contributes to the progress of atrophy that lasts for many years.
[2-a] Muscle fibers contract even in DMD patients. Relaxation is a prerequisite to contraction. At relaxation, cytoplasmic Ca++ ions should be kept below about 10−6 molar, where calpain cannot work. At contraction, they increase to the level of 5 × 10−5.98)
At first, calpain was found to be activated by millimolar level of calcium ions (m-calpain), levels not found in normal muscle. m-Calpain cannot work in the living cells. However, in the 1970’s, another type of calpain was discovered by Imahori that was activated by micromolar concentration of Ca++ ions (μ-calpain), which may work in vivo.7) However, how can μ-calpain in DMD muscle be regulated by a minute amount of Ca++ ions, where similar concentrations of free Ca++ ions wax and wane during the contraction–relaxation cycle? If μ-calpain is responsible for muscle atrophy, muscle proteins must be digested when the muscle fibers contract, even in the healthy muscle fibers in which calpain is contained. Why are only DMD muscle fibers amenable to atrophy that must be promoted by muscle contraction? I have sometimes put this question since the pre-dystrophin era.7) However, no satisfactory answer has so far been obtained.
[2-b] Since the pre-dystrophin era, it has repeatedly been reported that the content of calcium increases in DMD muscle.5) Upon contraction, Ca++ ions may be taken into muscle fibers through the microtears and also probably through calcium channels activated by membrane stretching as claimed by Iwata,99) both of which can occur simultaneously. Calcium can be present in muscle fibers as either free ions or calcium bound to some substance or organelle. Because almost all of the calcium in muscle fibers is present in the bound form, an increase in the total calcium concentration does not necessarily mean increase in free Ca++ ions, which is required for activation of calpain.
[2-c] Exogenous Ca++ ions must be sequestered rapidly after their entry into muscle fibers. DMD patients retain considerable contractile ability for years even after DMD symptoms appear. This means that DMD muscles can relax soon after contraction ceases. In other words, free Ca++ ion concentration must be sequestered to low level by sarcoplasmic reticulum and mitochondria immediately after the contraction, even when Ca++ ion entry occurs. Otherwise, prolonged contraction would happen like in the case of muscle fibers in malignant hyperthermia.
[2-d] Young DMD patients suffer from severe muscle cramps and pain mostly during the night after strenuous movements that are sometimes followed by myoglobinuria. This phenomenon may be one of the keys solving the question of when calpain works in DMD muscle fibers. Unfortunately, the space for explanation of these phenomena in relation to the calpain hypothesis has been exhausted.
[3] In addition, we must further consider whether the target of Ca++ ions is solely calpain or not, and whether calpain activity promoted by a minute amount of Ca++ ions is sufficient for inducing muscle atrophy that progresses during the chronic course of DMD. Ca++ ions might be closely related to muscular atrophy. However, without microtears, leakage of the soluble cytoplasmic proteins does not take place.
5.4. Discussion (6): The gene therapy of DMD from my viewpoints.
Although I did not work on this field, I will very briefly discuss them in relation to our works (see review51)).
[1] Gene transfer therapy: One of the most difficult problems in dystrophin gene transfer therapy is that its cDNA is too large to be carried by conventional viral vectors. In 1990, Davies100) proposed the use of mini-dystrophin gene derived from a BMD patient for this purpose. In 1995, I pointed out that its use is reasonable, because the gene contains the AB domain and DGBD.51) Feasibility of recombinant dystrophins having both binding domains to repair mdx mice was experimentally examined by Takeda45) in 1998 and more extensively by Chamberlain101) in 2002.
[2] Exon skipping method: In a patient with a specified out-of-frame deletion, a small number of exons close to the deletion can be artificially skipped to make an mRNA having a larger but in-frame deletion.102) Thus, this approach is usually accepted to change the clinical phenotype from DMD to BMD without discussing at molecular level. However, we should recollect the limit of application to the reading frame rule that I pointed out in Discussion (1). In the short term, the exon-skipping method should be effective on the mutations in the rod domain, which cause the majority of cases of DMD.
From my viewpoints, an effective exon skipping therapy must result in (1) genetically, avoidance of degradation of mRNA with NMD and at the same time (2) phenotypically, to construct the binding domain of dystrophin to β-DG (DGBD) by changing the reading frame to produce a functional dystrophin.
