Abstract
The nuclear cap-binding complex (CBC) is a heterodimer composed of CBP20 and CBP80 subunits and has roles in the biogenesis of messenger RNAs (mRNAs), small nuclear RNAs (snRNAs) and microRNAs. CBP20 is a phylogenetically conserved protein that interacts with the 7-methyl guanosine (m7G) cap added to the 5′ end of all RNA polymerase II transcripts. CBP80 ensures high affinity binding of the cap by CBP20 and provides a platform for interactions with other factors. Here we characterize an alternative splice variant of CBP20, termed CBP20S. The CBP20S transcript has an in-frame deletion, leading to the translation of a protein lacking most of the RNA recognition motif (RRM). We show that CBP20S is conserved among mammalian species and is expressed in human cell lines and bone marrow cells. Unlike the full-length CBP20, CBP20S does not bind CBP80 or the m7G cap. Nevertheless, CBP20S does bind mRNA, is localized to an active transcription site and redistributed to nucleolar caps upon transcription inhibition. Our results suggest that this novel form CBP20S plays a role in transcription and/or RNA processing independent of CBP80 or the cap.
Key words: CBC, CBP20, 7-methyl guanosine, alternative splicing, nucleolar caps
Introduction
A characteristic feature of most eukaryotic genes is their discontinuity: protein-coding sequences (exons) alternate with non-coding regions (introns). The process of intron removal and exon ligation—pre-mRNA splicing—is conducted by a multicomponent complex, the spliceosome. Prior to splicing catalysis, splicing factors recognize exon-intron boundaries, i.e., the 5′ and 3′ splice sites. In higher eukaryotes, splice sites are highly degenerate and additional sequence elements recognized by regulatory splicing factors are required.1 This can lead to the phenomenon of alternative splicing, in which exon extension, shortening or skipping modifies the mRNA product of a single gene. It is now estimated that 95–100% of human intron-containing genes are alternatively spliced.2 Although the functional significance of the majority of splicing events is not known, genetic switches based on alternative splicing have been shown to be important in many cellular processes, such as sex determination in Drosophila melanogaster.3 Changes introduced by alternative splicing can lead to three types of effects: introduction of a premature termination codon (PTC), variable untranslated regions (UTRs) and changes in the protein coding sequence.2,4 Alternative splicing-based introduction of PTCs leads to downregulation of gene expression through nonsense-mediated decay of transcripts; interestingly, this mechanism broadly affects the expression of regulatory splicing factors, such as polypyrimidine tract binding protein (PTB) and serine/arginine-rich (SR) proteins.5–7 Through changes in the coding region, several structurally and functionally distinct protein isoforms can be derived from a single gene. Alternative protein isoforms can act as dominant negative regulators, as in the case of Fox proteins, a family of factors controlling neuronal alternative splicing.8 Thus, alternative splicing-mediated regulation of splicing factor function is a common mechanism for modulation of gene expression.
The cap-binding complex (CBC) binds the 7-methyl guanosine (m7G) cap, a modification added to the 5′ end of all RNA polymerase II transcripts, including mRNAs, pre-snRNAs and primary microRNA transcripts. The CBC has been implicated in several RNA processing events: pre-mRNA splicing,9 mRNA 3′ end formation,10,11 nonsense-mediated decay,12,13 export of mRNAs and pre-snRNAs from the nucleus14,15 and miRNA processing.16–18 The CBC is a heterodimer composed of two subunits—CBP20 and CBP80. CBP20 is a protein of 156 amino acids that interacts directly with the m7G cap. It contains a classical RNA Recognition Motif (RRM), comprising RNP1 and RNP2 motifs, flanked by N- and C-terminal domains (Fig. 1A and B). Residues from all three domains contribute to m7G binding. The positively charged 7-methyl guanosine ring intercalates between two aromatic residues—Tyr 20 from the N-terminal domain and Tyr 43 from the RNP2 motif. Additionally, there are hydrogen bonds between residues from N- and C-terminal domains and the m7G moiety as well as stacking interactions between C-terminal Tyr 138 and the next nonmethylated base.19–21 Upon m7G cap binding, CBP20 undergoes conformational changes, due to its interaction with the larger subunit, CBP80. The structure of CBP80 is highly ordered and composed entirely of α-helices and loops.21 Apart from the CBP20 binding site, the CBP80 surface is poorly conserved across species. In contrast, CBP20 is highly conserved; 69% of its residues are identical or conservatively substituted in species from S. cerevisiae to human.14,19 Interestingly, two isoforms of human CBP20 (gene name NCBP2) are reported in the NCBI database, suggesting that multiple isoforms of CBP20 may be expressed and possess different activities. Here we report the expression, binding properties and subcellular distribution of the non-canonical CBP20 isoform.
Figure 1.
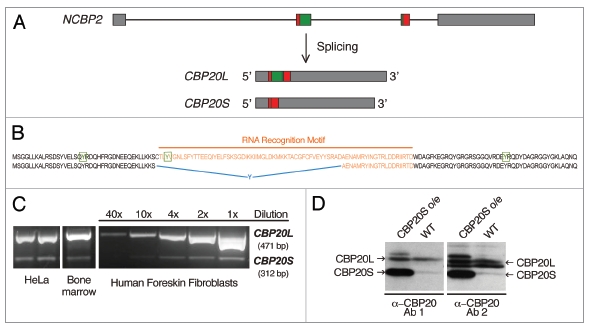
Human CBP20 isoforms. (A) Schematic representation of the NCBP2 gene and the CBP20L and CBP20S transcripts resulting from alternative splicing. Annotated introns and exons are depicted as lines and boxes, respectively. CBP20S mRNA is produced by an alternative combination of 5′ and 3′ splice sites embedded within exons 2 and 3, as indicated (Suppl. Fig. 1D). (B) Comparison of CBP20 protein isoforms. Tyrosine residues involved in m7G cap binding (Y20, Y43 and Y138) are marked in green and boxed. The RNA recognition motif (RRM) is marked in orange. In CBP20S, a tyrosine codon is introduced at the alternative exon-exon junction (marked in blue). (C) Detection of CBP20 isoforms in human culture cells and human tissue by RT-PCR. A dilution series of one of the RT-PCR reactions is included. (D) Detection of CBP20 isoforms by western blotting. Cells transiently transfected with CBP20S for overexpression (CBP20S o/e) provide a reference for CBP20S band localization. Two different polyclonal α-CBP20 antibodies (1: Abgent and 2: E. Izaurralde gift) were used.
