Abstract
Catechol-O-methyltransferase (COMT) is a key enzyme for dopamine catabolism and COMT is a candidate gene for human psychiatric disorders. In mouse it is located on chromosome 16 in a large genomic region of extremely low variation among the classical inbred strains, with no confirmed single nucleotide polymorphisms (SNPs) between strains C57BL/6J and DBA/2J within a 600-kB window. We found a B2 SINE in the 3′ untranslated region (UTR) of Comt1 which is present in C57BL/6J (Comt1B2i) and other strains including 129 (multiple sublines), but is not found in DBA/2J (Comt1+) and many other strains including wild-derived Mus domesticus, M. musculus, M. molossinus, M.castaneus and M. spretus. Comt1B2i is absent in strains closely related to C57BL/6, such as C57L and C57BR, indicating that it was polymorphic in the cross that gave rise to these strains. The strain distribution of Comt1B2i indicates a likely origin of the allele in the parental Lathrop stock. A stringent association test, using 670 highly outbred mice (Boulder Heterogeneous Stock), indicates that this insertion allele may be responsible for a difference in behavior related to exploration. Gene expression differences at the mRNA and enzyme activity level (1.7-fold relative to wild type) indicate a mechanism for this behavioral effect. Taken together, these findings show that Comt1B2i (a B2 SINE insertion) results in a relatively modest difference in Comt1 expression and enzyme activity (comparable to the human Val-Met polymorphism) which has a demonstrable behavioral phenotype across a variety of outbred genetic backgrounds.
Keywords: Behavior, comt, genetics, mouse, sine insertion
Catechol-O-methyltransferase (COMT) plays a regulatory role in catecholamine neurotransmission, particularly in the case of dopamine, by facilitating degradation (Tunbridge et al. 2004). Dopamine is known to play a role in reward-seeking behavior, cognition and motor activity (Goldman-Rakic et al. 2000; Schultz 2001; Yang et al. 2003). COMT is therefore an attractive candidate molecule for involvement in these processes.
The most frequently examined human polymorphism in COMT is a Valine to Methionine substitution. It has been ascertained that the Val158Met SNP results in a change in enzyme activity, but not mRNA levels, with Val158 homozygotes having approximately 1.4-fold greater COMT activity than Met158 homozygotes in the prefrontal cortex (Chen et al. 2004). COMT has been associated, with varying degrees of robustness, to a number of disorders, including schizophrenia (Shifman et al. 2002) and obsessive compulsive disorder (Pooley et al. 2007), and is also of interest in research into cognition (Egan et al. 2001; Tunbridge et al. 2006) and aggression (Rujescu et al. 2003).
The homologous mouse gene, previously called Comt, has been recently renamed Comt1. Research in mice has shown a link between Comt1 expression and cognitive (Papaleo et al. 2008) and aggressive phenotypes (Fernandes et al. 2004; Filipenko et al. 2001; Gogos et al. 1998). In Comt1 knockout mice a variety of phenotypic changes have been reported, including increased anxiety (Gogos et al. 1998), improved working memory, set-shifting performance and greater acoustic startle reactivity (Papaleo et al. 2008) and lower weight and greater motor activity (Haasio et al. 2003). An exploratory and habituation phenotype characterized by increased sifting and chewing has also been found in the mice heterozygous for the Comt1 deletion (Babovic et al. 2007). Mice overexpressing Comt1 also display a mild phenotype, being less active in the open field but showing no differences in prepulse inhibition (PPI) of the startle response (Stark et al. 2009).
In mouse, Comt1 is situated on chromosome 16, in an area with very little genetic variation between inbred mouse strains (Yang et al. 2007). However, a Comt1 expression difference in the nucleus accumbens and striatum between strains has been noted, with consistently higher expression in the C57BL/6J when compared to the DBA/2J mouse, except for probe sets at the far 3′ untranslated region (UTR) (Grice et al. 2007; Korostynski et al. 2006). An outbred highly recombinant mouse stock is an optimal way to stringently test mice for a phenotypic effect resulting from genotypic or expression differences (Chia et al. 2005). The Boulder Heterogeneous Stock (HS) mice (McClearn & Hofer 1999) were generated from 8 inbred progenitor strains and now have over 65 generations of accumulated recombination, creating a highly variable genetic background on which to examine phenotype. This allows phenotypic differences between the progenitor strains (including C57BL/6J and DBA/2J) to be associated with genetic loci.
We have identified a polymorphism in Comt1 that mediates the expression difference observed between strains and provides a possible resolution for the conflicting expression data between different probe sets. Additionally, we examine whether this Comt1 polymorphism is associated with a behavioral phenotype using the HS mouse stock.