Epilogue
Our studies, described above, are divided into those on the subsarcolemmal cytoskeleton networks that include the dystrophin–DAP complex and their application to the muscular dystrophy research. Although we deduce their functions only from the evidence obtained by short-term experiments, muscular dystrophy is a group of chronic diseases. Whether it is relevant to apply such evidence to understanding such chronic diseases stands as an unsolved problem. However, I consider that even though muscular dystrophy is a chronic progressive disease, its pathological process must be a long-lasting process with repeated short-term battles between destructive and repairing processes. To study active processes of the degenerating disease might give some clues for understanding the pathomechamisms of muscular dystrophy.
Acknowledgments
I express my special thanks to Dr. M. Yoshida for his trustworthy friendship, for his superb techniques of protein chemistry and for his painstaking works almost throughout the whole period of this study. I appreciate the hard work with excellent skills of my colleagues during their stay in my laboratory: Drs. K. Araishi, Y. Hagiwara, Y. Hama, K. Hayashi, K. Ikeya, M. Imamura, M. Ishikawa-Sakurai, Y. Mizuno, S. Noguchi, T. Sasaoka, A. Suzuki, H. Tanaka, E. Wakabayashi-Takai, H. Yamamoto and Y. Yoshimoto-Matsuda. I also thank all other members who worked in my laboratory, especially Ms. K. Koshigoe. I appreciate Dr. L. M. Kunkel for his long-lasting fruitful collaboration and warm friendship. My thanks are also due to late Dr. K. Arahata, and Drs. K. E. Davies, C. Eguchi, T. Ishiguro, I. Nonaka and H. Sugita for their collaboration.
I greatly thank Drs. J.S. Chamberlain, M.C. Malicdan, S. Noguchi and M. Yoshida for their reading and commenting on this article. Naturally, I hold myself accountable for erroneous information, if any, in this paper.
Abbreviations
- AA
amino acid
- AB-D
actin-binding domain
- AB-S
actin-binding site
- ARDMD
autosomal recessive Duchenne-like muscular dystrophy
- BMD
Becker muscular dystrophy
- CK
creatine kinase
- CMD
congenital muscular dystrophy
- CR-domain
cysteine-rich domain
- CRI
Central Research Institute of Ajinomoto Co.
- DAP
dystrophin-associated protein
- DAG
dystrophin-associated glycoprotein
- DGBD
dystroglycan-binding domain
- DMD
Duchenne muscular dystrophy
- DR
dihydropyridine-receptor
- DVIF
desmin–vimentin intermediate filament
- EM
electron microscopy
- IMP
integrated membrane protein
- mf
myofibril
- NMD
nonsense mediated decay
- PJA
Proceedings of Japan Academy
- SDS PAGE
sodium dodesyl-sulfate polyacryl amide gel electrophoresis
- SG-pathy
sarcoglycanopathy
- TFS
transverse fixation system
- WGA
wheat-germ agglutinin. For abbreviations of proteins
Profile
Eijiro Ozawa was born in 1934 in Fukushima Prefecture. In 1957, he left from University of Tokyo, Faculty of Science, Department of Physics, Geophysics course, before graduation, and in 1961, graduated from University of Tokyo, Faculty of Medicine. After one year of internship, he worked 3 years as a resident of internal medicine at the Toranomon General Hospital. In 1965, he started his research career in study on “Activation of glycogen phosphorylase kinase by a minute amount of Ca ion that activate the contractile system” under Professor S. Ebashi at University of Tokyo, Faculty of Medicine. That was the first work that described the physiological concentration of Ca ion working on the intracellular enzyme system “Ca activated protein kinase” except muscle contraction. His work solved the question how glycogen metabolism is triggered on muscle cell contraction, which was raised by Fletcher and Hopkins in the early 20th Century. On the other hand, his work became a forerunner of a numerous works on Ca ion as a second messenger, that regulate calmodulin and Ca activated protein kinases.
Then, he worked on the developmental biology of muscle at the Carnegie Institution of Washington in Baltimore, MD, USA and Tokyo Medical and Dental University, Fuculty of Medicine and the Institute of Medical Sciences. In 1978, he moved to the National Center of Nervous, Mental and Muscular Disorders that changed the name to the National Institute of Neuroscience, NCNP, in 1986. He concentrated on understanding the muscle cell growth and degeneration-regeneration problem using cultured muscle cells.
From 1987 to 2000, he worked on dystrophin on which he reviewed in this article. His work on dystrophin and its associated proteins covers the determination of their location on the cell membrane, constructed the architecture formed with these proteins and its function. At the same time, his works also covers the pathomechanism of Duchenne and related muscular dystrophies. He was appointed as the Director General of the Institute in 1993. He retired from the Institute in 2000, and is the Director General Emeritus. After retirement, he wrote several review articles on various journals and chapters of some large handbooks.