Results
We have termed the alternative isoform of CBP20 described in this report “CBP20S”, because it is shorter (103 amino acids) than the previously defined longer form (156 aa), which we term “CBP20L” for clarity. As shown in Figure 1A, CBP20S, arises through alternative 5′ and 3′ splice site choice during the excision of intron 2. CBP20S mRNA is missing a large part of NCBP2 exon 2 and a few nucleotides of exon 3. There is no frame shift that would lead to either altered amino acid sequence or a PTC(s). Intriguingly, this alternative splicing event leads to the elimination of a large part of the RRM. CBP20L RRM domain comprises residues 41–114 (based on SMART smart.embl-heidelberg. de/) and residues 40–93 are excluded from CBP20S sequence. This part of the sequence corresponds to conserved RNP motifs 1 (residues 81–88) and 2 (residues 41–46).21 Furthermore, a TAT codon is formed at the new splice junction, introducing a tyrosine residue in the amino acid sequence (Fig. 1B). These observations give rise to the following hypotheses. First, CBP20S could potentially have a regulatory function; for example, CBP20S could act as a dominant negative inhibitor of CBC function, if it interacts with CBP80 but fails to bind the m7G cap. Second, CBP20S might still retain RNA-binding capacity. Although CBP20S lacks a part of the RRM, one of the key tyrosine residues involved in the cap binding is still present (Tyr 20) and a new tyrosine (Tyr 40) is introduced in a similar position to the deleted (Tyr 43). Third, it is possible that CBP20S function is not related to cap metabolism but to another cellular process. Finally, the short isoform could represent splicing ‘noise’ and have no functional significance.22–25 We addressed these possibilities by investigating CBP20S expression, localization and protein- and RNA-binding properties.
It was of interest to know whether CBP20S is expressed in human cells and how abundant it is compared to CBP20L. RT-PCR experiments showed that HeLa cells, A431 cells and primary cultures of human foreskin fibroblasts (HFF) expressed CBP20S (Fig. 1C). From the dilution series of a PCR reaction, we estimate that CBP20S is approximately 20 times less abundant than CBP20L. Moreover, CBP20S was also detected in human bone marrow cells (Fig. 1C). The shorter RNA species corresponded to CBP20S, as determined by sequencing the PCR products (data not shown). Furthermore, the UCSC Genome Bioinformatics database was searched for CBP20S expression sequence tags (ESTs). Several ESTs corresponding to the short isoform from different human tissues were found and are summarized in Table 1. Strikingly, the vast majority of CBP20S-expressing cells or tissues were cancer-derived, suggesting that CBP20S expression may be either a feature or consequence of tumorigenesis. 50% of the ESTs in the EST database come from cancer cells.26 Interestingly, all CBP20S ESTs were identical with no variability around the alternative 5′ or 3′ splice sites, suggesting that they do not result from an unspecific lack of splicing fidelity within these two exons.
Table 1.
Human CBP20S ESTs
| EST | Tissue/cells |
| AL556084 | HeLa |
| BM459785 | Leiomyosarcoma |
| BE782414 | Retinoblastoma |
| BI754505 | n/a |
| BI197509 | Neuroblastoma |
| AU134209 | Ovary, tumor tissue |
| BE295628 | Rhabdomyosarcoma |
| CN421262 | Embryonic stem cells, cell line h1, h7 and h9 |
| BQ223411 | Large cell carcinoma |
| DB004179 | Breast tumor tissue |
| DA042574 | Bladder |
| DA596904 | n/a |
| BQ435070 | Melanotic melanoma |
CBP20S ESTs found in UCSC Genome Bioinformatics database.
Having established that CBP20S mRNA is present in various human cells, the question of CBP20S protein expression was addressed. CBP20L and CBP20S have predicted molecular weights of 18 and 12 kD, respectively. Western blot analysis of HeLa cell extracts showed the presence of a protein band with the same electrophoretic mobility as CBP20S, which was expressed from a plasmid as a marker (Fig. 1D). The expression level of the putative CBP20S protein was lower than that of CBP20L, likely reflecting the mRNA ratio between the two isoforms. Thus, both CBP20S mRNA and protein can be detected in human cells.
Next, we sought to determine whether other species express CBP20S. A BLAST search was performed, using both RNA and protein sequences. Six other mammalian species expressing both isoforms were identified (Suppl. Fig. 1A). The mRNA and protein sequences of CBP20S from all seven species were aligned. High levels of conservation were observed among the RNA sequence (Suppl. Fig. 1B). The proteins were identical in human, chimpanzee, rhesus macaque, horse and pig, while mouse and short-tailed opossum had a few mismatches (Suppl. Fig. 1C). 5′ and 3′ splice sites used in the CBP20S alternative splicing event were also aligned. These splice sites were similar and, with the exception of single mismatches in the case of Pan troglodytes and Monodelphis domestica, followed the general mammalian splice site profile (Suppl. Fig. 1D). The extremely high degree of conservation of the CBP20S splicing event and protein product strongly suggests that this alternative form of CBP20 has an important function.
In order to gain insight into the function of CBP20S, both CBP20 isoforms were tagged with GFP and transiently expressed in HeLa cells. Microscopy analysis of GFP-tagged isoforms revealed similarities and differences in their cellular localization (Fig. 2). Both proteins were detected in nuclei, excluding nucleoli. However, unlike CBP20L, CBP20S was only moderately enriched in the nuclei and did not display a speckled distribution, characteristic for splicing factors. The nuclear localization pattern of both CBP20 isoforms clearly differed from that of GFP alone. The cytoplasmic distributions of CBP20L-GFP, CBP20S-GFP and GFP alone were indistinguishable, indicating no specific localization to any cytosolic compartment. These observations show that CBP20S protein is stable enough to accumulate at readily detectable levels and exhibits specific nucleoplasmic enrichment as well as a cytosolic pool.