Materials and methods
Animals
DNAs from 44 different inbred strains of mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA; http://www.jax.org/dnares/). Male C57BL/6J and DBA/2J animals used to prepare hippocampal extracts for COMT1 activity assays were bred in the SPF facility at the Institute of Psychiatry, London, from original stocks obtained from the Jackson Laboratory via Charles River UK. Male, HS (McClearn & Hofer 1999) mice, excluding albinos, were obtained from the Institute for Behavioral Genetics, University of Colorado at Boulder (Boulder, USA) and shipped to the UK in 8 batches (80–100 mice per batch) at the age of approximately 8 weeks. The average age of HS mice at the start of open field testing was 90.24 ± 2.92 days (mean ± SD), and all HS mice were from generations 64–72. DNA was prepared from spleens of male C57BL/6J, DBA/2J and HS mice. RNA was prepared from hippocampi of male C57BL/6J and DBA/2J mice. The hippocampus was chosen as the source for the mRNA for this study as this is a key area of the brain involved in behaviors such as learning and memory, anxiety and aggression (Fernandes et al. 2004). All animal works were licensed under the Animals (Scientific Procedures) Act 1986, reviewed by the ethical review panel of the Institute of Psychiatry and the Home Office inspectorate, and are in accordance with the European Communities Council Directive of 24 November 1986 (86/609/EEC).
Tissue collection
Animals were killed by cervical dislocation and were immediately dissected and tissues snap frozen and stored at −80°C until use.
Primers, sequencing and genotyping
Primers shown in Fig. 1 were designed based on the RefSeq cDNA sequence NM_001111062, which is a composite of partial cDNA sequences of strain C57BL/6 origin, and were used to amplify a region of between 239 and 475 bp. PCR reactions contained 20 ng of template, 0.33 µm of each primer, 200 µm each nucleotide, 50 mm KCl, 0.1% Tween-20, 1.5 mm MgCl2, 35 mm Tris base and 15 mm Tris–HCl in a final volume of 30 µl, using a touchdown protocol with annealing temperature beginning at 55°C and stepping down to 50°C. PCR products were sequenced after cleanup with an Exo-SAP kit (USB Corporation, Staufen, Germany). The B2 insertion was identified using RepeatMasker (http://www.repeatmasker.org).
Figure 1.
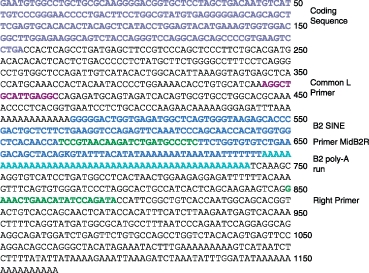
Annotated sequence for the C57BL/6J strain Comt1 gene. Sequence is a composite of RefSeq cDNA sequence NM_001111062 and sequence of the B2 insertion found in this study. Sequencing of additional strains shows identical sequence in those with the insertion, except for some differences in the length of the poly-A run at the end of the B2 element. Primers used for the sequencing and genotyping are displayed in color, as is the position of the B2 SINE insertion in the Comt1B2i allele.
Primer pair 5′-AGGCTGCATTGAGGC-3′ (Common L primer) and 5′-GAAACTGAACATATCCAGATA-3′ (Right primer) were used to assay a panel of inbred strain DNAs and 670 HS DNAs for Comt1B2i by agarose gel electrophoresis. To ensure correct calling of heterozygote genotype we used an additional PCR reaction with primers 5′-AGGCTGCATTGAGGC-3′ (Common L primer) and 5′-TCCGTAACAAGATCTGATGCCCTC-3′ (MidB2R, in B2 sequence) to assay the presence or absence of Comt1B2i.
Gene expression
Profiles of the hippocampi of 265 of the HS male mice were determined using the Affymetrix GeneChip® Mouse Exon 1.0 ST Array (Santa Clara, CA, USA) in experiments which will be fully described elsewhere. Briefly, RNA was extracted using TRIzol reagent (Invitrogen, Paisley, UK) and labeled and hybridized using the Affymetrix WT synthesis and labeling system according to the manufacturer's recommended protocols. The resulting data were normalized and summarized using the RMA sketch method of the Affymetrix power tools and further quality controlled and analyzed using the R packages ‘Affy’ (Gautier et al. 2004) and ‘Exonmap’ (Okoniewski et al. 2007).
3′ RACE
Length of the 3′ UTR of Comt1 mRNA in C57BL/6J mice was determined using the Invitrogen 3′ RACE system for rapid amplification of cDNA ends. Briefly, RNA was extracted using the Qiagen AllPrep DNA/RNA mini kit (Crawley, UK). Using the 3′ RACE kit, cDNA was synthesized according to the manufacturer's instructions. A ‘one-sided’ PCR was performed using 5′-AGGCTGCATTGAGGC-3′ (Common L primer) and a universal amplification primer that binds to the 3′ end of the cDNA. Length of the resulting PCR product was determined using agarose gel electrophoresis. All visible bands were extracted using Qiagen QIAquick gel extraction kit (Crawley, UK) and sequenced.