He was laurelled with the Takeda Medical Award in 1996, the Medical Prize of the Japan Medical Association in 1997, the Gaetano Conte Board Prize (Italy) in 2000 and Prize of the International Congress on Neuromuscular Diseases in 2006. He was awarded with the Order of the Sacred Treasure, Gold Rays with Neck Ribbon in 2009.
References
- 1).Gowers W.R. (1879) Pseudo-hypertrophic muscular paralysis. Lancet 2, 1–2, 37–39, 73–75 and 113–116 [Google Scholar]
- 2).Erb W. (1891) Dystrophia muscularis progressiva. Dtsch. Z. Nervenheilkd. 1, 13–94, 173–261 [Google Scholar]
- 3).Sibley J.A., Lehninger A.L. (1949) Aldolase in serum and tissue of tumor bearing animals. J. Natl. Cancer Inst. 9, 303–309 [PubMed] [Google Scholar]
- 4).Ebashi S., Toyokura Y., Momoi H., Sugita H. (1959) High creatine phosphokinase activity of sera of progressive muscular dystrophy. J. Biochem. 46, 103–104 [Google Scholar]
- 5).Engel, A.G. and Ozawa, E. (2004) Dystrophinopathies. In Myology 3rd ed. (eds. Engel, A.G. and Franzini-Armstrong, C). Vol. 2, McGraw-Hill, New York, pp. 961–1025. [Google Scholar]
- 6).Florence J.M., Fox P.T., Planer G.J., Brooke M.H. (1985) Activity, creatine kinase, and myoglobin in Duchenne muscular dystrophy: a clue to etiology? Neurology 35, 758–761 [DOI] [PubMed] [Google Scholar]
- 7).Imahori, K. (1980) Characterization of calcium-activated netral protease (CANP). In Muscular Dystrophy (ed. Ebashi, S.). University of Tokyo Press, Tokyo, pp. 253–264. [Google Scholar]
- 8).Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. (1987) Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50, 509–517 [DOI] [PubMed] [Google Scholar]
- 9).Koenig M., Monaco A.P., Kunkel L.M. (1988) The complete sequence of dystrophin predicts a rod-shaped cytoskeleton protein. Cell 53, 219–228 [DOI] [PubMed] [Google Scholar]
- 10).Rybakova I.N., Amann K.J., Ervasti J.M. (1996) A new model for the interaction of dystrophin with F-actin. J. Cell Biol. 135, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Love D.R., Hill D.F., Dickson G., Spurr N.K., Byth B.C., Marsden R.F., et al. (1989) An autosomal transcript in skeletal muscle with homology to dystrophin. Nature 339, 55–58 [DOI] [PubMed] [Google Scholar]
- 12).Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. (1989) The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science 244, 1578–1580 [DOI] [PubMed] [Google Scholar]
- 13).Sugita H., Arahata K., Ishiguro T., Suhara Y., Tsukahara T., Ishiura S., et al. (1988) Negative immunostaining of Duchenne muscular dystrophy (DMD) and mdx muscle surface membrane with antibody against synthetic peptide fragment predicted from DMD cDNA. Proc. Jpn. Acad. Ser. B 64, 37–39 [Google Scholar]
- 14).Arahata K., Ishiura S., Ishiguro T., Tsukahara T., Suhara Y., Eguchi C., et al. (1988) Immunostaining of skeletal and cardiac muscle surface membrane with antibody against Duchenne muscular dystrophy peptide. Nature 333, 861–863 [DOI] [PubMed] [Google Scholar]
- 15).Craig S.W., Pardo J.V. (1983) Gamma actin, spectrin, and intermedite filament proteins colocalize with vinculin at costameres, myofibril-to-sarcolemma attachment site. Cell Motil. 3, 449–462 [DOI] [PubMed] [Google Scholar]
- 16).Wakayama Y., Shibuya S. (1991) Gold-labelled dystrophin molecule in muscle plasmalemma of mdx control mice as seen by electron microscopy of deep etching replica. Acta Neuropathol. 82, 178–184 [DOI] [PubMed] [Google Scholar]
- 17).Cullen M.J., Walsh J., Nicholson L.V.B., Harris J.B., Zubrzycka-Gaarn E.E., Ray P.N., et al. (1991) Immunogold labelling of dystrophin in human muscle, using an antibody to the last 17 amino acids of the C-terminus. Neuromuscul. Disord. 1, 113–119 [DOI] [PubMed] [Google Scholar]
