Abstract
Unlike other retroviruses, human immunodeficiency virus type-1 (HIV-1) can infect terminally differentiated cells, due to the ability of its pre-integration complex (PIC) to translocate via the host nuclear pore complex (NPC). The PIC Nuclear import has been suggested to be mediated by the viral integrase protein (IN), via either the importin α or transportin 3 (TNPO3/transportin-SR2) pathways.
We show that in virus-infected cells, IN interacts with both importin α and TNPO3, simultaneously or separately, suggesting a multiple use of nuclear import pathways. Disruption of either the IN-importin α or IN-TNPO3 complexes in virus-infected cells by specific cell-permeable-peptides resulted in inhibition of IN and viral cDNA nuclear import. Here we show that peptides which disrupt either one of these complexes block virus infection, indicating involvement of both pathways in efficient viral replication. Formation of IN-importin α and IN-TNPO3 complexes has also been observed in IN-transfected cultured cells. Using specific peptides, we demonstrate that in transfected cells but not in virus infected cells the importin α pathway overrides that of TNPO3. The IN-importin α and IN-TNPO3 complexes were not observed in virus-infected Rev-expressing cells, indicating the Rev protein's ability to disrupt both complexes.
Our work suggests that IN nuclear import requires the involvement of both importin α and TNPO3. The ability to inhibit nuclear import of the IN-DNA complex and consequently, virus infection by peptides that interrupt IN's interaction with either importin α or TNPO3 indicates that for efficient infection, nuclear import of IN should be mediated by both nuclear-import receptors.
Key words: HIV-1, nuclear import, importin, transportin 3, integrase
Introduction
Integration of human immunodeficiency virus type 1 (HIV-1) cDNA into the host chromosomal DNA is one of the crucial events in the virus's life cycle, essentially controlling the entire replication and assembly process.1 This event is mediated by the viral integrase (IN) protein which is encapsidated within virions and released into the cell cytoplasm following virus cell fusion.2 Within the cytoplasm, IN attaches to the viral cDNA, forming an IN-DNA complex which is part of the viral pre-integration complex (PIC).3 For integration to occur, the IN-DNA complex has to be translocated into the infected cells' nuclei and reach the chromosomal DNA.4 Indeed, in contrast to other retroviruses, HIV-1 can infect terminally differentiated cells.5,6 HIV-1's capacity to infect cell cycle-arrested cells has been ascribed to the ability of its PIC7,8 to translocate across the nuclear envelope via the nuclear pore complex (NPC).9,10 However, despite extensive effort and numerous studies, our understanding of the events leading to nuclear import of the PIC is still very poor. In particular, the detailed mechanism of the PIC's nuclear-import pathway, as well as the viral or cellular factors that mediate its translocation into the nuclei of infected host cells, are highly controversial and hotly debated (reviewed in ref. 11–13).
Several cellular nuclear-import receptors have been suggested to be involved in the process of PIC nuclear import. Among them are the importin α/β heterodimer,14–18 importin 719,20 and transportin 3 (TPNO3/transportin-SR2, an importin β-like receptor).21–23 Various viral karyophilic proteins, such as the Matrix (MA), Vpr and IN,3,24–30 have been suggested to actively translocate the PIC into the host-cell nucleus. The cellular protein lens epithelium-derived growth factor p75 (LEDGF/p75), along with a short section of triple-stranded DNA present within the viral cDNA and known as the DNA flap, have also been implicated in promoting translocation of the PIC into nuclei of infected cells.30–32 In addition, the HIV capsid protein (CA) has been reported to play a crucial role in controlling nuclear import of the HIV genome.33,34 However, the exact mechanism governing nuclear import of the PIC remains unclear.3,11–14,25,27,28,30
Our previous works,17,18 as well as that of others,14,35–39 have demonstrated that the HIV-1 IN is a karyophilic protein that bears a nuclear-localization signal (NLS) and is actively translocated into the nuclei of IN-expressing cells.14,35–39 Based on the use of cell-permeable functional peptides, our work supports the view that IN has a dominant role in driving a functional PIC into infected cells' nuclei.18 This was inferred mainly from experiments showing that a peptide bearing a sequence suggested to be the IN NLS (NLS-IN, comprised of IN residues 161–173), as well as the SV40-T-antigen NLS peptide (SV40-NLS), block nuclear import of IN in IN-transfected and HIV-infected cells.14,17,18 Concurrent with inhibition of IN nuclear import, these two peptides blocked HIV-1 replication in virus-infected cells.18 Inhibition was due to these peptides' ability to disrupt the interaction of IN with importin α,17,18 supporting the view that this nuclear-import receptor is required for virus replication in cell cycle-arrested cells.
The results of the present work, based on co-immunoprecipitation (co-IP) and immunostaining experiments and the use of cell-permeable functional peptides, indicate that in addition to importin α, TNPO3 is involved in nuclear import of IN in virus-infected cells. Our work indicates that the requirement of one nuclear receptor, importin α, does not eliminate the involvement of a second one—TNPO3. It appears that both receptors are required to allow efficient translocation of IN, and thus of the entire PIC, into host-cell nuclei. This observation may assist in solving what has appeared, until now, to be a point of contention: nuclear import of IN via the importin α14–18 versus TNPO3,21–23 pathway.
Results
IN interacts with both importin α and TNPO3.
Whether nuclear import of HIV-1 IN is mediated by importin α14–18 or TNPO3,21–23 is unclear and under dispute. The co-IP experiments depicted in Figure 1A show that in virus-infected cells, IN interacts with both importin α and TNPO3. These results suggest that these two nuclear-import receptors are involved in the translocation of IN into nuclei of infected cells, either concurrently or alternately.