Figure 2.
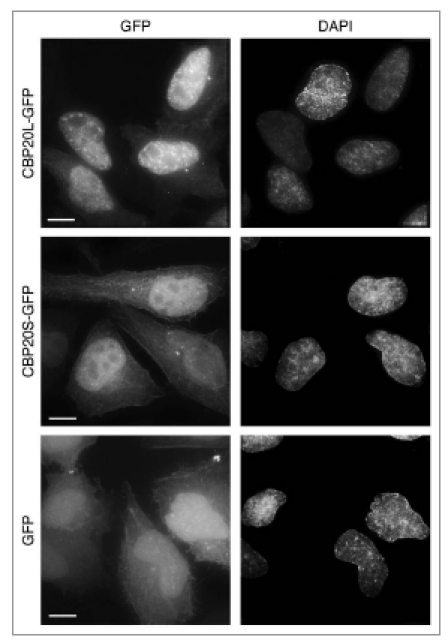
Cellular localization of GFP-tagged CBP20 isoforms. HeLa cells were transiently transfected with CBP20L-GFP, CBP20S-GFP or GFP alone, fixed after 48 hours and imaged. Nuclei were stained with DAPI. Projections of z-stacks are shown. Scale bar, 10 µm.
CBP20L is known to interact directly with CBP80 and the m7G cap present at the 5′ end of RNA polymerase II transcripts. It was of interest to know whether either of these characteristics is true for CBP20S. First, we tested whether CBP20S-GFP could co-immunoprecipitate CBP80 (Fig. 3). As expected, CBP20L pulled down CBP80. In contrast, CBP20S did not. We conclude from this that CBP20S, even when overexpressed, cannot assemble with CBP80 to produce an alternative form of the CBC. Second, the capacity of CBP20S to bind the m7G cap was tested directly in a cap-binding assay, in which HeLa cell extracts were incubated with 7-methyl-GTP (m7GTP) coupled to sepharose and the bound proteins analyzed by western blotting. Figure 4A shows that the m7GTP-sepharose specifically binds the cap-binding complex subunits CBP20L and CBP80 in extracts of untransfected HeLa cells, but not an unrelated protein, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). In transfected cells, m7GTP-sepharose selected CBP20L-GFP as well as endogenous CBP80, representing the canonical CBC complex (Fig. 4B). In contrast, m7GTP-sepharose failed to bind CBP20S-GFP and GFP alone. We conclude that CBP20S, unlike CBP20L, had no detectable m7GTP-binding capacity in this assay. These results indicate that CBP20S is not a component of the CBC and does not bind m7G.
Figure 3.
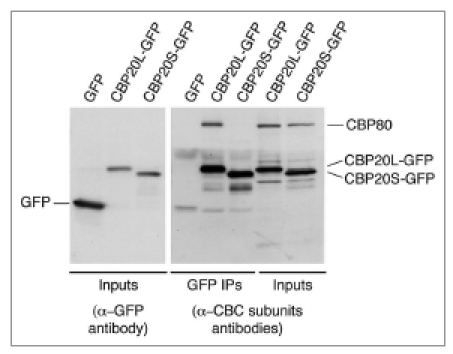
CBP20S does not interact with CBP80. HeLa cells were transiently transfected with CBP20L-GFP, CBP20S-GFP or GFP alone and harvested after 48 hours. Immunoprecipitation was carried out from cell extracts with a-GFP antibody. The immunoprecipitated and input material were analyzed by western blotting with α-GFP, α-CBP20 and α-CBP80. Bands corresponding to endogenous CBP80, CBP20L-GFP, CBP20S-GFP and GFP are indicated.
Figure 4.
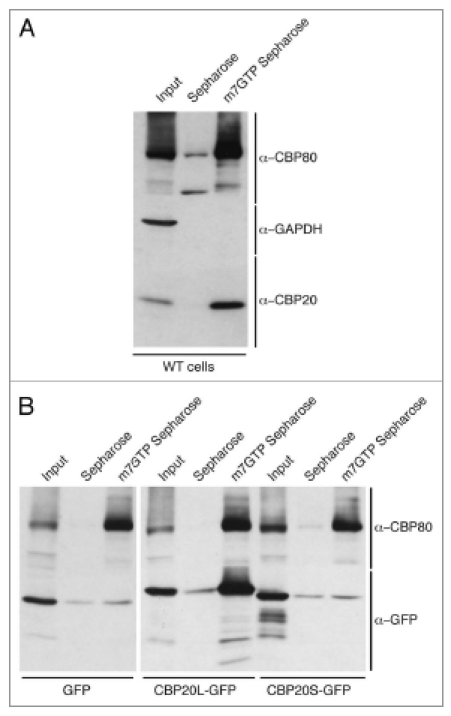
CBP20S does not bind m7GTP. (A) Extracts of untransfected HeLa cells were incubated with Sepharose 4B or m7GTP-Sepharose 4B. The selected and input samples were analyzed by western blotting with α-CBP80, α-CBP20 and α-GAPDH antibodies. Endogenous CBP80 and CBP20 bind to m7GTP-Sepharose 4B. (B) HeLa cells were transiently transfected with CBP20L-GFP, CBP20S-GFP or GFP alone and harvested after 48 hours. Cell extracts were incubated with Sepharose 4B or m7GTP-Sepharose 4B. The selected and input samples were analyzed by western blotting using α-CBP80 and α-GFP antibodies.
