Abstract
Staphylococcal nuclease (SNase) catalyzes the hydrolysis of DNA and RNA in a calcium-dependent fashion. We used AFM-based single-molecule force spectroscopy to investigate the mechanical stability of SNase alone and in its complex with an SNase inhibitor, deoxythymidine 3′,5′-bisphosphate. We found that the enzyme unfolds in an all-or-none fashion at ∼26 pN. Upon binding to the inhibitor, the mechanical unfolding forces of the enzyme-inhibitor complex increase to ∼50 pN. This inhibitor-induced increase in the mechanical stability of the enzyme is consistent with the increased thermodynamical stability of the complex over that of SNase. Because of its strong mechanical response to inhibitor binding, SNase, a model protein folding system, offers a unique opportunity for studying the relationship between enzyme mechanics and catalysis.
Introduction
Staphylococcal nuclease (SNase) is produced as an extracellular nuclease by certain strains of Staphylococcus aureus. It digests both DNA and RNA to oligo- and mononucleotides. SNase is a small globular protein composed of 149 amino acids forming three α-helices and five β-strands. Due to the absence of disulfide bridges, its thermodynamical stability, and small molecular weight (16.8 kDa), it has been a model system in protein-folding research (1–4). SNase shows both exo- and endo-5′-phosphodiesterase activities (3–7). The optimum pH range is between 8.6 and 10.3 and the enzyme activity varies inversely with Ca2+ concentration. Regardless of the pH level, a quite high Ca2+ concentration of ∼0.01 M, is typically required. The enzyme attacks phosphodiester bonds yielding a free 5′-hydroxyl group and a 3′-phosphate monoester (4–7). Its best-known inhibitor is pdTp (deoxythymidine 3′, 5′-bisphosphate) (4–11).
Interestingly, the thermodynamical stability of SNase increases after binding pdTp and calcium (4,12), which seem to stabilize the enzyme structure in the key region around the enzyme active site, near the β-barrel (13). Also, Taniuchi et al. (14) proposed that pdTp-induced resistance of staphylococcal nuclease against proteolysis results from its structural rigidification. In this report we hypothesize that (pdTp)-Ca2+ acts like a clamp mechanically bridging enzyme's domains around the active site. Therefore, we anticipate that not only the thermodynamical, but also the mechanical stability of SNase, increases significantly upon binding of this inhibitor. To verify this conjecture we use AFM-based single molecule force spectroscopy (15–27) to directly measure mechanical unfolding forces of SNase alone and in the complex with (pdTp)-Ca2+. Our results demonstrate that the insertion of the nucleotide inhibitor significantly enhances the mechanical stability of the enzyme.
Materials and Methods
Protein engineering and expression
A chimera protein composed of three SNase domains and four I27 domains of titin was constructed following the methods described previously (28). In this (I27-SNase)3-I27 construct, shown schematically in Fig. 1 a, the I27 domains serve as pulling handles and as a force spectroscopy reference for identifying single-molecule recordings (18,24,27–31). Staphylococcal nuclease (residues 1–149) was cloned into the pAFM1-8 vector (kindly provided by Dr. Jane Clarke) at positions 2, 4, and 6. The Strep-Tag II, WSHPQFEK, was synthesized and cloned into the pAFM1-8 plasmid at position 8 and was used for protein purification. The expression plasmids were transformed into Escherichia coli OverExpress C41 cells (Cat. No. 60442; Lucigen, Middleton, WI). The polyprotein (I27-SNase)3-I27 was expressed for 3 h at 37°C in the presence of 0.2 mM IPTG. Proteins were purified with a Strep-Tactin Sepharose column (Cat. No. 2-1202-001; IBA BioTAGnology, Göttingen, Germany) and eluted with Strep-Tag elution buffer (pH 8.0; IBA BioTAGnology) containing 2.5 mM desthiobiotin, 100 mM Tris-Cl, and 150 mM sodium chloride. Proteins were determined to be >95% pure by sodium dodecyl-sulfate polyacrylamide gel electrophoresis analysis.
Figure 1.
