Abstract
The nuclear envelope harbors numerous large proteinaceous channels, the nuclear pore complexes (NPCs), through which macromolecular exchange between the cytosol and the nucleoplasm occurs. This double-membrane nuclear envelope is continuous with the endoplasmic reticulum and thus functionally connected to such diverse processes as vesicular transport, protein maturation and lipid synthesis. Recent results obtained from studies in Saccharomyces cerevisiae indicate that assembly of the nuclear pore complex is functionally dependent upon maintenance of lipid homeostasis of the ER membrane. Previous work from one of our laboratories has revealed that an integral membrane protein Apq12 is important for the assembly of functional nuclear pores. Cells lacking APQ12 are viable but cannot grow at low temperatures, have aberrant NPCs and a defect in mRNA export. Remarkably, these defects in NPC assembly can be overcome by supplementing cells with a membrane fluidizing agent, benzyl alcohol, suggesting that Apq12 impacts the flexibility of the nuclear membrane, possibly by adjusting its lipid composition when cells are shifted to a reduced temperature. Our new study now expands these findings and reveals that an essential membrane protein, Brr6, shares at least partially overlapping functions with Apq12 and is also required for assembly of functional NPCs. A third nuclear envelope membrane protein, Brl1, is related to Brr6, and is also required for NPC assembly. Because maintenance of membrane homeostasis is essential for cellular survival, the fact that these three proteins are conserved in fungi that undergo closed mitoses, but are not found in metazoans or plants, may indicate that their functions are performed by proteins unrelated at the primary sequence level to Brr6, Brl1 and Apq12 in cells that disassemble their nuclear envelopes during mitosis.
Key words: endoplasmic reticulum, nuclear pore complex, nucleoporins, nucleocytoplasmic transport, lipid metabolism, membrane homeostasis
Introduction
The nuclear envelope (NE) defines the boundary of the nucleus in eukaryotic cells and is composed of two distinct membranes, the inner and outer nuclear membranes, enclosing a lumenal space. The inner and outer nuclear membranes become fused to form specialized membrane tunnels in which nuclear pore complexes are constructed. The outer nuclear membrane is continuous with the endoplasmic reticulum (ER) and thought to perform ER functions. In yeast, the NE accounts for 20–30% of the ER and is thus functionally closely related to the ER.1,2 The ER is the major site of lipid synthesis and most lipid biosynthetic enzymes are integral ER membrane proteins.3 How the NE-ER balances membrane expansion through lipid synthesis while maintaining its other functions as well as its characteristic shape, however, is not well understood. The recent characterization of three integral membrane proteins of the NE in Saccharomyces cerevisiae may provide clues to begin to understand how NPC assembly is coordinated with membrane expansion and lipid synthesis.
Nuclear pore complexes (NPCs) are large macromolecular assemblies through which all transport between the nucleus and the cytosol occurs (reviewed in refs. 4 and 5). NPCs show eightfold rotational symmetry perpendicular to the NE and two-fold symmetry in the plane of the NE. They are constructed using multiple copies of ∼30 proteins, termed nucleoporins (Nups), most of which are highly conserved in eukaryotes. The NPC itself can be divided roughly into three domains: the nuclear basket, the central core, and the cytoplasmic filaments. The basket and cytoplasmic filaments are composed of Nups that are found solely in those structures, whereas Nups of the central core are localized symmetrically on both the nuclear and cytoplasmic sides of the NE. Multiple integral membrane proteins are components of NPCs and these have been implicated in both the organization and proper assembly of NPCs. Although genetic and biochemical analyses have advanced the identification of Nups as well as their localizations and interactions within the NPC, the mechanism of NPC biogenesis is less well understood.