The results of the above m7GTP-sepharose binding assay, which require high affinity interactions, do not exclude the possibility that CBP20S associates with capped RNA polymerase II transcripts in vivo. Therefore, the binding of CBP20 isoforms to pre-snRNA and mRNA species was analyzed. First, GFP-specific antibodies were able to immunoprecipitate CBP20L-GFP together with metabolically labelled U1 and U2 pre-snRNAs, as expected (Fig. 5A). In parallel, immunoprecipitation of labelled snRNAs with a monoclonal antibody (K121) specific for hypermethylated (m2,2,7G) caps shows the gel positions of U1 and U2 pre-snRNAs (not yet trimmed at their 3′ ends) and mature snRNAs. Neither pre-snRNA was detected in complexes pulled down with CBP20S-GFP. Second, the ability of CBP20S to associate with mRNAs was tested by immunoprecipitation of CBP20S-GFP or CBP20L-GFP from cell lysates, followed by RT-qPCR detection of transcripts encoding c-myc (MYC) and β-actin (ACTB). In the case of CBP20L-GFP, both MYC and ACTB mRNAs were detected ∼30-fold above background, indicating strong association (Fig. 5B). Interestingly, CBP20S-GFP immunoprecipitated low but significant levels of both mRNAs, which were detected ∼4-fold above background. Thus, although CBP20S does not detectably interact with pre-snRNAs, it appears to associate with mRNA to a significant extent.
Figure 5.
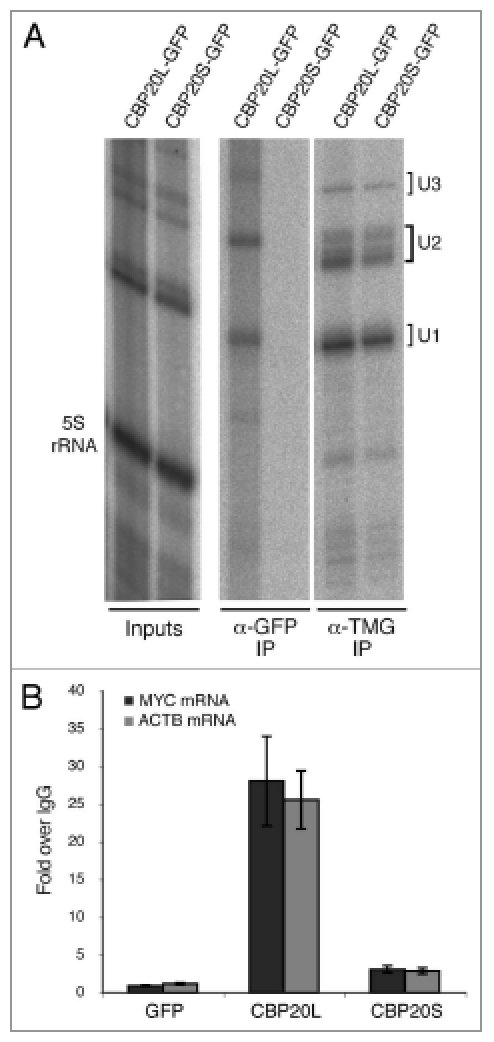
CBP20S pulls down mRNA. HeLa cells were transiently transfected with CBP20L-GFP, CBP20S-GFP or GFP alone and harvested after 48 hours. (A) 3-hour incubation with 32P-orthophosphate preceded cell extract preparation and immunoprecipitation. Cell extracts were immunoprecipitated with α-GFP or α-TMG antibodies; total “input” and immunoprecipitated RNAs were extracted, run on a 10% urea gel and detected by phosphorimager. (B) Cell extracts were immunoprecipitated with α-GFP or non-immune antibodies; immunoprecipitated RNAs were extracted and analyzed by RT-qPCR. The data represent the average of 3 independent experiments. Error bars represent SEM.
To pursue the detected association of CBP20S with mRNA, we examined CBP20S-GFP recruitment to a model transcription site consisting of a stably integrated gene array (similar to a previously published array27). The transcription unit can be activated by doxycycline and located in the nuclear landscape, using a lactose repressor (LacI) fused to a fluorescent protein (Fig. 6A). Interestingly, both CBP20L-GFP and CBP20S-GFP isoforms co-localized with RFP-LacI, indicating that both accumulated on the active transcription site (Fig. 6B). No accumulation was observed under non-induced conditions (data not shown). To measure the dynamic properties of the different CBC components in the nucleoplasm, we photobleached the nucleoplasmic CBP signal and followed the recovery of fluorescence over time (fluorescence recovery after photobleaching, FRAP). These experiments revealed that CBP20L-GFP and CBP80-GFP show similar recovery kinetics in the nucleoplasm compared to CBP20S-GFP, which displayed more rapid recovery. This suggests that CBP20L and CBP80 associate in a discrete, larger complex, while CBP20S diffuses within the nucleoplasm either alone or in a smaller complex. When we examined the association kinetics of these components with the active transcription unit, once again we identified pronounced and similar residency of CBP20L and CBP80 on the active transcription site, indicative of specific binding to the nascent transcripts. On the other hand, CBP20S showed very rapid association and disassociation rates, implying short-lived binding. This confirmed our previous evidence that CBP20S is not present in the CBC (Figs. 3 and 4), while it does have the ability to transiently associate with mRNA (Fig. 5). We conclude that, although CBP20S-GFP displays more dynamic mobility than CBP20L-GFP in living cells, it nevertheless accumulates specifically on an active RNA Polymerase II transcription unit.
Figure 6.