Enzyme assay
COMT1 enzyme activity was assayed in crude protein extracts as previously described (Tunbridge et al. 2007). Briefly, tissue was thawed on ice and homogenized in 25 mm Tris pH 7.4, 50% v/v glycerol and protease inhibitors (Complete Protease Inhibitor Cocktail Tablets, Roche, Burgess Hill, UK). Fifty micrograms of total protein was incubated at 37°C for 30 min in 100 mm Tris pH 7.4, 5 mm MgCl2, 100 mm catechol and 2 mm dithiothreitol, supplemented with 3.6 µCi per reaction of 3H-S-adenosylmethionine (specific activity: 5–15 Ci/mmol; Perkin Elmer, Waltham, MA, USA). Reactions were stopped with 1 volume of 1 n HCl and tritiated methylated catechol was extracted by mixing thoroughly with Monoflow 1 scintillation fluid (National Diagnostics, Atlanta, GA, USA). Samples were measured using a liquid scintillation counter. Each data point is the mean of four replicates, the individual results of which were highly correlated (R values between 0.955 and 0.992). Specific activity is expressed as counts per minute incorporated in 30 min/50 ug protein with background values (obtained by assaying protein extraction buffer only) subtracted.
Behavior
A comprehensive behavioral battery was conducted on males from a number of inbred strains as well as BXD and HS mice (Galsworthy et al. 2005, 2002; Lad et al. 2007, 2009). This large-scale experiment included 670 HS mice. The battery, which is described in detail in Lad et al. (2009), included eight behavioral tests: activity monitoring in the home-cage (1st and 23rd hour after transfer to a fresh cage); open field; novel object exploration; elevated plus-maze; light/dark box; puzzle box; Morris water maze; tail suspension test.
Statistical analysis
From our battery of eight behavior tests (Lad et al. 2009), we selected 54 measures for association testing with Comt1B2i. For details of the behavioral measures used for association and their selection see Lad et al. (2009). Association was tested by one-way analysis of variance (anova) with genotype as a categorical variable, implemented using the lm() function of R. Multiple testing was addressed using the false discovery rate approach (Storey & Tibshirani 2003).
Results
Novel Comt1 allele
No confirmed SNPs were identified in a survey of the Comt1 exons in 12 strains (A/J, AKR/J, BALB/cJ, C3H/HeJ, C57BL/10J, C57BL/6J, DBA/2J, ISCamEi, ISCamRK, RIIIDmMob, RIIIS/J and PWD/Ph). A length polymorphism of nearly 200 bp was identified in the 3′ UTR of Comt1 between C57BL/6J and DBA/2J, possessing the long and the short alleles, respectively. Sequence analysis of the PCR product from the above 12 inbred strains and two additional strains (NOD/LtJ and NON/LtJ) indicated that the length difference was because of a single insertion, consistent with the C57BL/6J sequence (RefSeq NM_001111062), found to be present in seven of the strains sequenced. RepeatMasker showed that the length difference is because of an insertion of a B2 SINE of family 1t (Jurka et al. 2005; Smit 2005; Smit et al. 1996–2004). The consensus sequence (Smit 2005) lacks 2 G residues at the 5′ end of our sequence and there are further probable discrepancies in the AT-rich 3′ portion. Excluding these regions, BLAT searching of the core 153 bp of the insertion (which deviates from the B2_Mm1t consensus at five positions) identifies a single perfect match which is at positions 136082980–136083132 on Mm 5, which would be a candidate parent for this insertion. By inspection the likely insertion site would be ATTT/A and the target site duplication would consist of a run of 15 As. We have named this allele Comt1B2i. Therefore, the presence of Comt1B2i was surveyed in the strains listed in Fig. 2.
Figure 2.
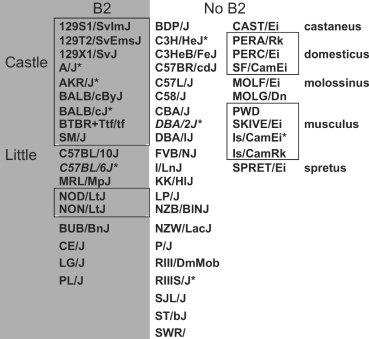
Strain distribution. Presence of Comt1B2i across inbred strains. Boxes group related strains, asterisks denote HS stock progenitors (or in the case of Is/CamEi and RIIIS/J, the probable nearest surviving relative).
Gene expression
Hippocampus consortium data (Overall et al. 2009; http://www.genenetwork.org), produced using the Affymetrix MOE430v2 array, shows very strong cis expression QTL (eQTL) signals for probe sets 1449183_at (LOD 7.3) and 1418701_at (LOD 30), but with opposite directions of effect. Comt1B2i is associated with increased expression for probeset 1449183_at, whereas the Comt1+ allele is associated with increased expression for probe set 1418701_at. A replication in the outbred HS animals, using a different array platform and population, showed an additive effect of genotype on expression. Genotype group means of standardized array signal intensities across the Comt1 gene are shown in Fig. 3.
Figure 3.