- 18).Watkins S.C., Hoffman E.P., Slayter H.S., Kunkel L.M. (1988) Immunoelectron microscopic localization of dystrophin in myofibres. Nature 333, 863–866 [DOI] [PubMed] [Google Scholar]
- 19).Hoffman E.P., Hudecki M.S., Rosenberg P.A., Pollina C.M., Kunkel L.M. (1988) Cell and fiber-type distribution of dystrophin. Neuron 1, 411–420 [DOI] [PubMed] [Google Scholar]
- 20).Hagiwara Y., Yoshida M., Nonaka I., Ozawa E. (1989) Developmental expression of dystrophin on the plasma membrane. Protoplasma 151, 11–18 [Google Scholar]
- 21).Tanaka H., Shimizu T., Ozawa E. (1989) Expression of a dystrophin-like protein on the surface membrane of muscle cells in mdx mice. Proc. Jpn. Acad., Ser. B 65, 238–241 [Google Scholar]
- 22).Tanaka H., Ishiguro T., Eguchi C., Saito K., Ozawa E. (1991) Expression of a dystrophin-related protein associated with the skeletal muscle cell membrane. Histochemistry 96, 1–5 [DOI] [PubMed] [Google Scholar]
- 23).Mizuno Y., Nonaka I., Hirai S., Ozawa E. (1993) Reciprocal expression of dystrophin and utrophin in muscles of Duchenne muscular dystrophy patients, female DMD-carriers and control subjects. J. Neurol. Sci. 119, 43–52 [DOI] [PubMed] [Google Scholar]
- 24).Mizuno Y., Yoshida M., Nonaka I., Hirai S., Ozawa E. (1994) Expression of utrophin (dystrophin-related protein) and dystrophin-associated glycoproteins in muscles from patients with Duchenne muscular dystrophy. Muscle Nerve 17, 206–216 [DOI] [PubMed] [Google Scholar]
- 25).Campbell K.P., Kahl S.D. (1989) Association of dystrophin and an integral membrane glycoprotein. Nature 338, 259–262 [DOI] [PubMed] [Google Scholar]
- 26).Yoshida M., Ozawa E. (1990) Glycoprotein complex anchoring dystrophin to sarcolemma. J. Biochem. 108, 748–752 [DOI] [PubMed] [Google Scholar]
- 27).Ervasti J.M., Ohlendieck K., Kahl S.D., Gaver M.G., Campbell K.P. (1990) Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature 345, 315–319 [DOI] [PubMed] [Google Scholar]
- 28).Ibraghimov-Beskrovnaya O., Ervasti J.M., Leveille C.J., Slaughter C.A., Sernett S.W., Campbell K.P. (1992) Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 355, 696–702 [DOI] [PubMed] [Google Scholar]
- 29).Ervasti J.M., Campbell K.P. (1991) Membrane organization of the dystrophin-glycoprotein complex. Cell 66, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 30).Tinsley J.M., Blake D.J., Zuellig R.A., Davies K.E. (1994) Increasing complexity of the dystrophin-associated protein complex. Proc. Natl. Acad. Sci. USA 91, 8307–8313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Yamamoto H., Hagiwara Y., Mizuno Y., Yoshida M., Ozawa E. (1993) Heterogeneity of dystrophin-associated proteins. J. Biochem. 114, 132–139 [DOI] [PubMed] [Google Scholar]
- 32).Yoshida M., Hama H., Ishikawa-Sakurai M., Imamura M., Mizuno Y., Araishi K., et al. (2000) Biochemical evidence for association of dystrobrevin with the sarcoglycan–sarcospan complex as a basis for understanding sarcoglycanopathy. Hum. Mol. Genet. 9, 1033–1040 [DOI] [PubMed] [Google Scholar]
- 33).Peters M.F., Adams M.E., Froehner S.C. (1997) Differential association of syntrophin pairs with the dystrophin complex. J. Cell Biol. 138, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Crosbie R.H., Heighway J., Venzke D.P., Lee J.C., Campbell K.P. (1997) Sarcospan, the 25-kDa transmembrane component of the dystrophin–glycoprotein complex. J. Biol. Chem. 272, 31221–31224 [DOI] [PubMed] [Google Scholar]
- 35).Levine B.A., Moir A.J., Patchell V.B., Perry S.V. (1992) Binding sites involved in the interaction of actin with the N-terminal region of dystrophin. FEBS Lett. 298, 44–48 [DOI] [PubMed] [Google Scholar]