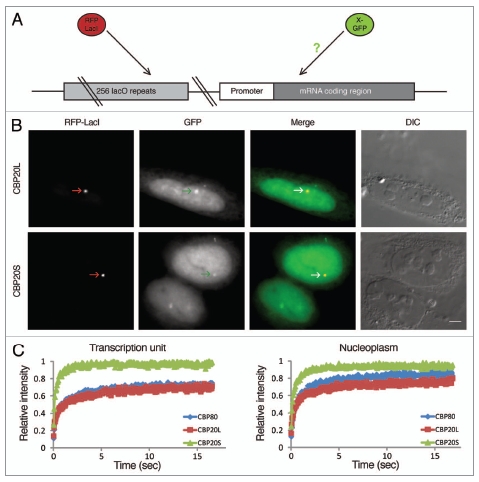
CBP20L and CBP20S accumulate in vivo on an active transcription site. (A) The recruitment of CBP20L-GFP and CBP20S-GFP was monitored on an active transcription unit driven by a tetracycline-inducible promoter. The gene locus is visualized via the co-integration of a plasmid containing 256 lacO repeats. Expression of RFP-LacI that binds to the lacO repeats allows the detection of the gene. Transcriptional induction is achieved by the addition of doxycycline to the medium. (B) U2OS Tet-On cells were co-transfected with expression constructs encoding GFP-tagged CBP20 isoforms and RFP-LacI; CBP20L and CBP20S accumulated at the transcription site marked by RFP-LacI in transcriptionally induced cells. Scale bar, 5 µm. DIC, Differential interference contrast microscopy. (C) Cells expressing either CBP80-GFP, CBP20L-GFP or CBP20S-GFP were photobleached in the nucleoplasm or at the active transcription unit, and the recovery of fluorescence (FRAP) was monitored over time indicating the rates at which these proteins roam the nucleus or associate with the active transcription unit.
Because CBP20S-GFP associated with mRNAs and localized to an active transcription site, we postulated that CBP20S might interact with a protein(s) involved in RNA processing. Therefore, we searched for CBP20S interactors by immunopurification followed by mass spectrometry (Table 2). CBP80 was not detected, consistent with our data that CBP20S is not a component of the CBC. Seven factors involved in RNA processing could be identified in the CBP20S interactome, among them hnRNP A2/B1 and PTB-associated splicing factor (PSF). Because both of these proteins are known to relocalize to the nucleolus or nucleolar caps upon transcriptional arrest,28,29 we tested the behavior of CBP20L and CBP20S after transcription inhibition by Actinomycin D (ActD). As shown in Figure 7, CBP20L-GFP and CBP20S-GFP relocalize to nucleolar caps after ActD treatment, adding them to the array of nucleoplasmic proteins that display this behavior. Both CBP20 isoforms segregated to PSF-positive nucleolar caps (termed dark nucleolar caps, DNC) but not to fibrillarin-positive caps (termed light nucleolar caps, LNC).29 Taken together, the data suggest that CBP20S has a specific function in nuclear RNA processing.
Table 2.
Proteins associated with CBP20S
| Protein name | Gene name | UniProt accession | MWa kDa | UPb 1 | UPb 2 | Comment |
| Proteins with RNA-related functions | ||||||
| MVP | MVP | Q14764 | 99 | 5 | 4 | Protein and mRNA transport |
| RNA-binding protein 8A | RBM8A | Q9Y5S9 | 20 | 1 | 2 | EJC component |
| hnRNP A2/B1 | HNRNPA2B1 | P22626 | 37 | 4 | 1 | mRNA processing |
| hnRNP A1 | HNRNPA1 | P09651 | 39 | 2 | 2 | mRNA processing |
| eIF-4A-I | EIF4A1 | P60842 | 46 | 5 | 1 | Translation |
| PSF | SFPQ | P23246 | 76 | 1 | 1 | mRNA processing |
| hnRNP A3 | HNRNPA3 | P51991 | 40 | 2 | 1 | mRNA processing |
| Proteins with other or unknown functions | ||||||
| GAPDH | GAPDH | P04406 | 36 | 7 | 2 | Glycolysis |
| Alpha-enolase | ENO1 | P06733 | 47 | 8 | 1 | Glycolysis |
| HD1 | HDAC1 | Q13547 | 55 | 2 | 5 | Chromatin remodeling |
| Peroxiredoxin-2 | PRDX2 | P32119 | 22 | 4 | 3 | Redox regulation |
| cDNA FLJ54957 | 69 | 6 | 2 | Highly similar to Transketolase | ||
| Fructose-bisphosphate aldolase A | ALDOA | P04075 | 39 | 7 | 1 | Glycolysis |
| TER ATPase | VCP | P55072 | 89 | 6 | 3 | Transport |
| KCTD12 | KCTD12 | Q96CX2 | 36 | 3 | 1 | Potassium ion transport |
| ATP5A1 | ATP5A1 | P25705 | 60 | 4 | 2 | ATP synthesis |
| UPF0027 protein C22orf28 | 55 | 2 | 1 | |||
| Prefoldin subunit 5 | GIM5 | Q04493 | 17 | 3 | 1 | Chaperone |
| CPVL | CPVL | Q9H3G5 | 54 | 3 | 3 | Proteolysis |
| Ezrin | EZR | P15311 | 69 | 6 | 1 | Cytoskeleton |
| Histone H1x | H1FX | Q92522 | 22 | 2 | 1 | Chromosomal protein |
| IMP dehydrogenase 2 | IMPDH2 | P12268 | 56 | 2 | 4 | GMP biosynthesis |
| V-ATPase subunit D | ATP6V1D | Q9Y5K8 | 28 | 2 | 3 | Ion transport |
| ALDH16A1 | ALDH16A1 | Q8IZ83 | 85 | 2 | 2 | Oxidation reduction |
| S29mt | DAP3 | P51398 | 46 | 1 | 2 | Mitochondrial ribosomal subunit |
Proteins associated with CBP20S were identified by mass spectrometry performed from solution (shot-gun). From the resulting list, we selected those proteins that were identified with at least one peptide, with more than 99% protein identification probability and that were absent in the control IPs (RAD51-GFP and GFP alone).
Calculated molecular weight in kDa.
Number of unique peptides sequenced for a given protein in two independent experiments (1 and 2).
Figure 7.
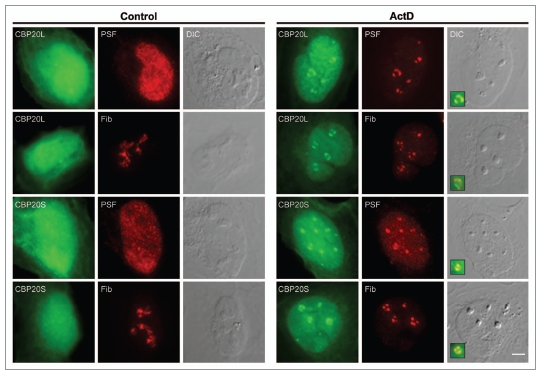
CBP20L and CBP20S redistribute to nucleolar caps after actinomycin D treatment. U2OS cells were transfected with GFP-tagged CBP20 isoforms, incubated with 5 µg/ml actinomycin D for 2 hours and fixed. Cells were stained with α-PSF and α-fibrillarin (Fib) antibodies (shown in red). Scale bar, 5 µm. DIC, Differential interference contrast microscopy.