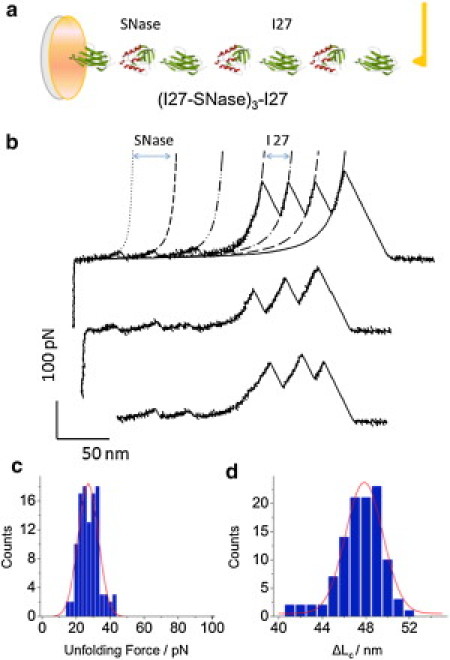
(a) Simplified structure of the chimera protein (I27-SNase)3-I27. (b) Representative unfolding force-extension traces of (I27-SNase)3-I27, the stretching speed of 200 nm/s. The AFM data are fitted with two families of WLC curves: the fits to small force peaks used the contour length increment ΔLc = 46.5 nm and the persistence length p = 0.57 nm (short dashed lines); the fits to the large force peaks used ΔLc = 28 nm and p = 0.36 nm (long dashed lines). (c) Histogram of unfolding forces of SNase domains. (Solid line) Gaussian fit to the data with 〈Funfolding〉 = 26.3 ± 0.5 pN (mean ± SE), the number of force peaks analyzed, n = 108). (d) Histogram of contour length increments ΔLc, attributed to the unfolding of SNase. (Solid line) Gaussian fit with 〈ΔLc〉 = 47.0 ± 0.2 nm (mean ± SE, n =108).
Enzymatic activity of SNase in the (I27-SNase)3-I27 construct
To verify that SNase, when flanked by I27 domains of titin in the (I27-SNase)3-I27 construct, maintains its enzymatic activity, we incubated the chimera protein with 1.5 kbp DNA and performed gel electrophoresis of the reaction products. Fig. 2 shows the results of this experiment. The lack of DNA fragments in lane 2 clearly indicates that the DNA was efficiently digested into fragments that are too small to be resolved by the gel. In lane 3 we show the products when the reaction was carried out in the presence of SNase inhibitor pdTp. It is evident that although SNase retains some activity under these conditions, this activity is significantly reduced in the presence of the inhibitor. Taken together, these results indicate that SNase in the (I27-SNase)3-I27 construct performs its function; therefore, it must be correctly folded.
Figure 2.
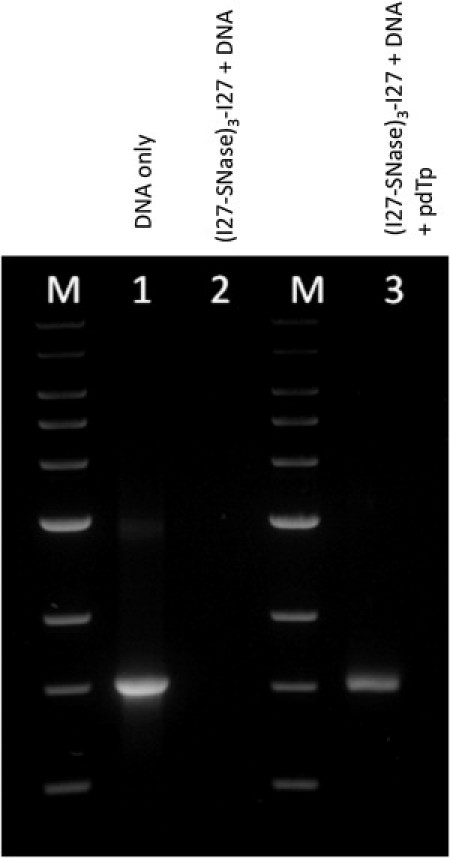
The enzymatic activity of (I27-SNase)3-I27 polyprotein documented by gel electrophoresis. (Lane 1) Agarose gel stained with ethidium bromide shows band of a DNA fragment in 1500 bp. (Lane 2) SNase in the (I27-SNase)3-I27 construct maintains its enzymatic activity against the DNA fragment and digests it completely. (Lane 3) The enzyme activity is partially inhibited by pdTp. (Lanes M) Molecular weight markers (Quick-Load 1 kB DNA ladder; New England BioLabs, Ipswich, MA). DNA was dissolved in the buffer (100 mM Tris-HCI, 10mM CaCI2, pH 7.4).