Most nucleocytoplasmic transport is mediated by members of the karyopherin family of receptors that include multiple importins, exportins and transportins, most of which are specialized for either import or export. These receptors recognize localization signals in their cargoes and are transported with their cargoes through the central channel of the NPC. mRNAs are exported in a complex with proteins, forming messenger RNP complexes, which are exported by binding to the mRNA export factor, Mex67.6 The NPC plays a mechanistic role in transport by providing docking sites for these transport complexes, thereby facilitating their translocation through the pore.7,8
In metazoan cells NPC assembly takes place by de novo assembly during interphase, when the number of NPCs doubles, and by re-assembly of NPCs when a dispersed nuclear envelope reassembles late in mitosis (reviewed in refs. 9 and 10). S. cerevisiae has a closed mitosis in which the nuclear envelope remains intact, and the majority of Nups appear to remain associated with the NPC during mitosis.11 Eukaryotes with closed mitosis also lack lamins, nuclear intermediate filament proteins that provide a structure to the nucleus and play roles in chromosome organization, DNA replication, maintenance of nuclear shape and distribution of the NPCs.12 In S. cerevisiae, segregation of NPCs into the daughter cell at anaphase is restricted at the bud neck by a septin-dependent lateral diffusion barrier.13 NPCs in the daughter cell are thus formed primarily through de novo insertion. Nups that are integral membrane proteins are thought to initiate NPC construction within the NE and coordinate the early steps of NPC biogenesis. Relatively little is known about how these Nups find one another or the extent to which Nups associate into subcomplexes before their assembly into NPCs,14 and nothing is known about the mechanism by which the inner and outer nuclear membranes become fused. In the filamentous fungus Aspergillus nidulans, which undergoes a closed mitosis, a subset of Nups, however are dispersed and this is coincident with an altered NPC permeability.15 This indicates that at least some fungal species have ways to alter the nuclear permeability barrier without going through an open mitosis and the associated NPC disassembly. Partial disassembly of NPCs via release of peripheral Nups has also been noted in starfish oocytes prior to nuclear envelope breakdown.16
Several factors in addition to Nups have been implicated in NPC biogenesis. These include components of the Ran GTPase system that also controls the directionality of karyopherin-mediated transport and regulates key events during cell division (reviewed in refs. 17 and 18). Cells carrying specific mutant alleles affecting these proteins showed mislocalization of nucleoporins as well as accumulation of nucleoporin-containing cytoplasmic vesicles.19–21 Proteins involved in ER to Golgi trafficking, including components of the COPII coat, have also been implicated in NPC assembly.20 In addition, proteins that affect membrane curvature of the ER, the reticulons and Yop1/DP1, are important for early stages of nuclear pore assembly, perhaps by stabilizing high curvature at the site of pore insertion and fusion of the inner and outer nuclear membranes.22
Several NPC components whose function in transport is documented have also been tied to cellular activities distinct from their roles at the NE. A wealth of recent data has documented the function of Nups in microtubule attachment to kinetochores.23,24 In addition, NPCs are involved in chromatin organization and gene expression.25 And a recent screen for essential genes that are required for maintaining proper localization of NPCs also uncovered components of the RSC chromatin-remodeling complex, suggesting that NE structure is functionally linked to proper chromatin architecture.26
Apq12, an Integral Membrane Protein Links NPC Assembly to Membrane Fluidity
APQ12 encodes an integral membrane protein of the nuclear envelope and ER and is required for efficient NPC biogenesis.27 Screens to identify non-essential yeast genes possibly involved in mRNA export or mRNA processing demonstrated that cells lacking APQ12 showed defects in these processes.28,29 Apq12-GFP localizes to the nuclear periphery and the ER, but it is not a Nup because its distribution is unaffected by mutations that cause NPCs to cluster.28 A role for Apq12 in cell division has been suggested because loss of APQ12 leads to synthetic growth defects with mutations affecting genes coding for spindle pole body (SPB) proteins and other proteins involved in cell division. In the absence of APQ12, anaphase is delayed, and re-replication of DNA before completion of cytokinesis is also observed.30