Discussion
The CBP20 subunit of the CBC is a highly conserved protein that binds the m7G cap. Here we have shown that CBP20 can be alternatively spliced, leading to the formation of a previously uncharacterized isoform (CBP20S) that lacks a large portion of the RNA binding domain present in the canonical (CBP20L) protein. CBP20S mRNA and protein were detected in several human cell culture lines and bone marrow cells. Moreover, this alternative splicing event is conserved in several mammalian species. The conservation of alternative splicing events across species and the absence of PTCs and/or frame shifts in the alternative isoforms constitute compelling evidence for the functionality of CBP20S.23,24,30 This raised the possibility that CBP20S might act as a dominant negative regulator of CBC function. Numerous examples of alternative splicing-induced changes in binding to RNA, DNA, and/or interacting proteins have been reported.4 Binding affinity can be either reduced or completely lost, the latter leading to dominant negative effects. For example, a truncated form of FOS-B retains the dimerization and DNA-binding capacity but cannot activate transcription because its trans-activation domain is missing.31,32 So far, the only reported regulation of CBC activity is the importin-β-mediated release from mRNA in the cytoplasm.33,34,35
Functional characterization of CBP20S ruled out two hypotheses concerning its cellular role. CBP20S is not an m7G cap-binding factor, as it did not display detectable binding to either m7GTP-Sepharose or m7G-capped pre-snRNAs (Figs. 4 and 5). Furthermore, CBP20S is not likely a dominant negative regulator of CBC function, because it did not detectably interact with either CBP80 or the m7G cap in contrast to the CBP20L isoform (Figs. 3 and 4). Taken together, these observations suggest that any activity of the CBP20S isoform must be independent of CBP80 or m7G binding. Because the role of CBC in nonsense-mediated decay requires CBP80 activity,12,13 these findings indicate that CBP20S is unlikely to participate in nonsense-mediated decay. However, other roles for CBP20S in RNA processing are not excluded by these data.
Further characterization of CBP20S yielded several lines of evidence that it plays a physiological role in RNA processing. First, CBP20S displayed specific subcellular distribution, with moderate enrichment in the nucleus except for nucleoli, similar to many proteins involved in splicing. Second, CBP20S bound weakly but specifically to two mRNAs tested. Third, CBP20S was detected on an active transcription site. Fourth, mass spectrometry analysis revealed the association of CBP20S with several proteins involved in RNA processing. Finally, both CBP20 isoforms specifically relocated to nucleolar caps after transcription inhibition, similar to several factors involved in RNA processing, like PSF, U1-70K, hnRNPK, the cleavage and polyadenylation factor CstF-64 or the SR protein ASF/SF2.29 Taken together, CBP20S has many properties common to RNA processing factors.
We speculate that CBP20S associates with mRNAs and active transcription sites due to transient interactions with the transcript itself or with proteins bound to it. A possible protein linking CBP20S to the mRNA is hnRNP A2/B1, which was identified among CBP20S-associated proteins by mass spectrometry. hnRNP family proteins have at least one RRM domain and assemble on nascent RNA, packaging the RNA into hnRNP particles.36 hnRNP A2/B1 was reported to redistribute to the nucleolus upon ActD treatment.28 Similarly, PSF—a factor containing two RRMs that is involved in pre-mRNA splicing—was also detected by mass spectrometry.37 Thus, hnRNP A2/B1 and/or PSF may bridge CBP20S to mRNA and/or transcription sites and may also direct it to nucleolar caps upon transcription inhibition. The precise cellular function of CBP20S remains elusive, yet the data presented here implicate CBP20S in aspects of RNA polymerase II transcription and/or co-transcriptional mRNA processing.
Materials and Methods
Cell culture.
Cells were cultured in media supplemented with penicillin and streptomycin (P/S, Gibco) and 10% fetal calf serum (FCS, Gibco) at 37°C and 5% CO2. HeLa cells (DSMZ ACC 57) were propagated in low glucose (1 g/L) DMEM (Gibco) and Human Foreskin Fibroblasts (HFF) were maintained in Eagle's Minimum Essential Medium (EMEM). Metabolic labeling of HeLa cells was carried out by incubation in phosphate-free medium with 1% FCS and 100 µCi/ml P32-orthophosphate for 3 h. Plasmid transfection of HeLa cells was carried out using FuGENE 6 (Roche) according to the manufacturer's guidelines; transfected cells were grown for 48 h and harvested for experiments. For selection of stable cell lines expressing GFP and GFP-tagged CBP20 isoforms, HeLa cells were transfected with pIRESpuro-based constructs and selected with 0.8 µg/ml puromycin (Merck Biosciences). GFP-positive cells were sorted using FacsAria cell sorter (Becton Dickinson).
Human U2OS osteosarcoma Tet-On cells were maintained in low glucose Dulbecco's modified Eagle's medium (DMEM, Biological Industries, Israel) containing 10% fetal bovine serum (FBS, HyClone, Logan, UT). For imaging experiments, cells were transfected with 1–5 µg of plasmid DNA and 40 µg of sheared salmon sperm DNA (Sigma) using electroporation (Gene Pulser Xcell, Bio-Rad, Hercules, CA). For examining recruitment to the active transcription unit, cells were co-transfected with RFP-LacI and CBP20S-GFP or CBP20L-GFP, and then transcriptionally induced with doxycycline (1 ug/ml) for several hours. To inhibit transcription, U2OS cells were incubated with 5 µg/ml actinomycin D (Sigma) for 2 h as previously described.29
RNA isolation and RT-PCR.