- 36).Brenman J.E., Chao D.S., Gee S.H., McGee A.M., Craven S.E., Santillano D.R., et al. (1996) Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell 84, 757–767 [DOI] [PubMed] [Google Scholar]
- 37).Ozawa, E. (2004) The muscle fiber cytoskeleton: The dystrophin sytem. In Myology 3rd ed. (eds. Engel, A.G. and Franzini-Armstrong). Vol. 1, McGraw-Hill, New York, pp. 455–470. [Google Scholar]
- 38).Sadoulet-Puccio H.M., Rajala M., Kunkel L.M. (1997) Dystrobrevin and dystrophin: an interaction through coiled-coil motifs. Proc. Natl. Acad. Sci. USA 94, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Yoshida M., Suzuki A., Shimizu T., Ozawa E. (1992) Proteinase-sensitive sites on isolated rabbit dystrophin. J. Biochem. 112, 433–439 [DOI] [PubMed] [Google Scholar]
- 40).Suzuki A., Yoshida M., Yamamoto H., Ozawa E. (1992) Glycoprotein-binding site of dystrophin is confined to the cysteine-rich domain and the first half of the carboxy-terminal domain. FEBS Lett. 308, 154–160 [DOI] [PubMed] [Google Scholar]
- 41).Koenig M., Beggs A.H., Moyer M., Scherpf S., Heindrich K., Bettecken T., et al. (1989) The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am. J. Hum. Genet. 45, 498–506 [PMC free article] [PubMed] [Google Scholar]
- 42).Suzuki A., Yoshida M., Hayashi K., Mizuno Y., Hagiwara Y., Ozawa E. (1994) Molecular organization at the glycoprotein-complex-binding site of dystrophin. Three dystrophin-associated proteins bind directly to the carboxy-terminal portion of dystrophin. Eur. J. Biochem. 220, 283–292 [DOI] [PubMed] [Google Scholar]
- 43).Jung D., Yang B., Meyer J., Chamberlain J., Campbell K.P. (1995) Identification and characterization of the dystrophin anchoring site of β-dystroglycan. J. Biol. Chem. 270, 27305–27310 [DOI] [PubMed] [Google Scholar]
- 44).Ishikawa-Sakurai M., Yoshida M., Imamura M., Davies K.E., Ozawa E. (2004) ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to β-dystroglycan. Hum. Mol. Genet. 13, 693–702 [DOI] [PubMed] [Google Scholar]
- 45).Yuasa K., Miyagoe Y., Yamamoto K., Nabeshima Y., Dickson G., Takeda S. (1998) Effective restoration of dystrophin-associated proteins in vivo by adenovirus-mediated transfer of truncated dystrophin cDNAs. FEBS Lett. 425, 329–336 [DOI] [PubMed] [Google Scholar]
- 46).Rafael J.A., Cox G.A., Corrado K., Jung D., Campbell K.P., Chamberlain J.S. (1996) Forced expression of dystrophin deletion constructs reveals structure–function correlations. J. Cell Biol. 134, 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Monaco A.P., Bertelson C.J., Liechti-Gallati S., Moser H., Kunkel L.M. (1988) An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2, 90–95 [DOI] [PubMed] [Google Scholar]
- 48).Tuffery-Giraud S., Beroud C., Leturcq F., Yaou R.B., Hamroun D., Michel-Calemard L., et al. (2009) Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum. Mutat. 30, 934–945 [DOI] [PubMed] [Google Scholar]
- 49).Malhotra S.B., Hart K.A., Klamut H.J., Thomas N.S., Bodrug S.E., Burghes A.H., et al. (1988) Frame-shift deletions in patients with Duchenne and Becker muscular dystrophy. Science 242, 755–759 [DOI] [PubMed] [Google Scholar]
- 50).Ozawa E., Yoshida M., Suzuki A., Mizuno Y., Hagiwara Y., Noguchi S. (1995) Dystrophin-associated proteins in muscular dystrophy. Hum. Mol. Genet. 4, 1711–1716 [DOI] [PubMed] [Google Scholar]
- 51).Ozawa, E. (2006) The functional biology of dystrophin: structural components and the pathogenesis of Duchenne muscular dystrophy. In Duchenne Muscular Dystrophy (eds. Chamberlain, J.S. and Rando, T.A.). Taylor & Francis, New York, pp. 21–53. [Google Scholar]