Cells lacking APQ12 are cold sensitive for growth and display a temperature-dependent defect in NPC assembly. At the restrictive temperature of 16°C, several Nups, including all Nups that are components of the cytoplasmic filaments (CF) of the NPC, mislocalize to cytoplasmic foci. Components of the nuclear basket and most components of the central structural framework of the NPC, including the integral membrane Nups, however, are not mislocalized. Because relocalization of Nups in APQ12 mutant cells depends on ongoing protein synthesis, Apq12 affects NPC biogenesis rather than the stability of pre-existing NPCs. These observations suggest that the cytoplasmic foci observed in APQ12 mutant cells contain Nup subcomplexes that are unable to be assembled into functional NPCs.27
Remarkably, proper localization of CF Nups in APQ12 mutant cells is restored upon addition of low levels of benzyl alcohol to the medium. Benzyl alcohol is thought to increase the fluidity and flexibility of membranes.31 Addition of benzyl alcohol not only prevents Nup mislocalization but also restores proper localization of Nups that had accumulated in cytoplasmic foci. Since normal Nup localization is restored even when protein synthesis is blocked by cycloheximide, restoration of the normal NPC distribution reflects assembly of pre-existing and mislocalized Nups or Nup complexes into functional NPCs. The restored NPCs in APQ12 mutant cells treated with benzyl alcohol are functional because the mRNA export phenotype is also rescued. Thus it is likely that the defects in mRNA export, pre-mRNA processing, and cell division of APQ12 mutant cells are indirect consequences of altered membrane fluidity. On the other hand, defects in nuclear transport can affect phospholipid biosynthesis and thus membrane properties. One of the enzymes involved in phosphatidylcholine biosynthesis, for example, requires importin β/Kap95 mediated nuclear import for its full in vivo activity.32 Since mutants that affect nuclear import do not generally display NPC assembly defects, the function of Apq12 in the dynamics of the NE appears to be independent of its defect in mRNA export.27
Interestingly, increasing the amount of benzyl alcohol beyond the level used to suppress apq12 phenotypes causes Nup mislocalization in wild-type cells.33 This suggests that NPC assembly is sensitive to the fluidity of the NE and that the observed defects in NPC assembly in APQ12 mutants could result from improper regulation of the lipid composition of the nuclear membrane in response to changes in temperature.
Brr6, an Essential Integral Membrane Protein Genetically Interacts with Apq12 and Links NPC Assembly to Lipid Homeostasis of the ER Membrane
BRR6 was identified as a dosage suppressor of the cold-sensitive growth defect of APQ12 mutant cells and overexpression of APQ12 partially suppresses the growth defect of cells bearing a conditional allele of BRR6, brr6-1, indicating that Apq12 and Brr6 share some common function.34 Unlike APQ12, however, BRR6 is essential for viability. The gene was originally identified in a screen for cold-sensitive mutants defective in mRNA export and, like Apq12, encodes an integral membrane protein of the NE and ER.35 Like Apq12, Brr6 is not found in NPC-containing subcellular fractions and does not cluster in yeast mutant strains where NPCs cluster.35,36 Cells carrying the conditional brr6-1 allele show mislocalization of CF Nups but not nuclear basket Nups, similar to what is observed in cells lacking APQ12. In addition, brr6-1 mutants are hypersensitive to drugs that inhibit sterol or fatty acid synthesis, whereas the APQ12 mutant is sensitive only against the fatty acid synthesis inhibitor. Consistent with this broader drug sensitivity, brr6-1 displays strong genetic interactions with mutants that affect sterol biosynthesis or fatty acid elongation. Biochemical analyses revealed that brr6-1, but not APQ12 mutant cells contain dramatically elevated levels of two classes of neutral lipids, steryl esters and triacylglycerols. Because brr6-1 cells, but not wild-type cells, are non-viable if neutral lipid synthesis is blocked, overproduction of these storage lipids is functionally essential in these mutant cells. This observation is particularly intriguing and suggests that these cells accumulate elevated levels of free sterols in the ER that need to be “neutralized” by esterification so that they do not adversely affect ER and NE function. Elevated levels of free sterols in the ER of animal cells result in depletion of ER calcium stores, induction of the unfolded protein response (UPR) and ultimately apoptosis.37 These observations indicate that Brr6 has an essential function in regulating lipid homeostasis in the NE-ER, thereby impacting NPC formation and nucleocytoplasmic transport.