Figure 1.
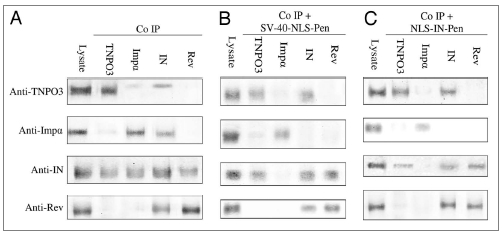
NLS-IN and SV40-NLS peptides disrupt the IN-importin α but not IN-TNPO3 complex in virus infected cells. SupT1 lymphocytes were infected with wild-type HIV-1 and incubated in the presence or absence of the indicated peptides. Following infection, cells were lysed and half of the cells' lysate volume was subjected to SDS-PAGE, then immunoblotted with either anti-importin α (anti-Impα), anti-TNPO3, anti-IN or anti-Rev antibodies. The complementary HRP-conjugated antibodies were used as the second antibody. The remaining lysate was subjected to co-IP with either the anti-Impα, anti-TNPO3, anti-IN or anti-Rev antibodies. Following SDS-PAGE, the gels were immunoblotted by each of the above antibodies and then incubated with the complementary HRP-conjugated antibodies as second antibody. (A) No peptide; (B) SV40-NLS-Pen peptide; (C) NLS-IN-Pen peptide. When peptides were used, cells were incubated with 150 µM of the indicated peptide.
We have previously shown (reviewed in ref. 18 and see also Figure 1B and C) that cell-permeable peptides bearing the SV40-NLS or the putative NLS of IN (NLS-IN) can disrupt the complex formed between IN and importin α and inhibit translocation of IN into the nuclei of infected cells. As shown in Figure 1B and C, these two peptides failed to promote dissociation of the IN-TNPO3 complex, indicating that different domains mediate the interaction of IN with these two nuclear-import receptors. The results in Figure 1 also show that neither peptide had any effect on the interaction between Rev and IN, shown by us previously to occur in virus-infected cells40–44 and indicating specificity of function.
Disruption of the IN-TNPO3 complex by IN-interacting peptides blocks nuclear import of IN and viral cDNA.
Three IN-interacting cell permeable peptides were previously selected by us.45–48 These peptides, termed IN-1,45,47 Rev 53–67 (Fig. 2A and B) and Rev 13–23,46,48 (not shown respectively), have been shown to promote dissociation of the IN-TNPO3 complex but not of the IN-importin α complex. Specificity of these peptides' functions should be inferred from results showing that a peptide bearing the NLS sequence of the Rev protein [also known as the arginine-rich motif (ARM)44,49] fails to disrupt IN interactions with either importin α or TNPO3 (Fig. 2C). However, as expected, the Rev ARM peptide was able to promote dissociation of the Rev-importin β complex (Fig. 2D). Also, and as shown previously,44 only Rev 53–67 (Fig. 2B) and Rev 13–23 (not shown) peptides promoted dissociation of the Rev-IN complex formed in virus-infected cells.
Figure 2.

Disruption of IN-TNPO3 but not IN-importin α complex by the Rev 53–67 and IN-1 peptides. SupT1 lymphocytes were infected with wild-type HIV-1, incubated with the indicated peptides and subjected to co-IP and immunoblotting as described in the legend to Figure 1 and Methods. (A) IN-1 peptide; (B) Rev 53–67 peptide; (C) Rev ARM peptide; (D) similar to (B) and (C) but with the anti-Rev and anti-importin β (anti-Impβ) antibodies and with IN-expressing SupT1 cells in the right lane. When peptides were used, cells were incubated with 150 µM of the indicated peptide. Other experimental details are described in Methods.
The three above-described IN-interacting peptides inhibited nuclear import of IN in infected cells (Fig. 3, Table 1 and reviewed in ref. 42), most likely due to their ability to disrupt the IN-TNPO3 complex. The same three peptides were also found to inhibit nuclear import of the viral cDNA (Fig. 4A), as well as the appearance of 2LTR (long terminal repeat) circles. These circles can be formed only in the nuclei of infected cells following the PIC nuclear import24 (Fig. 4B). The Rev-NES peptide,50 which has been used as a negative control, did not have any effect on either nuclear import of viral DNA (Fig. 4A) or the formation of the 2LTR circles (Fig. 4B). It should be noted that in virus-infected cells, the Rev ARM peptide, as expected, blocked nuclear import of Rev. However, the Rev ARM peptide also partially inhibited nuclear import of IN (Fig. 3 and Table 1) despite its inability to disrupt the interaction between IN and its nuclear receptors (Fig. 2C). This may be expected since in virus-infected cells, a large proportion of the IN molecules are present as Rev-IN complexes (reviewed in ref. 40–44 and 48 and see also Figs. 1 and 2), thus inhibition of Rev nuclear import is expected to lead to cytoplasmic retention of IN as well. Although the Rev-IN complex can be observed in the PICs of the infected cells,40,44 a large portion of it is localized within the cytoplasm42 and is not necessarily associated with the PIC. This is probably due to the fact that Rev promotes dissociation of the IN from the PIC localized viral cDNA.44
Figure 3.