- 52).Yoshida M., Suzuki A., Yamamoto H., Noguchi S., Mizuno Y., Ozawa E. (1994) Dissociation of the complex of dystrophin and its associated proteins into several unique groups by n-octyl β-d-glucoside. Eur. J. Biochem. 222, 1055–1061 [DOI] [PubMed] [Google Scholar]
- 53).Ozawa E., Noguchi S., Mizuno Y., Hagiwara Y., Yoshida M. (1998) From dystrophinopathy to sarcoglycanopathy: evolution of a concept of muscular dystrophy. Muscle Nerve 21, 421–438 [DOI] [PubMed] [Google Scholar]
- 54).Nigro V., Piluso G., Belsito A., Politano L., Puca A.A., Papparella S., et al. (1996) Identification of a novel sarcoglycan gene at 5q33 encoding a sarcolemmal 35 kDa glycoprotein. Hum. Mol. Genet. 5, 1179–1186 [DOI] [PubMed] [Google Scholar]
- 55).Yoshida M., Noguchi S., Wakabayashi E., Piluso G., Belsito A., Nigro V., et al. (1997) The fourth component of the sarcoglycan complex. FEBS Lett. 403, 143–148 [DOI] [PubMed] [Google Scholar]
- 56).Ozawa E., Mizuno Y., Hagiwara Y., Sasaoka T., Yoshida M. (2005) Molecular and cell biology of the sarcoglycan complex. Muscle Nerve 32, 563–576 [DOI] [PubMed] [Google Scholar]
- 57).Suzuki A., Yoshida M., Ozawa E. (1995) Mammalian α1- and β1-syntrophin bind to the alternative splice-prone region of the dystrophin COOH terminus. J. Cell Biol. 128, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Ahn A.H., Kunkel L.M. (1995) Syntrophin binds to an alternatively spliced exon of dystrophin. J. Cell Biol. 128, 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Ozawa E., Nishino I., Nonaka I. (2001) Sarcolemmopathy: muscular dystrophies with cell membrane defects. Brain Pathol. 11, 218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Rafael J.A., Tinsley J.M., Potter A.C., Deconinck A.E., Davies K.E. (1998) Skeletal muscle-specific expression of a utrophin transgene rescues utrophin–dystrophin deficient mice. Nat. Genet. 19, 79–82 [DOI] [PubMed] [Google Scholar]
- 61).Mizuno Y., Noguchi S., Yamamoto H., Yoshida M., Suzuki A., Hagiwara Y., et al. (1994) Selective defect of sarcoglycan complex in severe childhood autosomal recessive muscular dystrophy muscle. Biochem. Biophys. Res. Commun. 203, 979–983 [DOI] [PubMed] [Google Scholar]
- 62).Chan Y., Kunkel L.M. (1997) In vitro expressed dystrophin fragments do not associate with each other. FEBS Lett. 410, 153–159 [DOI] [PubMed] [Google Scholar]
- 63).Rybakova I.N., Ervasti J.M. (1997) Dystrophin–glycoprotein complex is monomeric and stabilizes actin filaments in vitro through a lateral association. J. Biol. Chem. 272, 28771–28778 [DOI] [PubMed] [Google Scholar]
- 64).Matsumura K., Tome F.M., Collin H., Azibi K., Chaouch M., Kaplan J.C., et al. (1992) Deficiency of the 50K dystrophin-associated glycoprotein in severe childhood autosomal recessive muscular dystrophy. Nature 359, 320–322 [DOI] [PubMed] [Google Scholar]
- 65).Ben Othmane K., Ben Hamida M., Pericak-Vance M.A., Ben Hamida C., Blel S., Carter S.C., et al. (1992) Linkage of Tunisian autosomal recessive Duchenne-like muscular dystrophy to the pericentromeric region of chromosome 13q. Nat. Genet. 2, 315–317 [DOI] [PubMed] [Google Scholar]
- 66).Passos-Bueno M.R., Oliveira J.R., Bakker E., Anderson R.D., Marie S.K., Vainzof M., et al. (1993) Genetic heterogeneity for Duchenne-like muscular dystrophy (DLMD) based on linkage and 50 DAG analysis. Hum. Mol. Genet. 2, 1945–1947 [DOI] [PubMed] [Google Scholar]
- 67).Roberds S.L., Leturcq F., Allamand V., Piccolo F., Jeanpierre M., Anderson R.D., et al. (1994) Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell 78, 625–633 [DOI] [PubMed] [Google Scholar]
- 68).Bonnemann C.G., Modi R., Noguchi S., Mizuno Y., Yoshida M., Gussoni E., et al. (1995) β-sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat. Genet. 11, 266–273 [DOI] [PubMed] [Google Scholar]