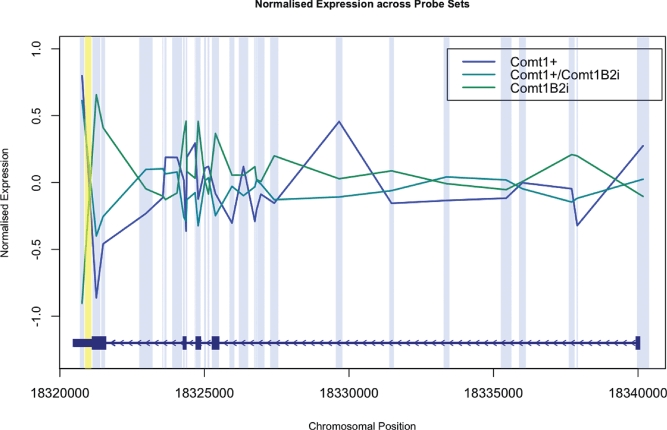
Exon array Comt1 gene expression data. Genotype group means of standardized array signal intensities are plotted by genomic position. Comt1+/+ expression means are plotted in dark blue, Comt1+/B2i in teal and Comt1B2i/B2i in green. The structure of the gene is shown at the bottom, in genomic orientation (the 3′ end is at the left). Probe set positions are marked in light blue. The position of the B2 SINE insertion is shown in yellow. Probe sets flank, but do not span, the insertion site. Data presented are based on NCBI36/mm8.
3′ RACE
PCR of the 3′ UTR of Comt1 mRNA in C57BL/6J produced a band of approximate length 300 bp. Preliminary sequencing of the product indicates that the 3′ UTR includes the B2 SINE. Furthermore, the 3′ UTR ends at the 3′ end of the B2 insertion.
Enzyme activity
COMT1 activity was assayed in hippocampal protein from adult male C57BL/6J and DBA/2J mice. The specific activity of COMT1 in C57BL/6J hippocampus shows a 1.7-fold difference compared to DBA/2J (t = 3.43, df = 14, P = 0.008, two-tailed test, Fig. 4).
Figure 4.
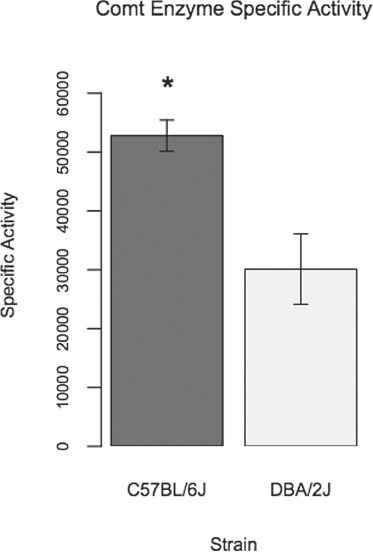
Comt enzyme activity. Means ± standard error of the mean for specific Comt enzyme activity. Enzyme activity is around 1.7-fold higher in C57BL/6J (Comt1B2i) compared to DBA/2J (Comt1+) (t = 3.43, df = 14, P = 8 × 10−3).
Outbred behavior and genotyping
We surveyed Comt1B2i and behavior of 670 male mice from the outbred HS population (McClearn & Hofer 1999). The progenitors of this population are eight inbred strains (A, C57BL/6, BALB/c, AKR, DBA, C3H, Is/Bi and RIII), and all these are represented in Fig. 2 except for the last two, of which the probable nearest surviving relative is shown. Four of the eight progenitor strains are thus known to be Comt1B2i and therefore substantial numbers of each allele should be present in the population. We found 53 homozygotes for Comt1+, 333 Comt1+/Comt1B2i and 284 Comt1B2i homozygotes, giving an allele frequency of 0.672 for Comt1B2i in the whole population.
The novel object exploration test was the only behavioral measure to show a significant association with Comt1B2i (Duration: F2,572 = 8.7, P = 1 × 10−4; Frequency: F2,572 =6.1, P = 2 × 10−3, Fig. 5). Both the duration and the frequency of exploration of the novel object were greater in Comt1+ compared to the Comt1+/Comt1B2i and Comt1B2i genotype groups. The Comt1B2i is therefore dominant, at least with respect to the behavioral phenotype. There were no differences in anxiety measures of the open field test, performed on the previous day in the same arena, or in any of the other behavioral tasks in our battery.
Figure 5.