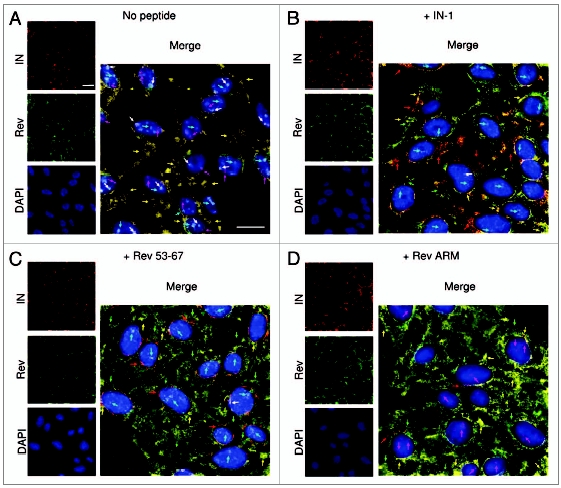
Inhibition of IN nuclear import in virus-infected cells by Rev 53–67 and IN-1. TZM-bl cells were infected (with wild-type HIV-1) (A), incubated with the indicated peptides (B–D) and immunostained as described in Methods: IN (red), Rev (green), DAPI (blue); the entire merged picture was magnified for a better view of IN localization within the infected cell. Bar = 10 µm. Arrows indicate the presence of intranuclear IN (magenta) or Rev (cyan) molecules or Rev-IN complex (white) and of cytoplasmic IN (red), Rev (green) and Rev-IN complex (orange-yellow).
Table 1.
The effect inhibitory peptides on the extent of nuclei containing IN in infected cells
| Treatment | Percentage of nuclei containing IN from all infected cells [%]* |
| No peptide | 94.11 ± 3.75 |
| IN-1 | 7.14 ± 1.26 |
| Rev 53–67 | 5.17 ± 1.07 |
| Rev ARM | 72.14 ± 4.67 |
The average percentage was calculated from 3 repeats of 10 random fields. For each experiment the error represent the standard deviation between each of the repeats.
Figure 4.
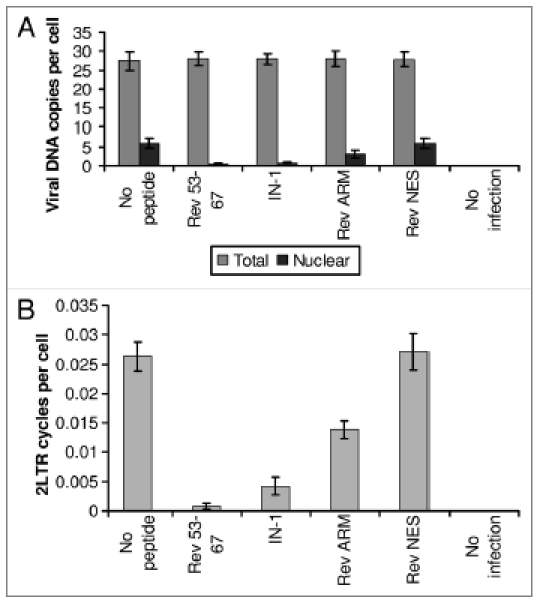
Inhibition of cDNA nuclear import and 2LTR circle formation in virus-infected cells by Rev 53–67 and IN-1. SupT1 lymphocytes were infected with wild-type HIV-1 at a MOI of 1 in the presence or absence of the indicated peptides as described in Methods. (A) Following infection, the nuclear fraction was isolated from half of the cells and the amount of viral DNA was estimated by real-time PCR. (B) The amount of 2LTR circles was estimated in the DNA fraction isolated from whole cells by real-time PCR. Other experimental details are described in Methods. Error bars represent standard deviation, ca. ±5%
The effect of the IN-interacting peptides on nuclear import of IN protein in transfected cultured cells.
In light of the above and previous results18,42 observed in virus-infected cells, it was of interest to study the effect of the IN-interacting peptides on the intracellular localization of IN in transfected cultured cells. In transfected cells, as opposed to virus-infected cells, Rev 53–67 (and Rev 13–23, not shown), as well as IN-1, only partially inhibit nuclear import of the expressed IN (Fig. 5A). Specificity of inhibition could be inferred from the fact that nuclear import of the Rev protein was not affected by these peptides (Fig. 5B). On the other hand, the Rev ARM peptide, as expected, blocked nuclear import of Rev but not of IN (Fig. 5A and B).
Figure 5.
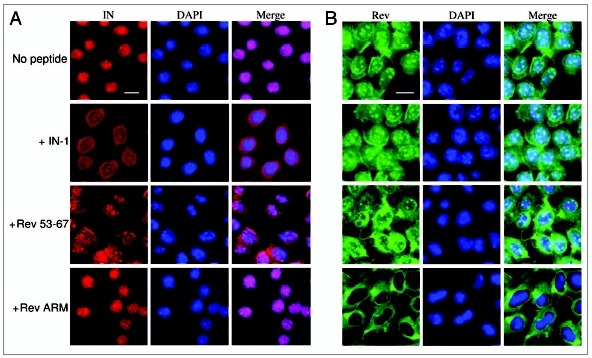
Rev 53–67 and IN-1 peptides only partially inhibit IN nuclear import in transfected cells. TZM-bl cells expressing either IN (A) or Rev (B) were generated by stable transfection into TZM-bl cells of pcDNA3.1 plasmid bearing the full wild-type IN or Rev genes, respectively. Cells were fixed and immunostained using 1:100 rabbit anti-IN or 1:50 rat anti-Rev antibodies and Cy3-conjugated anti-rabbit or Cy2-conjugated anti-rat antibodies, respectively, as second antibody. IN (red), Rev (green) and DAPI (blue) were observed under confocal microscope. Bar = 10 µm. Other experimental details are described in Methods.