Brl1, a Third Nuclear Envelope Protein is Functionally and Structurally Related to Brr6
Mutations in a third gene, BRL1 (BRR6-like 1) also result in mislocalization of Nups and mRNA-export defects similar to those observed in BRR6 and APQ12 mutants.38 BRL1 was identified as a suppressor of a temperature sensitive exportin 1 (Xpo1) mutant, a major karyopherin required for nuclear export of most proteins and ribosomal subunits.38,39 Brl1 appears to be functionally related to Brr6, because overexpression of BRR6 suppresses the growth defect of BRL1 mutants, and conditional mutations in BRL1 are synthetically lethal with brr6-1. However, neither can restore viability to strains lacking the other gene entirely.38 Moreover, unlike BRR6, overexpression of BRL1 cannot suppress defects resulting from the absence of APQ12.34
Like BRR6, BRL1 is essential, and encodes an integral membrane protein of the NE. The C-terminal half of Brl1 shows substantial homology to Brr6 and deletion analysis of Brl1 indicates that this conserved domain is functionally important. The domain contains four conserved cysteines and mutations at one of these cysteines renders Brl1 temperature sensitive, suggesting that disulfide bridges could play a role in forming a putative Brr6/Brl1 complex.38 Whether mutations to replace the other cysteines would show a similar phenotype is not known.
Interestingly, Schizosaccharomyces pombe has only a single Brr6/Brl1 family protein. SpBrl1 fulfills the functions of both Brl1 and Brr6 when expressed in S. cerevisiae suggesting that (1) Brr6 and Brl1 arose from an ancestral gene or genome duplication event and (2) evolved to perform distinct essential functions, which may be in addition to their roles in maintenance of nuclear envelope functionality. Although Brl1 and Brr6 may function independently in membrane homeostasis, the observation that they interact with each other in a two-hybrid analysis suggests that they assemble into a single complex that functionally corresponds to SpBrl1.38 Disruption of either Brr6 or Brl1 may abolish or reduce the functionality of this putative complex in S. cerevisiae.
Intriguingly, Brr6/Brl1 homologues are found throughout the fungi and lower eukaryotes that carry out a closed mitosis, including Candida glabarata and Cryptococcus neoformans. Most of these organisms encode a single protein that is more closely related to Brl1 than to Brr6. No homologues have been so far found in metazoans or plants, in which the nuclear lamina formed from lamins is disassembled early during mitosis. During the evolution of metazoans, lamins evolved around the same time that the transition from closed to open mitosis occurred. It has been suggested that open mitosis may have co-evolved with lamins to circumvent a possible interference of lamins in chromosome segregation under conditions where the nuclear envelope would not dissasemble.40 The presence of lamins may confer a selective advantage on metazoan cells, allowing for greater complexity in nuclear organization or efficiency in gene expression or nuclear signaling. In S. cerevisiae where lamins are absent, it may be that nuclear envelope proteins such as the Brr6/Brl1 family contribute to gene expression through interactions with the nuclear pores and the transport machinery.38 In support of this hypothesis, increased membrane fluidity with benzyl alcohol treatment prevents structural defects and NPC mislocalization in a conditional mutant of the RSC chromatin-remodeling complex.26
Lipid Composition and Synthesis in the ER Affect Nuclear Structure and NPC Distribution/Assembly
A number of earlier studies indicate that the shape of the nucleus and the function of NPCs are related to lipid composition of the ER membrane. A conditional mutation in ACC1/MTR7, which encodes acetyl-CoA carboxylase, was isolated in a screen for mRNA-export mutants.41,42 Acc1 is essential and catalyzes the synthesis of malonyl-CoA, the key building block for synthesis and elongation of fatty acids. mtr7-1/acc1-7-1 mutant cells have abnormal NPCs and mislocalized Nups, and are defective in the synthesis of very-long-chain fatty acids. These observations suggested that very-long chain fatty acids might have a role in NPC assembly, perhaps by helping to bring together the INM and ONM at points where fusion is to occur when NPC are assembled.42–44