Sample preparation for AFM experiments
Purified protein solutions composed of 1.5 mg/mL of (I27-SNase)3-I27 were dialyzed in a solution containing 100 mM Tris-HCl buffer at the pH of 7.4. 50 μL of a diluted protein solution (1–10 μg/mL) was incubated on the gold substrate for 30 min at room temperature. For AFM stretching measurements on the inhibited SNase, the protein solution was diluted with the buffer that was supplemented with 200 μM inhibitor, deoxy-thymidine 3′, 5′-diphosphate (pdTp) (Toronto Research Chemicals, North York, Ontario, Canada) and with 10 mM calcium. This composition allowed the formation of SNase-pdTp-Ca+2 complex in the (I27-SNase)3-I27 chimera protein. After incubation on gold substrate, the sample was washed 2–3 times with the same buffer and then used for pulling experiments. This procedure is known to remove weakly-bound proteins but leaves proteins that are nonspecifically adsorbed to a gold surface, allowing AFM measurements to be conducted on them (32).
AFM-based single molecule force spectroscopy
All stretching measurements were carried out on custom-built AFM instruments (32,33) equipped with an AFM detector head (Veeco Instruments, Plainview, NY) and high-resolution piezoelectric stages from Physik Instrumente (Auburn, MA), equipped with capacitive or strain-gauge position sensors (vertical resolution of 0.1 nm). The spring constant of the cantilever was calibrated in solution using the energy equipartition theorem as described (34). Proteins were picked up for stretching measurements by gently touching the substrate with the AFM cantilever tip, allowing proteins to adhere to its gold coating (32). Force-extension measurements were performed with Biolevers cantilevers (OBL from Veeco Instruments; typical spring constant, kc ≈ 6 pN/nm) at the pulling speed of 0.2 nm/ms, in solution, and at room temperature. To determine contour length increments of the protein after forced-domains unfolding, the force peaks in the force-extension curves were fitted with the worm-like chain (WLC) model of polymer elasticity (35).
Results and Discussion
SNase displays a unique mechanical unfolding fingerprint
The thermodynamical stability and folding of wild-type SNase and its numerous mutants alone and in complexes with inhibitors were thoroughly examined by bulk methodologies (7,12,14,36–38). Because thermodynamical stability of proteins does not always correlate with their mechanical stability (39,40), it is difficult to predict whether SNase, not being a protein with a clear mechanical function, is mechanically stable. For new proteins, studying their multiple tandem repeats in a single polyprotein may help us to identify their mechanical fingerprint in the force-extension relationship (force spectrogram). In addition, it is sometimes desirable to be able to compare their force spectrograms with the reference force spectrogram of a protein that already has been mechanically well characterized. Specifically for these reasons we engineered a chimera protein, (I27-SNase)3-I27, in which three SNase modules are flanked by the best mechanically characterized protein so far, the I27 domain of titin (Fig. 1 a).
We checked that SNase is enzymatically active in this construct (Fig. 2, and see Materials and Methods), and therefore must be correctly folded. In Fig. 1 b, we show typical force-extension curves obtained by stretching the chimera protein in an AFM. It is evident that these force extension curves can be divided into two regions. At low protein extensions, two-to-three small, regular force peaks of ∼20–30 pN that are separated by ∼47 nm (based on the WLC fits to the data) are recorded. At higher extensions, much larger force peaks of ∼200 pN, separated by 28 nm, are registered. The latter reports the mechanical unfolding of individual I27 domains of titin (18,24).
Based on the design of the chimera protein, we conclude that the small force peaks must report the mechanical unfolding of individual SNase proteins in the polyprotein. This conclusion is reinforced by the observation that the spacing between these small force peaks is consistent with the unfolded length of SNase. This contour length increment-based argument is based on the following estimation: each of the 149 amino acids of SNase contributes 0.365 nm (21) to the unfolded length of the polyprotein, yielding 54 nm. The relaxed length of the folded SNase (that contributes to the initial length of the polyprotein) is ∼4 nm. Thus, the expected increase in the contour length of SNase upon its forced unfolding, ΔLC, is 54–4 nm = 50 nm—which is close to the WLC-obtained value of 47 nm.
In Fig. 1, c and d, we show the histograms of unfolding forces and of ΔLC, determined by analyzing the small force peaks in Fig. 1 b and other similar force extension curves. From these histograms, the average unfolding force of SNase determined at the stretching speed of 200 nm/s is
the number of force peaks analyzed, n = 108). The average contour length increment is
and the number of force peaks fitted is n = 108. Taken together, these results indicate that SNase displays its unique mechanical unfolding fingerprint that can be easily differentiated from that of I27, which is characterized by significantly higher unfolding forces (200 pN vs. 26 pN) and a significantly shorter contour length increment (28 nm vs. 47 nm).