The Rev protein disrupts both IN-importin α and IN-TNPO3 complexes.
Following our results regarding the ability of the Rev-derived peptides to promote dissociation of the IN-TNPO3 complex and our previous observation that the full-length Rev protein blocks IN nuclear import,42 it was essential to study the effect of Rev itself on the interactions between IN and the nuclear-import receptors. When Rev-expressing cells were infected with HIV-1, IN failed to interact with either importin α or TNPO3 (Fig. 6). This was inferred from the co-IP experiment showing that, in contrast to the Rev-derived peptides, the Rev protein promotes disruption of both the IN-importin α and IN-TNPO3 complexes.
Figure 6.
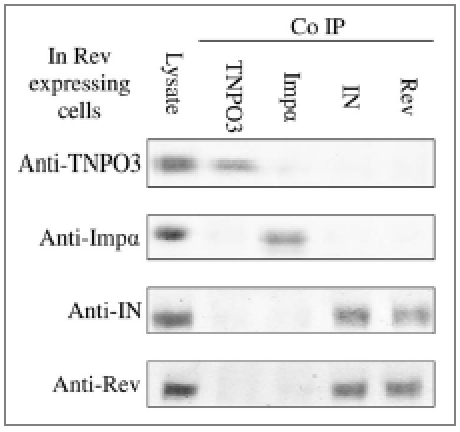
Disruption of both IN-TNPO3 and of IN-importin α complexes by the Rev protein. Rev-expressing SupT1 lymphocytes were infected with wild-type HIV-1 and then half the volume of cell lysate was subjected to SDS-PAGE and immunoblotting with either anti-importin α (anti-Impα), anti-TNPO3, anti-IN or anti-Rev antibodies. Complementary HRP-conjugated antibodies were used as second antibodies. The remaining lysate was subjected to co-IP with anti-Impα, anti-TNPO3, anti-IN or anti-Rev antibodies and the complementary HRP-conjugated antibodies as second antibodies. Other experimental details are described in Methods.
Discussion
The results of the present work show that in virus-infected cells IN interacts with both importin α and TNPO3. Our previous observation showed that nuclear import of HIV-1 IN is mediated by a specific NLS domain (amino acids 161–173, NLS-IN), which mediates its translocation via the importin α pathway.15,17,18,35 This was demonstrated by import of BSA molecules conjugated to the NLS-IN peptides into nuclei of permeabilized cultured cells.17 Moreover, we have showed that cell-permeable peptides bearing NLS-IN, as well as those bearing SV40-NLS, were able to inhibit nuclear import of IN in transfected and virus-infected cultured cells.18 As would be expected, the same peptides inhibit HIV-1 infection of TZM-bl and lymphocyte-cultured cells.18 Recently, our observations regarding the interaction between the viral IN and importin α(3) in virus infected cells have been confirmed in independent studies using in-vitro pull-down assay and cell-based co-immunoprecipitation method in 293T cells and HIV-1 infected cells.51 Furthermore, a short hairpin RNA (ShRNA) was employed to knockdown Impα3 in dividing C8166T, HeLa cells and non-dividing monocyte-derived macrophages (MDM). HIV-1 replication in Impα3 knockdown cells was significantly downregulated, (reviewed in ref. 51 and 52).
The use of cell-permeable peptides allowed us to demonstrate that TNPO3, in addition to importin α, is required for mediation of IN nuclear import and virus replication in infected cells. However, our co-IP experiments did not allow us to determine whether IN interacts directly with TNPO3, as was demonstrated recently using an in-vitro assay system,53 or if its interaction is mediated by a third component, such as t-RNA.22 The identification of TNPO3 as a binding partner of IN has recently been confirmed independently and the IN binding site for TNPO3 has been elucidated.54
The requirement of TNPO3 for HIV replication was based previously21–23 mainly on the significantly low level of HIV-1 replication in RNAi TNPO3-knockdown cultured cells. Our present study, demonstrating inhibition of HIV-1 infection following disruption of the IN-TNPO3 complex by the Rev-derived peptides as well as by IN-1, strongly supports involvement of TNPO3 in the HIV-1 infection process. However, the way in which TNPO3 participates in the HIV-1 replication cycle or is involved in nuclear import of the PIC is still under debate. Christ et al.21 and Luban22 suggest that TNPO3 is required to mediate nuclear import of HIV-1 IN and thus for translocation of the PIC into nuclei of infected cells and consequently, for virus infection. Our demonstration of Rev-derived and IN-1 peptides disruption of the IN-TNPO3 interaction and virus infection supports this view. On the other hand, the use of MuLV/HIV chimera viruses failed to demonstrate the requirement of IN-TNPO3 interaction for HIV replication.53
Krishnan et al.53 used in vitro binding assays to study the interaction between TNPO3 and several IN proteins from various retroviruses, such as HIV, MuLV and SIV. A comparison of their results to the infectivity profiles of HIV chimera viruses led them to conclude that the dominant protein dictating TNPO3 dependency during virus infection is the HIV-1 CA and not IN.53 This conclusion was further supported by a work published during the preparation of this ms.34 However, no direct or indirect interaction between TNPO3 and CA has yet been demonstrated. Furthermore, rapid removal of CA from the RTC (reverse transcriptase complex)/PIC has been suggested to be required for nuclear import of the HIV-1 PIC.33 Thus, a functional interaction between TNPO3 and CA remains to be proven.