Cells lacking an ER-NE associated protein phosphatase implicated in regulation of lipid biosynthetic genes have abnormal nuclear morphology including extensions of the NE that contain NPCs similar to what is observed in brr6-1 cells.45 This heterodimeric phosphatase is composed of the catalytic subunit Nem1 and the regulatory subunit Spo7. One identified target of this phosphatase is Pah1/Smp2, which is a homologue of mammalian lipin, a gene that is expressed at high levels in adipose tissue.46,47 Pah1 has phosphatidic acid phosphatase activity and thereby controls conversion of phospholipids into triacylglycerols.48 Overproduction of Pah1 restores normal nuclear membrane structure to NEM1 and SPO7 mutant cells, indicating that lipid homeostasis is crucial for maintaining a proper structure of the NE. The role of NEM1, SPO7 and PAH1 in regulating nuclear membrane structure and expansion is conserved from yeast to humans.49–51
Do Apq12, Brr6 and Brl1 Affect the Functioning of a Membrane-Fluidity Sensor?
An interesting possibility to account for defects in NPC assembly, NE structure and lipid synthesis in cells lacking APQ12, or carrying mutant alleles of BRR6, or BRL1 is that these mutations impact the ability of cells to sense environmental changes that normally trigger modifications in membrane composition needed to maintain membrane homeostasis (reviewed in refs. 52–54). How fluidity and other biophysical properties of membranes are sensed and how this information is transduced to adjust lipid synthesis and membrane composition is not well understood. Fluidity sensors are thought to respond to decreased temperature in part through activating fatty acids desaturases, which introduce double-bonds into acyl chains of fatty acids and phospholipids.55 The double bond introduces a kink that disrupts acyl chain packing in the lipid bilayer, thereby helping to maintain normal fluidity. Increased incorporation of oleate into phospholipids therefore would be expected to increase membrane fluidity.56
Yeast has a single essential fatty acid desaturase, Ole1, whose transcription is tightly regulated.57 Mga2 and Spt23, two homologous membrane-associated transcription factors related to mammalian NFκB, are proteolytically released from their membrane association when cells sense the need to activate OLE1.58 Neither Mga2 nor Spt23 is essential, but cells lacking both are not viable.59 Consistent with a role in regulating membrane fluidity, brr6-1 is synthetic lethal with mutants lacking either MGA2 or SPT23.34 Morphological defects in the NE observed following a shift of an mga2 spt23-ts mutant to non-permissive temperature indicate that membrane fluidity impacts NE structure.59 Cells lacking Ole1 are able to grow when supplemented with unsaturated fatty acids (UFA) but the membrane abnormalities seen following depletion of UFA are substantially more severe than those seen in mga2 spt23-ts cells.59
If APQ12 or BRR6 mutant cells were defective in their ability to induce modifications in membrane composition in response to a shift to lower temperature, it might be expected that supplementing the medium with oleic acid would partially suppress the defects. Oleic acid supplementation, however, does not suppress the growth phenotype of BRR6 and APQ12 mutants.34 Rather, both mutants are hyper-sensitive to oleic acid. This suggests that the observed effects of these mutations in NE function might reflect an excess of membrane fluidity at lower temperatures, which is consistent with the hypersensitivity of brr6-1 cells to benzyl alcohol. If this were the case, then the primary defect might be the inability of cells to sense when proper membrane composition and dynamics had been restored following a temperature shift (Fig. 1). Further studies are now required to understand the mechanisms by which APQ12, BRR6 and BRL1 affect lipid metabolism and nuclear membrane homeostasis, and how this impacts NPC biogenesis.