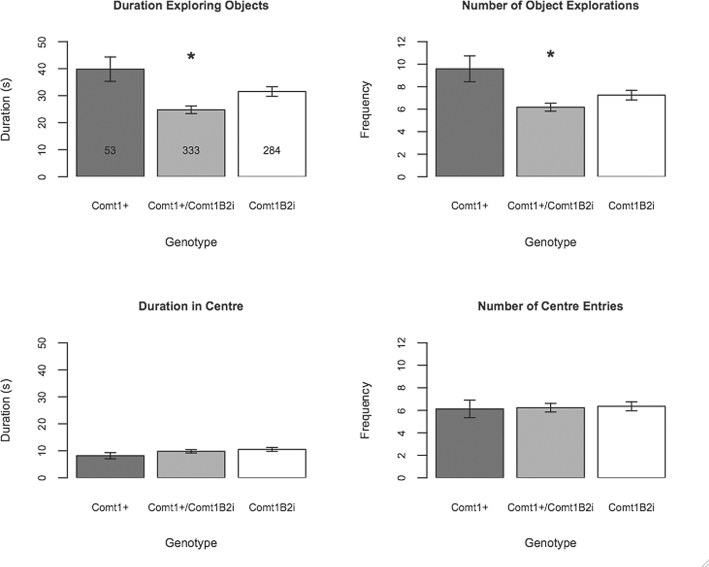
Behavior. Means ± standard error of the mean for measures from the novel object task (NO) and the open field task (OF) in the HS mice grouped by Comt1B2i genotype. Duration and frequency of exploration in the novel object are greater in Comt1+ than the other genotype classes, and least in the Comt1+/Comt1B2i genotype (Duration: F2,572 = 8.7, P = 1 × 10−4; Frequency: F2,572 = 6.1, P = 2 × 10−3). No significant differences are seen in the OF.
Discussion
We have found that a B2 SINE in Comt1 (Comt1B2i) present in some inbred strains but not others is the likely cause of the expression difference between these strains, being the only confirmed variation within the gene between C57BL/6J and DBA/2J. Comt1B2i is associated with an increase in specific enzyme activity, as well as changes in behavior related to exploration. These findings suggest that a modest difference in Comt1 expression levels can have a significant behavioral phenotype, in line with previous findings (Fernandes et al. 2004).
Sequencing indicates that the insertion site and the B2 SINE insert sequence are identical across Comt1B2i strains, except for possible length variation in the flanking poly A runs, suggesting identical origin of Comt1B2i. Comparison of the strain distribution of Comt1B2i with what is known of the breeding history of the inbred strains (Beck et al. 2000) makes it immediately evident that although C57BL/6J is Comt1B2i, several very closely related strains (C57L, C57BR) are not. Therefore, Comt1 must have been polymorphic in the cross between Miss Lathrop's female 57 and male 52 that gave rise to these strains (Beck et al. 2000). Comt1B2i is not present in our sampling of wild-derived Mus domesticus, M. musculus, M. molossinus and M. castaneus strains. Those classical inbred strains that do contain the insertion are almost all known to have ancestry from the Castle and Little stocks, suggesting that Comt1B2i actually arose in the Lathrop stock around the start of the 20th century. The exception is the pair of strains NOD/LtJ and NON/LtJ, which are of ‘Swiss’ origin, without a known connection to Castle's stocks.
The frequency of insertional mutagenesis by SINEs in mouse is unknown, but at least one similar case has been reported in the literature: Alas1 contains a B2 SINE insertion in DBA/2J but not C57BL/6J (Chernova et al. 2008). Research into the most common human SINE, Alu, hypothesizes a retrotransposition rate of 1 new insertion per 20 births (Cordaux & Batzer 2009). Additionally, transposition is much more active in the mouse genome when compared to the human genome, with transposons being found to be responsible for about 10% of spontaneous mutations (Guenet 2005).
Using Affymetrix exon array data, we showed in the hippocampi of the HS that Comt1B2i is associated with high gene expression signal for probes 5′ of the insertion site but low expression for 3′ located probes. The opposite relationship is seen in Comt1+ mice, while Comt1+/Comt1B2i mice have intermediate expression at these loci. Microarray analysis using the Affymetrix MG_U74Av2 microarray with a single Comt1 probe set (98535_at) found variation between eight strains in the hippocampus (Fernandes et al. 2004). C57BL/6J had the highest Comt1 expression and DBA/2J the lowest, and this difference correlated with an aggressive phenotype. Subsequent microarray studies showed similar strain differences in Comt1 expression in the nucleus accumbens (Grice et al. 2007) and striatum (Korostynski et al. 2006). In the latter study it was noted that a probe set located further 3′ in the gene showed a strain difference in the opposite direction. This relationship is also clearly visible in hippocampus across the BXD recombinant inbred panel (Overall et al. 2009; http://www.webQTL.org) which shows strong (Mendelian) cis-genetic effect for the probe set 1418701_at (DBA/2J allele increasing expression) and probe set 1449183_at (C57BL/6J allele increasing expression). One possible explanation for the expression difference is that the B2 insertion may lead to a new polyadenylation site resulting in a modified 3′ UTR. A 3′ RACE conducted on C57BL/6J mRNA shows a transcript ending at the 3′ end of the B2 insertion, 460 bp shorter than the reference sequence (NM_ 001111062). Polyadenylation of the transcript occurs at the polyadenylation signal (AATAAA) contained with the B2 sequence. No evidence was found for a longer 3′ UTR in strains bearing the insertion, suggesting that the shorter 3′ UTR is the dominant transcript. In animals with the insertion, the shorter 3′ end could result in no signal from the furthest out probe sets, accounting for the low signal seen in Comt1B2i mice.