- 69).Lim L.E., Duclos F., Broux O., Bourg N., Sunada Y., Allamand V., et al. (1995) β-sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat. Genet. 11, 257–265 [DOI] [PubMed] [Google Scholar]
- 70).Noguchi S., McNally E. M., Ben Othmane K., Hagiwara Y., Mizuno Y., Yoshida M., et al. (1995) Mutations in the Dystrophin-associated protein γ-sarcoglycan in chromosome 13 muscular dystrophy. Science 270, 819–822 [DOI] [PubMed] [Google Scholar]
- 71).Passos-Bueno M.R., Moreira E.S., Vainzof M., Marie S.K., Zatz M. (1996) Linkage analysis in autosomal recessive limb-girdle muscular dystrophy (AR LGMD) maps a sixth form to 5q33-34 (LGMD 2F) and indicates that there is at least one more subtype of AR LGMD. Hum. Mol. Genet. 5, 815–820 [DOI] [PubMed] [Google Scholar]
- 72).Nigro V., de Sa Moreira E., Piluso G., Vainzof M., Belsito A., Politano L., et al. (1996) Autosomal recessive limb-girdle muscular dystrophy, LGMD 2F, is caused by a mutation in the δ-sarcoglycan gene. Nat. Genet. 14, 195–198 [DOI] [PubMed] [Google Scholar]
- 73).Vainzof M., Passos-Bueno M.R., Canovas M., Moreira E.S., Pavanello R.C., Marie S.K., et al. (1996) The sarcoglycan complex in the six autosomal recessive limb-girdle muscular dystrophies. Hum. Mol. Genet. 5, 1963–1969 [DOI] [PubMed] [Google Scholar]
- 74).Noguchi S., Wakabayashi E., Imamura M., Yoshida M., Ozawa E. (1999) Developmental expression of sarcoglycan gene products in cultured myocytes. Biochem. Biophys. Res. Commun. 262, 88–93 [DOI] [PubMed] [Google Scholar]
- 75).Wakabayashi-Takai E., Noguchi S., Ozawa E. (2001) Identification of myogenesis-dependent transcriptional enhancers in promoter region of mouse γ-sarcoglycan gene. Eur. J. Biochem. 268, 948–957 [DOI] [PubMed] [Google Scholar]
- 76).Noguchi S., Wakabayashi E., Imamura M., Yoshida M., Ozawa E. (2000) Formation of sarcoglycan complex with differentiation in cultured myocytes. Eur. J. Biochem. 267, 640–648 [DOI] [PubMed] [Google Scholar]
- 77).Sakamoto A., Ono K., Abe M., Jasmin G., Eki T., Murakami Y., et al. (1997) Both hypertrophic and dilated cardiomyopathies are caused by mutation of the same gene, δ-sarcoglycan, in hamster: an animal model of disrupted dystrophin-associated glycoprotein complex. Proc. Natl. Acad. Sci. USA 94, 13873–13878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).Araishi K., Sasaoka T., Imamura M., Noguchi S., Hama H., Wakabayashi E., et al. (1999) Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in β-sarcoglycan-deficient mice. Hum. Mol. Genet. 8, 1589–1598 [DOI] [PubMed] [Google Scholar]
- 79).Sasaoka T., Imamura M., Araishi K., Noguchi S., Mizuno Y., Takagoshi N., et al. (2003) Pathological analysis of muscle hypertrophy and degeneration in muscular dystrophy in γ-sarcoglycan-deficient mice. Neuromuscul. Disord. 13, 193–206 [DOI] [PubMed] [Google Scholar]
- 80).Hagiwara Y., Sasaoka T., Araishi K., Imamura M., Yorifuji H., Nonaka I., et al. (2000) Caveolin-3 deficiency causes muscle degeneration in mice. Hum. Mol. Genet. 9, 3047–3054 [DOI] [PubMed] [Google Scholar]
- 81).Ozawa E., Hagiwara Y., Yoshida M. (1999) Creatine kinase, cell membrane and Duchenne muscular dystrophy. Mol. Cell. Biochem. 190, 143–151 [PubMed] [Google Scholar]
- 82).Ozawa, E. and Nonaka, I. (2002) Limb-girdle muscular dystrophy and related disorders. In Neuromuscular Disorders in Clinical Practice (eds. Katirji, B., Kaminski, H.J. etal). Butterworth-Heinemann, Boston, pp. 1049–1071. [Google Scholar]
- 83).Lazarides E. (1980) Intermediate filaments as mechanical integrators of cellular space. Nature 283, 249–256 [DOI] [PubMed] [Google Scholar]