Our conclusion that nuclear import of IN requires the involvement of both impotin α and TNPO3 and plays a dominant role in HIV-1 infection is based mainly on results obtained using two groups of peptides (Fig. 7A). One group, which includes SV40-NLS as well as NLS-IN,18 disrupted only—and specifically—the interaction between IN and importin α (Fig. 7A). The second group, which includes the two Rev-derived peptides and IN-1, disrupted only the interaction between IN and TNPO3 (Fig. 7A). Interestingly, the Rev protein which, similar to the Rev-derived peptides, has been shown to block nuclear import of IN,42 prevented interaction of IN with both receptors. The disruption of the IN-TNPO3 complex by the Rev derived and IN-1 peptides is not necessarily due to direct masking of the TNPO3 binding sites. This can also be due to the peptides inducing a change in the IN oligomeric state45–47 which may prevent binding of TNPO3.
Figure 7.
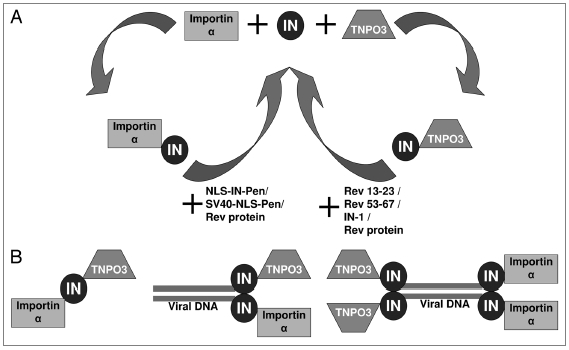
Schematic view of IN-impotin α and IN-TNPO3 interactions. (A) Disruption of IN-impotin α and IN-TNPO3 interactions by two different groups of peptides. (B) Three alternative possibilities for interaction of IN with both impotin α and TNPO3.
The ability to block virus infection by a peptide belonging to either the first or second group clearly indicates that for efficient infection, nuclear import of IN should be mediated at the same time by both nuclear receptors. However, it appears that in IN-expressing cells, as opposed to virus-infected cells, nuclear import via the importin α pathway is dominant. This is inferred from the results showing significant inhibition of IN nuclear import by a peptide belonging to the first group, whereas inhibition was only partial by a peptide belonging to the second group. Thus it appears, as also indicated in our previous observations,42 that results obtained in transfected cells are not necessarily relevant to those obtained in infected ones.
Our results demonstrate that IN, a karyophilic protein, is translocated into the cell nucleus by two distinct and different nuclear-import pathways. Confirming previous observations,18,42 we show that blocking IN nuclear import simultaneously inhibits import of viral cDNA and the formation of 2LTR circles. These results strengthen the view that IN is an integral part of the PIC.3 It is thus conceivable that participation of the two nuclear-import receptors is required to allow efficient nuclear import of the relatively large PIC, whose main component is the IN-DNA complex: the function of only one of these receptors does not appear to be sufficient to drive the translocation of this IN-DNA complex via the NPC.
It is still unclear why nuclear import of IN requires the involvement of both impotin α and TNPO3. Furthermore, we still do not know whether the same IN molecule interacts, via different domains, with both receptors or if different IN molecules—or IN oligomers—interact individually with each receptor. The scheme in Figure 7B summarizes our view of the three alternative interaction possibilities between IN and the two nuclear-import receptors impotin α and TNPO3.
In addition to impotin α and TNPO3, there are two additional pathways that have been implicated in PIC nuclear transport. These include the nuclear import receptor importin 7,19,20,55 and the direct interaction with Nup153,34,56 which is one of the NPC proteins.57 The current results as well as previous results20,21,34,52,56,58 may imply that these two pathways could also be required to ensure PIC nuclear transport as a multi nuclear import complex. Thus, inhibition of only one of these nuclear import pathways should result in partial decrease in the level of the HIV PIC nuclear import but should not block it completely.
Materials and Methods
Mammalian cells.
Monolayer adherent HeLa TZM-bl cells (obtained through the NIH AIDS Research and Reference Reagent Program) expressing the β-galactosidase gene under regulation of a transactivation response element59 were grown in Dulbecco's modified Eagle's medium. The T-lymphocyte cell line SupT1 was grown in RPMI 1640. All media were supplemented with 10% (v/v) fetal calf serum, 0.3 g/l L-glutamine, 100 units/ml penicillin and 100 units/ml streptomycin (Biological Industries, Beit Haemek, Israel). Cells were incubated at 37°C in a 5% CO2 atmosphere and re-cultured every 4 days. IN- and Rev-expressing HeLa TZM-bl cells50 were generated by stable transfection60 into TZM-bl cells of plasmid pcDNA3.1 bearing the full-length wild-type IN or Rev gene, respectively. Selection was carried out for 4 weeks with 400 µg/ml hygromycin B.
Viruses.
Wild-type HIV-1 was generated by transfection of HEK293T cells with pSVC21 plasmid containing the full-length HIV-1 HXB2 viral DNA. Wild-type viruses were harvested from HEK293T cells 48 and 72 h post-transfection. The viruses were stored at −75°C.
Virus stock titration.
Quantitative titration of HIV-1 was carried out using the MAGI assay, as described by Kimpton and Emerman.61 Briefly, TZM-b1 cells were grown in 96-well plates at 1 × 104 cells per well. The cells were then infected with 50 µl of serially diluted virus as described.61 Two days post-infection (PI), cultured cells were fixed and β-galactosidase was estimated exactly as described previously.61 Blue cells were counted under a light microscope at 200X magnification.
Synthesis of peptides.