Based on our current data and that of previous studies it is clear that there is a difference in transcript structure and abundance between inbred strains with Comt1+ compared to Comt1B2i. We have showed that this polymorphism is associated with functional consequences (Fig. 4) as hippocampal COMT1 enzyme activity is substantially greater in C57BL/6J (Comt1B2i) than in DBA/2J (Comt1+). This may be mediated to some extent by the presence of a shorter 3′ UTR in Comt1B2i. This difference in enzyme activity is particularly worth noting in comparison to the most studied polymorphism in humans, the Val/Met, which produces a 1.4-fold difference in protein activity with significant behavioral differences. As the difference we found is at a magnitude of around 1.7-fold, we would expect to see similar levels of behavioral differences in an outbred population of mice.
Given the key role played by COMT1 in dopamine metabolism, we tested whether Comt1B2i might have a behavioral effect. We used HS outbred mice to perform a stringent association test, in which the effect of Comt1B2i locus is examined against a large panel of different highly heterozygous genetic backgrounds, generated by over 65 generations of accumulated recombination from eight inbred progenitor strains (Boulder Heterogeneous Stock, McClearn & Hofer 1999). We investigated the behavioral phenotypes of the HS mice using a test battery (Lad et al. 2009) which includes tests of baseline activity, and measures relating to anxiety, depression and cognition. The majority of these measures did not show an effect of Comt1B2i; however, there was an association with novel object exploration. Comt1B2i/B2iand Comt1+/B2i mice spent less time exploring, and made fewer visits to a novel object than do Comt1+/+ mice. Comt1B2i does not seem to alter anxiety levels as no differences were seen in any of the classical anxiety tasks used in the battery (open field, elevated plus-maze or light/dark box). However, we cannot definitively exclude an anxiety effect as the novel object task was performed in a potentially aversive environment, given that mice had only one previous exposure to the open field and may not have fully habituated to the novel arena. Further experiments testing novel object exploration in a familiar (home-cage) environment could be used to address this issue.
Behavioral differences were to be expected in the light of previous research, where several transgenic Comt1 mice have been engineered. A knockout of the gene produces a remarkably mild phenotype in terms of basic behavior (Babovic et al. 2007; Gogos et al. 1998), although associations with cognitive phenotypes are more robust (Papaleo et al. 2008). More recently, strains overexpressing Comt1 have been produced by bacterial artificial chromosome (B AC) transgenesis (Stark et al. 2009) and transgenic overexpression of the higher activity Val allele of the heavily studied Val158Met human polymorphism (Papaleo et al. 2008). In the Stark et al. study, PPI was the main phenotype of interest and no effect of Comt1 overexpression was noted, although these findings are inconsistent with those of Papaleo et al. (2008) who showed a reduction in PPI in COMT-Val-tg overexpressors and an increase in acoustic startle reactivity, but no change in PPI in Comt1 knockout mice, compared with their respective wild-types. Additionally, open field was studied and a small difference in total distance traveled noted. Papaleo et al. also found impairments in several cognitive measures, including attentional set shifting and working memory in COMT-Val-tg overexpressors, compared with wild-type controls, although these mice were created on a mixed genetic background.
Given the results obtained in the Comt1 and COMT-Val-tg transgenic mice, it is perhaps surprising that we found associations only with object exploration. Impaired emotional reactivity was observed in Comt1 knockout mice (Gogos et al. 1998) but this effect was only seen in female mice and the present study used males. However, the behavioral effect we have observed is no doubt just part of the phenotype attributable to Comt1B2i, but without more specific phenotyping of higher cognitive functions and behavioral analysis in both male and female mice, our results should be considered preliminary. Given our previous observation of a correlation across inbred strains between Comt1 expression with intermale aggression, it would be of interest to test these mice in the resident-intruder paradigm. Furthermore, the results of Papaleo et al. (2008), and the associations between cognitive function and COMT in humans, suggest that it would be worthwhile to investigate the phenotype of these mice in detailed cognitive tasks. However, given that cognitive tests are generally labour-intensive, and the high-stress nature of the resident-intruder paradigm, studies of this type require specific testing, rather than as part of a test battery as used here.
Our data taken together provide evidence that the Comt1B2i is itself linked to the changes in object exploration. The association of Comt1B2i with decreased exploration holds across a large panel of outbred genetic backgrounds. If the responsible allele were at another locus, even one quite closely linked to Comt1, it is likely that recombination would have occurred between the causative locus and the Comt1 gene, and that the association with Comt1B2i would therefore have been lost. Furthermore, our survey of the exons of the Comt1 gene showed no other confirmed polymorphisms in a set of strains that represent as far as possible the progenitors of the Comt1 locus, which is in an extensive region of identity-by-descent across the classical inbred strains (Yang et al. 2007). Between C57BL/6J and DBA/2J, there is no known polymorphism within the Comt1 gene.