Total RNA was isolated using TRIZOL (Invitrogen) and treated with DNA-free™ DNase Treatment and Removal Reagent (Ambion). Reverse transcription was performed using the SuperScript III kit (Invitrogen) with an oligo(dT)18 reverse primer. “No RT” controls were included. cDNA samples from human bone marrow were a kind gift of Dr. Christian Thiede (Universitätsklinikum, TU, Dresden). The cDNA amplified by conventional PCR with primers flanking the CBP20 coding region (20F: 5′-CCC AAG CTT CAC CAT GTC GGG TGG CCT C-3′and 20R: 5′-CGC GGA TCC TCA CTG GTT CTG TGC CAG-3′) was subjected to agarose gel analysis. Digital images of agarose gels were acquired using a Biostep trans-illuminator and ArgusX1 v.2 software.
Protein extraction and western blot analysis.
Whole cell extract from HeLa cells was prepared in NEST-2 buffer (50 mM Tris-HCl pH 6.8, 20 mM EDTA, 5% SDS) supplemented with complete protease inhibitor cocktail (Roche). Protein concentration was determined using the Amido Black Protein Assay.37 For western blot analysis, 30 µg of total protein extract was resolved on a Novex® 4–20% Tris-glycine gel (Invitrogen). The following primary antibodies were used: α-CBP80 serum9 at 1:2,000, α-CBP20 serum14 at 1:500, α-NCBP2 (Abgent) at 2.5 µg/ml, α-GAPDH (Novus Biologicals) at 0.1 µg/ml and goat α-GFP (a kind gift of D. Drechsel, MPI-CBG) at 1.5 µg/ml. As secondary antibodies, HRP-conjugated IgGs α-rabbit (GE Healthcare) at 1:10,000, α-goat (Sigma) at 1:80,000 and α-mouse (Sigma) at 1:80,000 were used.
Cloning.
CBP20L and CBP20S cDNAs were amplified by PCR with primers flanking the coding region and introducing BamHI and HindIII restriction sites (20F, 20R and 20RnS: 5′-CGC GGA TCC TCT GGT TCT GTG CCA GTT-3′). CBP20 isoforms were cloned untagged into pCS2+ (http://sitemaker.umich.edu/dlturner.vectors), a kind gift of Andrew C. Oates (MPI-CBG, Dresden, Germany) and as C-terminally tagged fusions into pEGFP-N2 (Clontech). To clone EGFP-tagged CBP20 sequences into pIRES puro (Clontech), CBP20L-EGFP, CBP20S-EGFP and EGFP were excised from pEGFP-N2 based plasmids using Eco47III and NotI (NEB) and cloned into pIRES puro. Constructs were verified by sequencing.
Fluorescence microscopy.
HeLa cells transfected with GFP-tagged protein constructs were fixed in 4% formaldehyde (PFA), permeabilized and mounted in DAPI/DABCO, as described.39 Images were acquired with a 60× oil immersion objective (NA 1.42, PlanApoN, Olympus), using the DeltaVision microscope system (Applied Precision) coupled with Olympus IX70 fluorescence microscope. For each image, 20–30 z-sections were collected in 200 nm steps. Image stacks were subjected to deconvolution using the SoftWoRx software (Applied Precision). The images presented are projections of the deconvolved z-stacks.
U2OS cells grown on coverslips were fixed for 20 min in 4% PFA and permeabilized in 0.5% Triton X-100 for 3 min. After blocking with 5% BSA, cells were immunostained for 1 h with anti-PSF (clone B92, Sigma) and anti-fibrillarin (Abcam). Wide-field fluorescence images were obtained using the Cell^R system based on an Olympus IX81 fully motorized inverted microscope (60X PlanApo objective, 1.42 NA) fitted with an Orca-AG CCD camera (Hamamatsu).
Fluorescence recovery after photobleaching (FRAP).
For FRAP, cells were maintained in Leibovitz's L-15 phenol red-free (Invitrogen) containing 10% FCS at 37°C. FRAP image sequences were obtained on an Olympus FV1000 inverted scanning confocal microscope equipped with a heated chamber and objective heater (37°C) and a 60×, 1.35 NA oil objective. Cells were scanned using a 488 laser for detection of GFP-labeled CBP components and a 561 nm laser for the detection of RFP-lacI. The active transcription site was bleached using the 488 nm laser. 5 pre-bleach images were acquired. Post-bleach images were acquired at a frequency of 29 images every 2 sec. For analysis of the fluorescence recovery, FRAP data were normalized and calculated as previously described.29
Immunoprecipitation of ribonucleoproteins (RNP IP).
This assay was conducted as described.39 Briefly, whole HeLa cell extracts were prepared in NET-2 buffer supplemented with protease inhibitors (Roche). Immunoprecipitation was carried out at 4°C with GammaBind™ G Sepharose beads™ (GE Healthcare) coupled to 12 µg α-GFP (gift of David Drechsel), 1 µg α-TMG (K121, Calbiochem) or control [non-immune mouse or goat IgG (Sigma)] antibodies. For protein detection, 33% of the immunoprecipitate and 0.7% of starting material were analyzed by western blotting. For RNA detection, the immunoprecipitated RNA was isolated by phenol/chloroform extraction, precipitated with ethanol, washed twice with 70% ethanol and resuspended in water. The resulting RNA was either subjected to reverse transcription (mRNA levels determination) or resolved on 10% denaturing polyacrylamide urea gel (snRNA levels determination, P32-labelled samples). Reverse transcription was performed using the SuperScript III kit (Invitrogen) with an oligo(dT)18 reverse primer. “No RT” controls were included. Fold enrichment of α-GFP antibody-precipitated mRNA over non-immune IgG IP was determined by quantitative real-time PCR (qPCR). The following equation was used: 2(Ct(IgG) − Ct(GFP)), where Ct stands for threshold cycle value, IgG for non-immune IgG IP and GFP for GFP IP. Primers used in qPCR were: mycF: 5′-GCG ACT CTG AGG AGG AAC AAG AAG-3′, mycR: 5′-ACT CTG ACC TTT TGC CAG GAG C-3′, actBF: 5′-ACT GGA ACG GTG AAG GTG ACA G-3′ and actBR: 5′-GTG GCT TTT AGG ATG GCA AGG-3′. Urea gel-resolved P32-labelled RNAs were detected using a PhosphorImager FLA-3000 film (Fuji).