Figure 1.
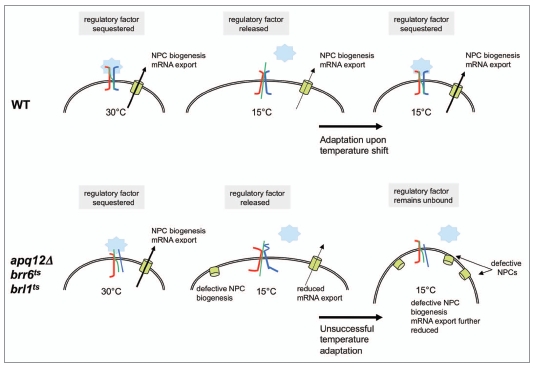
Possible function of Apq12, Brr6 and Brl1 in temperature adaptation. Under constant temperature conditions, the membrane-sensing complex composed of Apq12, Brr6 and Brl1 (colored in green, red and blue) would sequester a putative regulatory factor and thus turn off adaptive changes (e.g., altered expression of lipid modifying enzymes). Upon a temperature shift to 15°C, the membrane-sensing complex releases this regulatory factor to initiate the adaptive response. Upon successful temperature adaptation, the functionally restored membrane-sensing complex sequesters the regulatory factor again and thus turns off the adaptive change. Defects in Apq12, Brr6 or Brl1 would result in unproductive initiation and/or termination of the adaptive response.
Acknowledgements
This work was supported by grants from the Swiss National Science Foundation (3100-120650) to R.S. and from the National Institute of General Medical Sciences, National Institutes of Health (GM33998) to C.N.C.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/12333
References
- 1.Strambio-de-Castillia C, Blobel G, Rout MP. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, et al. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- 3.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 5.Lim RY, Ullman KS, Fahrenkrog B. Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol. 2008;267:299–342. doi: 10.1016/S1937-6448(08)00632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, et al. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 8.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 9.Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- 10.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makhnevych T, Lusk CP, Anderson AM, Aitchison JD, Wozniak RW. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 2003;115:813–823. doi: 10.1016/s0092-8674(03)00986-3. [DOI] [PubMed] [Google Scholar]
- 12.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 13.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 14.Rexach M. Piecing together nuclear pore complex assembly during interphase. J Cell Biol. 2009;185:377–379. doi: 10.1083/jcb.200904022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 16.Lenart P, Rabut G, Daigle N, Hand AR, Terasaki M, Ellenberg J. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Biol. 2003;160:1055–1068. doi: 10.1083/jcb.200211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 18.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 19.Ryan KJ, McCaffery JM, Wente SR. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J Cell Biol. 2003;160:1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan KJ, Wente SR. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 2002;3:17. doi: 10.1186/1471-2156-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan KJ, Zhou Y, Wente SR. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol Biol Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan GK, Liu ST, Yen TJ. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol Biol Cell. 2010;21:1072–1087. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarcelli JJ, Hodge CA, Cole CN. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol. 2007;178:799–812. doi: 10.1083/jcb.200702120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker KE, Coller J, Parker R. The yeast Apq12 protein affects nucleocytoplasmic mRNA transport. RNA. 2004;10:1352–1358. doi: 10.1261/rna.7420504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montpetit B, Thorne K, Barrett I, Andrews K, Jadusingh R, Hieter P, et al. Genome-wide synthetic lethal screens identify an interaction between the nuclear envelope protein, Apq12p, and the kinetochore in Saccharomyces cerevisiae. Genetics. 2005;171:489–501. doi: 10.1534/genetics.105.045799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon LM, Sauerheber RD, Esgate JA, Dipple I, Marchmont RJ, Houslay MD. The increase in bilayer fluidity of rat liver plasma membranes achieved by the local anesthetic benzyl alcohol affects the activity of intrinsic membrane enzymes. J Biol Chem. 1980;255:4519–4527. [PubMed] [Google Scholar]