The molecular mechanism for the altered COMT1 enzyme activity remains unclear. Given the complexity of the microarray results, it is unlikely to be because of a simple difference in mRNA abundance. The longer insertion-bearing transcript is polyadenylated at the 3′ end of the insertion, resulting in a shorter 3′ UTR. This may lead to alternate processing by miRNAs, or may result in an altered ratio of COMT1 protein isoforms. Other instances of insertional mutagenesis by retrotransposition have been associated with physical phenotypes (Duhl et al. 1994; Ho et al. 2004), and the mechanism for these events is not always clear. Further dissection of the effects of this allele, both in terms of behavioral phenotype and molecular biology, will be informative about the normal function of the Comt1 gene.
Acknowledgments
This work was supported by grants from the UK Medical Research Council (MRC G0000170) and the European Commission (NEST 028594) to L.S. and MRC DTA studentship to R.K. C.F. is supported by a Research Councils UK Fellowship. E.M.T. is supported by a Royal Society Research Fellowship.
References
- Babovic D, O'Tuathaigh CM, O'Sullivan GJ, Clifford JJ, Tighe O, Croke DT, Karayiorgou M, Gogos JA, Cotter D, Waddington JL. Exploratory and habituation phenotype of heterozygous and homozygous COMT knockout mice. Behav Brain Res. 2007;183:236–239. doi: 10.1016/j.bbr.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MFW, Fisher EMC. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, Kolachana BS, Hyde TM, Herman MM, Apud J, Egan MF, Kleinman JE, Weinberger DR. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mrna, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75:807–821. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T, Higginson FM, Davies R, Smith AG. B2 SINE retrotransposon causes polymorphic expression of mouse 5-aminolevulinic acid synthase 1 gene. Biochem Biophys Res Commun. 2008;377:515–520. doi: 10.1016/j.bbrc.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Chia R, Achilli F, Festing MF, Fisher EM. The origins and uses of mouse outbred stocks. Nat Genet. 2005;37:1181–1186. doi: 10.1038/ng1665. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl DMJ, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes C, Paya-Cano JL, Sluyter F, D’Souza U, Plomin R, Schalkwyk LC. Hippocampal gene expression profiling across eight mouse inbred strains: towards understanding the molecular basis for behaviour. Eur J Neurosci. 2004;19:2576–2582. doi: 10.1111/j.0953-816X.2004.03358.x. [DOI] [PubMed] [Google Scholar]
- Filipenko ML, Beilina AG, Alekseenko OV, Kudryavtseva NN. Changes in Catechol-O-Methyltransferase gene expression associated with agonistic behavior in male mice. Doklady Biol Sci. 2001;377:125–128. [Google Scholar]
- Galsworthy MJ, Paya-Cano JL, Monleon S, Plomin R. Evidence for general cognitive ability (g) in heterogeneous stock mice and an analysis of potential confounds. Genes Brain Behav. 2002;1:88–95. doi: 10.1034/j.1601-183x.2002.10204.x. [DOI] [PubMed] [Google Scholar]
- Galsworthy MJ, Paya-Cano JL, Liu L, Monleon S, Gregoryan G, Fernandes C, Schalkwyk LC, Plomin R. Assessing reliability, heritability and general cognitive ability in a battery of cognitive tasks for laboratory mice. Behav Genet. 2005;35:675–692. doi: 10.1007/s10519-005-3423-9. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gogos JA, Morgan M, Luine V, Santha M, Ogawa S, Pfaff D, Karayiorgou M. Catechol-O-methyltransferase-deficient mice exhibit sexually dimorphic changes in catecholamine levels and behavior. Proc Natl Acad Sci U S A. 1998;95:9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS, Muly IIIEC, Williams GV. D1 receptors in prefrontal cells and circuits. Brain Res Rev. 2000;31:295–301. doi: 10.1016/s0165-0173(99)00045-4. [DOI] [PubMed] [Google Scholar]
- Grice DE, Reenila I, Mannisto PT, Brooks AI, Smith GG, Golden GT, Buxbaum JD, Berrettini WH. Transcriptional profiling of C57 and DBA strains of mice in the absence and presence of morphine. BMC Genomics. 2007;8:76. doi: 10.1186/1471-2164-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenet JL. The mouse genome. Genome Res. 2005;15:1729–1740. doi: 10.1101/gr.3728305. [DOI] [PubMed] [Google Scholar]
- Haasio K, Huotari M, Nissinen E, Mannisto PT. Tissue histopathology, clinical chemistry and behaviour of adult Comt-gene-disrupted mice. J Appl Toxicol. 2003;23:213–219. doi: 10.1002/jat.909. [DOI] [PubMed] [Google Scholar]