7-methyl GTP-sepharose affinity assay.
Whole HeLa cell extracts were prepared in NET-2 buffer supplemented with complete protease inhibitor cocktail (Roche) and pre-cleared with 30 µl of Sepharose 4B blocked with 0.5 mg/ml BSA. A 1:1 mixture of 7-methyl GTP-Sepharose™ 4B (GE Healthcare) and Sepharose 4B (GE Healthcare) was used for specific, i.e., 7-methyl GTP, selection. Sepharose 4B alone was used as a negative control. Affinity selection was conducted for 4 h at 4°C on a rotary shaker. The beads were then washed 5 times with NET-2 buffer and bead-selected material was analyzed by western blot.
Immunopurification and mass spectrometry.
Immunopurification was conducted, using the GFP tag as an epitope,41 omitting the tobacco etch virus protease cleavage step. Briefly, approximately 8 × 107 HeLa cells expressing CBP20S-GFP, CBP20L-GFP, RAD51-GFP (kind gift of Mikolaj Slabicki) or GFP were harvested in Freezing Buffer (50 mM Hepes pH 7.5, 1 mM EGTA, 1 mM MgCl2, 100 mM KCl, 10% glycerol) and lysed in Lysis Buffer (75 mM Hepes pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 150 mM KCl, 0.075% NP-40, 10% glycerol) supplemented with complete protease inhibitor cocktail (Roche) and 90 U/ml Benzonase (Merck Biosciences). Following centrifugation, cell extracts were incubated with 40 µl GFP-trap® (ChromoTek), washed with Wash Buffer (50 mM Hepes pH 7.5, 1 mM EGTA, 1 mM MgCl2, 150 mM KCl, 0.5% NP-40, 10% glycerol) and eluted in 0.1 M glycine pH 2.0, which was subsequently neutralized with 1.5 M Tris pH 8.5. The eluate was subjected to trypsin digestion and mass spectrometry analysis, as described.41 LC-MS/MS analysis of peptide mixtures was performed on an Ultimate nanoLC system (Dionex, Amsterdam, The Netherlands) interfaced on-line to a LTQ Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) via a robotic nanoflow ion source TriVersa (Advion BioSciences Ltd., Ithaca, NY) as described.43,44 Data validation and organization was carried out with Scaffold 2 (Proteome Software, Inc., Portland, OR), which is based on the PeptideProphet™ algorithm.45
Acknowledgements
We thank Christian Thiede for providing us with human bone marrow cDNA (Universitätsklinikum, TU, Dresden), D. Drechsel and E. Izuarralde for antibody gifts and Mikolaj Slabicki for sharing the RAD51-GFP stable HeLa cell line. We are grateful to the members of our labs for helpful discussions. This work was supported by the Max Planck Society and grants from the German Research Foundation (NE909/2-1 to K.M.N.) and the German-Israeli Foundation (929-192.13 to K.M.N. and Y.S.-T.). Y.S.-T. is the Jane Stern Lebell Family Fellow in Life Sciences at BIU.
Abbreviations
- CBC
cap-binding complex
- CBP20
cap-binding protein 20
- RNP
ribonucleoprotein
- RRM
RNA recognition motif
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/12839
Supplementary Material
References
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol. 2007;623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- 7.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 8.Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 10.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, et al. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 13.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 15.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 16.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, et al. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazza C, Ohno M, Segref A, Mattaj IW, Cusack S. Crystal structure of the human nuclear cap binding complex. Mol Cell. 2001;8:383–396. doi: 10.1016/s1097-2765(01)00299-4. [DOI] [PubMed] [Google Scholar]
- 20.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 22.Graveley BR. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 23.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 24.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–4886. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baranova AV, Lobashev AV, Ivanov DV, Krukovskaya LL, Yankovsky NK, Kozlov AP. In silico screening for tumour-specific expressed sequences in human genome. FEBS Lett. 2001;508:143–148. doi: 10.1016/s0014-5793(01)03028-9. [DOI] [PubMed] [Google Scholar]
- 27.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, et al. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 29.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, et al. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nurtdinov RN, Artamonova II, Mironov AA, Gelfand MS. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum Mol Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- 31.Nakabeppu Y, Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991;64:751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- 32.Wisdom R, Yen J, Rashid D, Verma IM. Transformation by FosB requires a trans-activation domain missing in FosB2 that can be substituted by heterologous activation domains. Genes Dev. 1992;6:667–675. doi: 10.1101/gad.6.4.667. [DOI] [PubMed] [Google Scholar]
- 33.Gorlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey RA, et al. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 34.Dias SM, Wilson KF, Rojas KS, Ambrosio AL, Cerione RA. The molecular basis for the regulation of the cap-binding complex by the importins. Nat Struct Mol Biol. 2009;16:930–937. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dias SM, Cerione RA, Wilson KF, et al. Unloading RNAs in the cytoplasm: An “importin” task. Nucleus. 2010;1:139. doi: 10.4161/nucl.1.2.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 38.Schaffner W, Weissmann C. A rapid, sensitive and specific method for the determination of protein in dilute solution. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 39.Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell. 2006;17:4972–4981. doi: 10.1091/mbc.E06-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 41.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;11 doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 42.Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 2009;28:1453–1465. doi: 10.1038/emboj.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Junqueira M, Spirin V, Santana Balbuena T, Waridel P, Surendranath V, Kryukov G, et al. Separating the wheat from the chaff: unbiased filtering of background tandem mass spectra improves protein identification. J Proteome Res. 2008;7:3382–3395. doi: 10.1021/pr800140v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shevchenko A, Roguev A, Schaft D, Buchanan L, Habermann B, Sakalar C, et al. Chromatin Central: Towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008;9:167. doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