Peptides were synthesized on Rink amide resin using a model 433A Applied Biosystems peptide synthesizer as described.62 The Pen peptide,63 with the following sequence: RQIKIWFQNRRMKWKK (Ant 43–58), was added to the SV40-NLS- and NLS-IN-bearing peptides to allow their cell permeability as described previously.18
Immunostaining studies.
Localization of IN and Rev in transfected cultured cells.
TZM-bl IN- or Rev-expressing cells were grown on chamber slides (Nunc). After reaching 70–80% confluence, cell cycle arrest was obtained by treatment with 5 µg/ml of aphidicolin. The cells were then incubated with 150 µM of the indicated peptide for 6 h and fixed and immunostained essentially as described previously64 with the following modifications. After fixation, cells were blocked with 5% (w/v) BSA (IgG free) (Jackson) in PBS for 60 min. For detection of HIV-1 IN, cells were incubated with 1:100 rabbit α-IN (NIH AIDS Research and Reference Reagent Program, cat. no. 758) or with 1:50 α-Rev65 at room temperature for 60 min. Cells were washed five times with PBS + 0.05% (v/v) Tween 20. Then the cells were incubated with a second antibody, Cy3-conjugated anti-rabbit antibody (Jackson) (1:200) or Cy2-conjugated anti-rat antibody (Jackson) (1:200) at room temperature for 60 min, followed by another five washes with PBS + 0.05% Tween 20. For detection of DNA, cells were stained with DAPI according to the manufacturer's protocol (KPL,USA). The slides were prepared with mounting media (Bio-Rad) and immunofluorescent cells were detected with a confocal microscope.
Localization of IN and Rev in HIV-1-infected cultured cells.
TZM-bl cells were grown on chamber slides. Cells were arrested as described in A and then incubated with 150 µM of the indicated peptide for 2 h. After incubation with the peptides, cells were infected with wild-type HIV-1 at a multiplicity of infection (MOI) of 25. Cells were fixed and stained as described above with the following modifications: fixation was performed at 12 h PI and the first antibody was used at a dilution of 1:50, the second antibody at a dilution of 1:100 for the detection of IN. For the detection of Rev, the first antibody was used at a dilution of 1:50, the second antibody at a dilution of 1:50.
In both cases 10 fields were selected from each experiment and one shown is a representative of the image obtained
Co-IP studies of in-vivo protein-protein interactions.
Cells were infected with the indicated viruses at a MOI of 15, harvested at 12 h PI, washed three times with PBS and lysed by the addition of PBS containing 1% (v/v) Triton X-100. Half of the lysate volume was subjected to SDS-PAGE, then immunoblotted with either antiserum raised against IN amino acids 276–288 (anti-IN) (NIH AIDS Research and Reference Reagent Program, cat. no. 758), anti-importin α (anti-Impα) antibody (Santa Cruz), anti importin β (anti-Impβ) antibody (Santa Cruz), anti-TNPO3 antibody (Abcam) or anti-Rev antibody.65 The complementary HRP-conjugated antibodies (Jackson) were used as the second antibody.
The remaining lysate or isolated fractions were incubated for 1 h at 4°C with anti-Impα, anti-Impβ, anti-TNPO3, anti-IN or anti-Rev antibodies. Following 3 h incubation with protein G-agarose beads (Santa Cruz) at 4°C, the samples were washed three times with PBS containing 1% (v/v) Nonidet P-40. SDS buffer was added to the samples and after boiling and running on an SDS polyacrylamide gel, the membranes were immunoblotted with anti-Impα, anti-Impβ, anti-TNPO3, anti-IN or anti-Rev antibodies and then with the complementary HRP-conjugated antibodies (Jackson) as second antibodies.
When peptides were used, cells were incubated with 150 µM of the indicated peptide for 2 h prior to infection.
Isolation of cytoplasm and nuclei from infected cells.
The various fractions were obtained from virus-infected cells essentially as described previously44,66 with several modifications. Briefly, cells were harvested and washed twice in buffer A (20 mM Hepes pH 7.3, 150 mM KCl, 5 mM MgCl2, 1 mM DTT and 0.1 mM PMSF). Cells were then suspended in 200 µl of buffer A containing 0.025% (w/v) digitonin, incubated at room temperature for 10 min and then centrifuged for 3 min at 1,000 g at room temperature. The supernatant was then centrifuged at 8,000 g and separated into supernatant (cytoplasm) and pellet (nuclei) and stored at −70°C.
Quantitation of total and nuclear viral DNA.
Total viral DNA was estimated, using SYBR green real-time quantitative PCR at 10 h PI, from the total cell lysate or nuclear-isolated fractions of the infected cells. DNA was isolated by phenol chloroform method. DNA samples (1 µg) were added to 95 µl containing 1X Hot-Rescue Real Time PCR Kit-SG (Diatheva s.r.l, Fano, Italy) and 100 nM of each primer-binding site primer: F5 (5′ primer, 5′-TAG CAG TGG CGC CCG A-3′) and R5 (3′ primer, 5′-TCT CTC TCC TTC TAG CCT CCG C-3′). All amplification reactions were carried out using an ABI Prism 7700 Sequence Detection System (Applied Biosystems) under the following program: 1 cycle at 95°C for 10 min, followed by 45 cycles of 15 s at 95°C and 35 s at 68°C. Three replicates were performed for each PCR run. All other details are exactly as described in Casabianca et al.67
Quantitation of 2LTR circles.
Quantification of 2LTR circles was estimated exactly as described in Butler et al.68
All experiments were repeated three to four times and the quantitative differences between the experiments never exceeded ±10%.