- 32.MacKinnon MA, Curwin AJ, Gaspard GJ, Suraci AB, Fernandez-Murray JP, McMaster CR. The Kap60-Kap95 karyopherin complex directly regulates phosphatidylcholine synthesis. J Biol Chem. 2009;284:7376–7384. doi: 10.1074/jbc.M809117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izawa S, Takemura R, Inoue Y. Gle2p is essential to induce adaptation of the export of bulk poly(A)+ mRNA to heat shock in Saccharomyces cerevisiae. J Biol Chem. 2004;279:35469–35478. doi: 10.1074/jbc.M403692200. [DOI] [PubMed] [Google Scholar]
- 34.Hodge CA, Choudhary V, Wolyniak MJ, Scarcelli JJ, Schneiter R, Cole CN. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J Cell Sci. 2010;123:141–151. doi: 10.1242/jcs.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Bruyn Kops A, Guthrie C. An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J. 2001;20:4183–4193. doi: 10.1093/emboj/20.15.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 38.Saitoh YH, Ogawa K, Nishimoto T. Brl1p—a novel nuclear envelope protein required for nuclear transport. Traffic. 2005;6:502–517. doi: 10.1111/j.1600-0854.2005.00295.x. [DOI] [PubMed] [Google Scholar]
- 39.Maurer P, Redd M, Solsbacher J, Bischoff FR, Greiner M, Podtelejnikov AV, et al. The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p) Mol Biol Cell. 2001;12:539–549. doi: 10.1091/mbc.12.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- 41.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, et al. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneiter R, Kohlwein SD. Organelle structure, function and inheritance in yeast: a role for fatty acid synthesis? Cell. 1997;88:431–434. doi: 10.1016/s0092-8674(00)81882-6. [DOI] [PubMed] [Google Scholar]
- 44.Schneiter R, Brugger B, Amann CM, Prestwich GD, Epand RF, Zellnig G, et al. Identification and biophysical characterization of a very-long-chain-fatty-acid-substituted phosphatidylinositol in yeast subcellular membranes. Biochem J. 2004;381:941–949. doi: 10.1042/BJ20040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 48.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tange Y, Hirata A, Niwa O. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci. 2002;115:4375–4385. doi: 10.1242/jcs.00135. [DOI] [PubMed] [Google Scholar]
- 50.Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JCJ, Dixon JE. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci USA. 2007;104:6596–6601. doi: 10.1073/pnas.0702099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siniossoglou S. Lipins, lipids and nuclear envelope structure. Traffic. 2009;10:1181–1187. doi: 10.1111/j.1600-0854.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- 52.Murata N, Los DA. Membrane fluidity and temperature perception. Plant Physiol. 1997;115:875–879. doi: 10.1104/pp.115.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Los DA, Murata N. Membrane fluidity and its roles in the perception of environmental signals. Biochim Biophys Acta. 2004;1666:142–157. doi: 10.1016/j.bbamem.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 55.Vigh L, Los DA, Horvath I, Murata N. The primary signal in the biological perception of temperature: Pd-catalyzed hydrogenation of membrane lipids stimulated the expression of the desA gene in Synechocystis PCC6803. Proc Natl Acad Sci USA. 1993;90:9090–9094. doi: 10.1073/pnas.90.19.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hazel JR. Thermal adaptation in biological membranes: is homeoviscous adaptation the explanation? Annu Rev Physiol. 1995;57:19–42. doi: 10.1146/annurev.ph.57.030195.000315. [DOI] [PubMed] [Google Scholar]
- 57.Stukey JE, McDonough VM, Martin CE. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- 58.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 59.Zhang S, Skalsky Y, Garfinkel DJ. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]