- Ho M, Post CM, Donahue LR, Lidov HGW, Bronson RT, Goolsby H, Watkins SC, Cox GA, Brown JRH. Disruption of muscle membrane and phenotype divergence in two novel mouse models of dysferlin deficiency. Hum Mol Genet. 2004;13:1999–2010. doi: 10.1093/hmg/ddh212. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Korostynski M, Kaminska-Chowaniec D, Piechota M, Przewlocki R. Gene expression profiling in the striatum of inbred mouse strains with distinct opioid-related phenotypes. BMC Genomics. 2006;7:146. doi: 10.1186/1471-2164-7-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad HV, Liu L, Paya-Cano JL, Fernandes C, Schalkwyk LC. Quantitative traits for the tail suspension test: automation, optimization, and BXD RI mapping. Mamm Genome. 2007;18:482–491. doi: 10.1007/s00335-007-9029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad HV, Liu L, Paya-Cano JL, Parsons MJ, Kember R, Fernandes C, Schalkwyk LC. Behavioural battery testing: evaluation and behavioural outcomes in 8 inbred mouse strains. Physiol Behav. 2009;99:301–316. doi: 10.1016/j.physbeh.2009.11.007. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Hofer SM. Genes as gerontological variables: genetically heterogeneous stocks and complex systems. Neurobiol Aging. 1999;20:147–156. doi: 10.1016/s0197-4580(99)00046-9. [DOI] [PubMed] [Google Scholar]
- Okoniewski MJ, Yates T, Dibben S, Miller CJ. An annotation infrastructure for the analysis and interpretation of Affymetrix exon array data. Genome Biol. 2007;8:R79. doi: 10.1186/gb-2007-8-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall RW, Kempermann G, Peirce J, Lu L, Goldowitz D, Gage FH, Goodwin S, Smit AB, Airey DC, Rosen GD, Schalkwyk LC, Sutter TR, Nowakowski RS, Whatley S, Williams RW. Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front Neurogenomics. 2009;1:55. doi: 10.3389/neuro.15.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo F, Crawley JN, Song J, Lipska BK, Pickel J, Weinberger DR, Chen J. Genetic dissection of the role of catechol-O-methyltransferase in cognition and stress reactivity in mice. J Neurosci. 2008;28:8709–8723. doi: 10.1523/JNEUROSCI.2077-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooley EC, Fineberg N, Harrison PJ. The met158 allele of catechol-O-methyltransferase (COMT) is associated with obsessive-compulsive disorder in men: Case-control study and meta-analysis. Mol Psychiatry. 2007;12:556–561. doi: 10.1038/sj.mp.4001951. [DOI] [PubMed] [Google Scholar]
- Rujescu D, Giegling I, Gietl A, Hartman AM, Möller HJ. A functional single nucleotide polymorphism (V158M) in the COMT gene is associated with aggressive personality traits. Biol Psychiatry. 2003;54:34–39. doi: 10.1016/s0006-3223(02)01831-0. [DOI] [PubMed] [Google Scholar]
- Schultz W. Reward signaling by dopamine neurons. Neuroscientist. 2001;7:293–302. doi: 10.1177/107385840100700406. [DOI] [PubMed] [Google Scholar]
- Shifman S, Bronstein M, Sternfeld M, et al. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. B2_Mm1t - a subfamily of SINEs from mouse. 2005. Repbase Update, 11-NOV-2005.
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996. 2004 URL http://www.repeatmasker.org Date accessed 1 June 2010.
- Stark KL, Burt RA, Gogos JA, Karayiorgou M. Analysis of prepulse inhibition in mouse lines overexpressing 22q11.2 orthologues. Int J Neuropsychopharmacol. 2009;12:983–989. doi: 10.1017/S1461145709000492. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbridge EM, Bannerman DM, Sharp T, Harrison PJ. Catechol-O-methyltransferase inhibition improves set-shifting performance and elevates stimulated dopamine release in the rat prefrontal cortex. J Neurosci. 2004;24:5331–5335. doi: 10.1523/JNEUROSCI.1124-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbridge EM, Harrison PJ, Weinberger DR. Catechol-o-Methyltransferase, cognition, and psychosis: Val158Met and beyond. Biol Psychiatry. 2006;60:141–151. doi: 10.1016/j.biopsych.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Tunbridge EM, Weickert CS, Kleinman JE, Herman MM, Chen J, Kolachana BS, Harrison PJ, Weinberger DR. Catechol-o-methyltransferase enzyme activity and protein expression in human prefrontal cortex across the postnatal lifespan. Cerebral Cortex. 2007;17:1206–1212. doi: 10.1093/cercor/bhl032. [DOI] [PubMed] [Google Scholar]
- Yang YK, Chiu NT, Chen CC, Chen M, Yeh TL, Lee IH. Correlation between fine motor activity and striatal dopamine D2 receptor density in patients with schizophrenia and healthy controls. Psychiatry Res Neuroimaging. 2003;123:191–197. doi: 10.1016/s0925-4927(03)00066-0. [DOI] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel De Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]