Acknowledgements
This work was supported by the Israeli Science Foundation (A. Loyter) and by a starting grant from the European Research Council (ERC) (to A.F.).
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/12903
Authors' Contributions
A. Levin designed and performed the experiments, analyzed data and contributed to writing the paper; Z.H. performed peptide synthesis and purification; A.F. designed the study and contributed to the writing; A. Loyter designed the study, contributed to the writing of the paper and coordinated the study. All authors have read and approved the manuscript.
References
- 1.Esposito D, Craigie R. HIV integrase structure and function. Adv Virus Res. 1999;52:319–333. doi: 10.1016/s0065-3527(08)60304-8. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- 3.Sherman MP, Greene WC. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 2002;4:67–73. doi: 10.1016/s1286-4579(01)01511-8. [DOI] [PubMed] [Google Scholar]
- 4.Furtado MR, Callaway DS, Phair JP, Kunstman KJ, Stanton JL, Macken CA, et al. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 5.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz RA, Greger JG, Boimel P, Skalka AM. Human immunodeficiency virus type 1 DNA nuclear import and integration are mitosis independent in cycling cells. J Virol. 2003;77:13412–13417. doi: 10.1128/JVI.77.24.13412-13417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki Y, Craigie R. The road to chromatin—nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5:187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- 9.Bukrinsky M. A hard way to the nucleus. Mol Med. 2004;10:1–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 11.Goff SP. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J Gene Med. 2001;3:517–528. doi: 10.1002/1521-2254(200111)3:6<517::AID-JGM234>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 12.Greene WC, Peterlin BM. Charting HIV's remarkable voyage through the cell: Basic science as a passport to future therapy. Nat Med. 2002;8:673–680. doi: 10.1038/nm0702-673. [DOI] [PubMed] [Google Scholar]
- 13.Nisole S, Saib A. Early steps of retrovirus replicative cycle. Retrovirology. 2004;1:9. doi: 10.1186/1742-4690-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hearps AC, Jans DA. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem J. 2006;398:475–484. doi: 10.1042/BJ20060466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ao Z, Fowke KR, Cohen EA, Yao X. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology. 2005;2:62. doi: 10.1186/1742-4690-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armon-Omer A, Graessmann A, Loyter A. A synthetic peptide bearing the HIV-1 integrase 161–173 amino acid residues mediates active nuclear import and binding to importin alpha: characterization of a functional nuclear localization signal. J Mol Biol. 2004;336:1117–1128. doi: 10.1016/j.jmb.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 18.Levin A, Armon-Omer A, Rosenbluh J, Melamed-Book N, Graessmann A, Waigmann E, et al. Inhibition of HIV-1 integrase nuclear import and replication by a peptide bearing integrase putative nuclear localization signal. Retrovirology. 2009;6:112. doi: 10.1186/1742-4690-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, et al. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J Biol Chem. 2007;282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- 20.Zaitseva L, Cherepanov P, Leyens L, Wilson SJ, Rasaiyaah J, Fassati A. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology. 2009;6:11. doi: 10.1186/1742-4690-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, et al. Transportin-SR2 imports HIV into the nucleus. Curr Biol. 2008;18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 22.Luban J. HIV-1 infection: going nuclear with TNPO3/Transportin-SR2 and integrase. Curr Biol. 2008;18:710–713. doi: 10.1016/j.cub.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 23.Rain JC, Cribier A, Gerard A, Emiliani S, Benarous R. Yeast two-hybrid detection of integrase-host factor interactions. Methods. 2009;47:291–297. doi: 10.1016/j.ymeth.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of integrase, matrix and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Depienne C, Roques P, Creminon C, Fritsch L, Casseron R, Dormont D, et al. Cellular distribution and karyophilic properties of matrix, integrase and Vpr proteins from the human and simian immunodeficiency viruses. Exp Cell Res. 2000;260:387–395. doi: 10.1006/excr.2000.5016. [DOI] [PubMed] [Google Scholar]
- 26.Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haffar OK, Popov S, Dubrovsky L, Agostini I, Tang H, Pushkarsky T, et al. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J Mol Biol. 2000;299:359–368. doi: 10.1006/jmbi.2000.3768. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins Y, McEntee M, Weis K, Greene WC. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143:875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 31.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, et al. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouyac-Bertoia M, Dvorin JD, Fouchier RA, Jenkins Y, Meyer BE, Wu LI, et al. HIV-1 infection requires a functional integrase NLS. Mol Cell. 2001;7:1025–1035. doi: 10.1016/s1097-2765(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 36.Depienne C, Mousnier A, Leh H, Le Rouzic E, Dormont D, Benichou S, et al. Characterization of the nuclear import pathway for HIV-1 integrase. J Biol Chem. 2001;276:18102–18107. doi: 10.1074/jbc.M009029200. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda T, Nishitsuji H, Zhou X, Nara N, Ohashi T, Kannagi M, et al. Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J Virol. 2004;78:11563–11573. doi: 10.1128/JVI.78.21.11563-11573.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pluymers W, Cherepanov P, Schols D, De Clercq E, Debyser Z. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology. 1999;258:327–332. doi: 10.1006/viro.1999.9727. [DOI] [PubMed] [Google Scholar]