- 84).Granger B.L., Lazarides E. (1979) Desmin and vimantin coexist at the periohery of the myofibril Z disc. Cell 18, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 85).Shear C.R., Bloch R.J. (1985) Vinculin in subsarcolemmal densities in chicken skeletal muscle: localization and relationship to intracellular and extracellular structures. J. Cell Biol. 101, 240–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Elliott G.F. (1963) A small-angle optically focusing X-ray diffraction camera in biological research. II. J. Ultrastruct. Res. 9, 171–176 [DOI] [PubMed] [Google Scholar]
- 87).Ohtsuki I., Ozawa E. (1977) Difference in sapanin sensitivity between myotubes and mononucleated cells from chick breast muscle. Cell Struct. Funct. 2, 367–370 [Google Scholar]
- 88).Matsubara I., Umazume Y., Yagi N. (1984) Lateral shrinkage of the myofilament lattice in chemically skinned muscle during contraction. Adv. Exp. Med. Biol. 170, 711–720 [DOI] [PubMed] [Google Scholar]
- 89).Noakes T.D. (1987) Effect of exercise on serum activities in humans. Sports Med. 4, 245–267 [DOI] [PubMed] [Google Scholar]
- 90).Cai C., Masumiya H., Weisleder N., Matsuda N., Nishi M., Hwang M., et al. (2009) MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol. 11, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91).Hutter O.F., Burton F.L., Bovell D.L. (1991) Mechanical properties of normal and mdx mouse sarcolemma: bearing on function of dystrophin. J. Muscle Res. Cell Motil. 12, 585–589 [DOI] [PubMed] [Google Scholar]
- 92).Iwata Y., Nakamura H., Mizuno Y., Yoshida M., Ozawa E., Shigekawa M. (1993) Defective association of dystrophin with sarcolemmal glycoproteins in the cardiomyopathic hamster heart. FEBS Lett. 329, 227–231 [DOI] [PubMed] [Google Scholar]
- 93).Chiba A., Matsumura K., Yamada H., Inazu T., Shimizu T., Kusunoki S., et al. (1997) Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve α-dystroglycan. J. Biol. Chem. 272, 2156–2162 [DOI] [PubMed] [Google Scholar]
- 94).Endo, T. (2007) Dystroglycans. In Comprehensive Glycoscience. From Chemistry to System Biology (eds. Kamerling, J.P., Boons, G.-J. etal.). Elsevier, Oxford, pp. 285–296. [Google Scholar]
- 95).Tome T. (1999) The saga of congenital muscular dystrophy. Neuropediatrics 30, 55–65 [DOI] [PubMed] [Google Scholar]
- 96).Hayashi Y.K., Ishihara T., Domen K., Hori H., Arahata K. (1997) A benign allelic form of laminin α2 chain deficient muscular dystrophy. Lancet 349, 1147. [DOI] [PubMed] [Google Scholar]
- 97).Sher R.B., Aoyama C., Huebsch K.A., Ji S., Kerner J., Yang Y., et al. (2006) A rostrocaudal muscular dystrophy caused by a defect in choline kinase b, the first enzyme in phosphatidylcholine biosynthesis. J. Biol. Chem. 281, 4938–4948 [DOI] [PubMed] [Google Scholar]
- 98).Ebashi S., Endo M. (1968) Calcium ion and muscle contraction. Prog. Biophys. Mol. Biol. 18, 123–183 [DOI] [PubMed] [Google Scholar]
- 99).Iwata Y., Katanosaka Y., Arai Y., Kamakura K., Miyatake K., Shigekawa M. (2003) A novel mechanism of myocyte degenertion involving the Ca2+-permeable growth factor regulated channel. J. Cell Biol. 161, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100).England S.B., Nicholson L.V., Johnson M.A., Forrest S.M., Love D.R., Zubrzycka-Gaarn E.E., et al. (1990) Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature 343, 180–182 [DOI] [PubMed] [Google Scholar]
- 101).Harper S.Q., Hauser M.A., DelloRusso C., Duan D., Crawford R.W., Phelps S.F., et al. (2002) Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 8, 253–261 [DOI] [PubMed] [Google Scholar]
- 102).Matsuo M., Masumura T., Nakajima T., Kitoh Y., Takumi T., Nishio H., et al. (1990) A very small frame-shifting deletion within exon 19 of the Duchenne muscular dystrophy gene. Biochem. Biophys. Res. Commun. 170, 963–967 [DOI] [PubMed] [Google Scholar]