- 39.Tsurutani N, Kubo M, Maeda Y, Ohashi T, Yamamoto N, Kannagi M, et al. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J Virol. 2000;74:4795–4806. doi: 10.1128/jvi.74.10.4795-4806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levin A, Hayouka Z, Brack-Werner R, Volsky DJ, Friedler A, Loyter A. Novel regulation of HIV-1 replication and pathogenicity: Rev inhibition of integration. Protein Eng Des Sel. 2009;22:753–763. doi: 10.1093/protein/gzp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin A, Hayouka Z, Friedler A, Brack-Werner R, Volsky DJ, Loyter A. A novel role for the viral Rev protein in promoting resistance to Super-infection by Human Immunodeficiency Virus type 1. J Gen Virol. 2010;91:1503–1513. doi: 10.1099/vir.0.019760-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin A, Hayouka Z, Friedler A, Loyter A. Nucleocytoplasmic shuttling of HIV-1 integrase is controlled by the viral Rev protein. Nucleus. 2010;1:190–201. doi: 10.4161/nucl.1.2.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin A, Hayouka Z, Helfer M, Brack-Werner R, Friedler A, Loyter A. Peptides derived from HIV-1 integrase that bind Rev stimulate viral genome integration. PLoS ONE. 2009;4:4155. doi: 10.1371/journal.pone.0004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levin A, Rosenbluh J, Hayouka Z, Friedler A, Loyter A. Integration of HIV-1 DNA is regulated by interplay between viral Rev and cellular LEDGF/p75 proteins. Mol Med. 2010;16:34–44. doi: 10.2119/molmed.2009.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armon-Omer A, Levin A, Hayouka Z, Butz K, Hoppe-Seyler F, Loya S, et al. Correlation between shiftide activity and HIV-1 integrase inhibition by a peptide selected from a combinatorial library. J Mol Biol. 2008;376:971–982. doi: 10.1016/j.jmb.2007.11.095. [DOI] [PubMed] [Google Scholar]
- 46.Hayouka Z, Rosenbluh J, Levin A, Maes M, Loyter A, Friedler A. Peptides derived from HIV-1 Rev inhibit HIV-1 integrase in a shiftide mechanism. Biopolymers. 2008;90:481–487. doi: 10.1002/bip.20930. [DOI] [PubMed] [Google Scholar]
- 47.Maes M, Levin A, Hayouka Z, Shalev DE, Loyter A, Friedler A. Peptide inhibitors of HIV-1 integrase: From mechanistic studies to improved lead compounds. Bioorg Med Chem. 2009;17:7635–7642. doi: 10.1016/j.bmc.2009.09.053. [DOI] [PubMed] [Google Scholar]
- 48.Rosenbluh J, Hayouka Z, Loya S, Levin A, Armon-Omer A, Britan E, et al. Interaction between HIV-1 Rev and integrase proteins: a basis for the development of anti-HIV peptides. J Biol Chem. 2007;282:15743–15753. doi: 10.1074/jbc.M609864200. [DOI] [PubMed] [Google Scholar]
- 49.Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 50.Levin A, Hayouka Z, Friedler A, Loyter A. Overexpression of the HIV-1 Rev promotes death of non-dividing eukaryotic cells. Virus Genes. 2010;40:341–346. doi: 10.1007/s11262-010-0458-7. [DOI] [PubMed] [Google Scholar]
- 51.Ao Z, Jayappa KD, Wang B, Zheng Y, Kung S, Rassart E, et al. Importin {alpha}3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J Virol. 2010;84:8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nitahara-Kasahara Y, Kamata M, Yamamoto T, Zhang X, Miyamoto Y, Muneta K, et al. Novel nuclear import of Vpr promoted by importin alpha is crucial for human immunodeficiency virus type 1 replication in macrophages. J Virol. 2007;81:5284–5293. doi: 10.1128/JVI.01928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, et al. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J Virol. 2010;84:397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thys W, De Houwer S, Demeulemeester J, Taltynov O, De Rijck J, Gijsbers R, et al. Cold Spring Harbor Laboratory: Retroviruses meeting. Cold Spring Harbor Laboratory; 2010. Is transportin-SR2 mediated nuclear import of HIV mediated by integrase or capsid? [Google Scholar]
- 55.Fassati A, Gorlich D, Harrison I, Zaytseva L, Mingot JM. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 2003;22:3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodward CL, Prakobwanakit S, Mosessian S, Chow SA. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J Virol. 2009;83:6522–6533. doi: 10.1128/JVI.02061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 59.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, et al. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cullen BR. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 61.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, et al. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Natl Acad Sci USA. 2007;104:8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pietersz GA, Li W, Apostolopoulos V. A 16-mer peptide (RQIKIWFQNRRMKWKK) from antennapedia preferentially targets the Class I pathway. Vaccine. 2001;19:1397–1405. doi: 10.1016/s0264-410x(00)00373-x. [DOI] [PubMed] [Google Scholar]
- 64.Levin A, Kutznetova L, Kahana R, Rubinstein-Guini M, Stram Y. Highly effective inhibition of Akabane virus replication by siRNA genes. Virus Res. 2006;120:121–127. doi: 10.1016/j.virusres.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Kramer-Hammerle S, Ceccherini-Silberstein F, Bickel C, Wolff H, Vincendeau M, Werner T, et al. Identification of a novel Rev-interacting cellular protein. BMC Cell Biol. 2005;6:20. doi: 10.1186/1471-2121-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Scadden DT, Crumpacker CS. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casabianca A, Gori C, Orlandi C, Forbici F, Federico Perno C, Magnani M. Fast and sensitive quantitative detection of HIV DNA in whole blood leucocytes by SYBR green I real-time PCR assay. Mol Cell Probes. 2007;21:368–378. doi: 10.1016/j.mcp.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]