It is somewhat surprising that SNase, despite its rich β-strand content (five strands), unfolds at low forces that are more typical of proteins composed primarily of α-helices (Spectrin (41–43), repeat protein (23,29,44,45)). A visual inspection of the SNase fold indicates that it may be the C-terminal part of the protein, composed of α-helical segments, that renders it mechanically weak. It seems that the stretching forces applied to the N- and C-termini of SNase may quite easily detach the two C-terminal α-helices, exposing the hydrogen bonds within the β-barrel to unzipping forces, which then easily break these bonds one by one. This scenario, if true, would contrast the shear topology of hydrogen bonds within the I27 domain, which must be broken simultaneously, leading to high unfolding forces (46–48). These speculations need to be substantiated by steered molecular dynamics simulations of SNase, which will clarify structural rearrangements within the protein under force.
pdTp enhances the mechanical stability of SNase
pdTp in the presence of calcium was proposed to stabilize and rigidify the enzyme's structure around the active site (4,12,14). We hypothesized that these interactions between the inhibitor and the enzyme may also increase the enzyme's resistance to mechanical unfolding. To test this conjecture we carried out single-molecule force spectroscopy measurements on individual (I27-SNase)3-I27 molecules in the presence of 200 μM pdTp and 10 mM calcium.
In Fig. 3 b, we show typical force-extension curves obtained at the same 200 nm/s stretching speed that was used during the measurements of SNase in the absence of the inhibitor. As before (compare Fig. 1 b), the force curves display two sets of force peaks. The large force peaks at high extensions can be assigned to the I27 domains and the small force peaks at low extensions are expected to report the unfolding of SNase. This is verified by the analysis of the contour length increment, ΔLC, upon SNase unfolding, which at 46.3 ± 0.2 nm (Fig. 3 d, n =142) remained practically unaffected by the presence of the inhibitor. We note that the I27 force peaks are similar to those obtained in the absence of inhibitor. However, the SNase unfolding force peaks in the presence of the inhibitor increased to 50.7 ± 1.5 pN (Fig. 3 c, number of force peaks analyzed, n = 142) as compared to 26.3 ± 0.5 pN measured without the inhibitor (Fig. 1 c). Thus, the binding of pdTp ad calcium to SNase increases its mechanical stability almost twofold.
Figure 3.
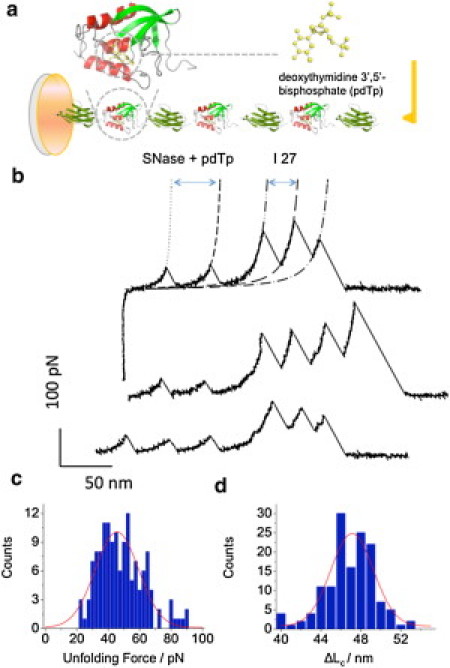
The mechanical stability of staphylococcal nuclease increases in the presence of its inhibitor, pdTp-Ca2+. (a) Schematic of the SNase-pdTp-Ca+2 complex in (I27-SNase)3-I27 polyprotein. (Inset, left) Magnified structure of SNase with pdTp (shown on the right). (b) Representative unfolding force-extension traces of (I27-SNase)3-I27 in the presence of 200 μM pdTp, the stretching speed of 200 nm/s. The AFM data is fitted with two families of WLC curves with ΔLc = 45.5 nm and p = 0.6 nm (short dashed lines), and ΔLc = 28 nm, p = 0.36 nm (long dashed lines). (c) Histogram of unfolding forces of SNase domains in the presence of pdTp. (Solid line) Gaussian fit to the data with Funfolding = 50.7 ± 1.5 pN (mean ± SE, n = 142). (d) Histogram of ΔLc, attributed to the unfolding of SNase (in the presence of pdTp). (Solid line) Gaussian fit with ΔLc = 46.3 ± 0.2 nm (mean ± SE, n =142).
What is the origin of this increased mechanical stability of SNase?
Studies of crystal structure of the staphylococcal nuclease-pdTp complex revealed extensive interactions between the inhibitor, pdTp, calcium, and the active site of the enzyme (Fig. 4 b) (4).
Figure 4.
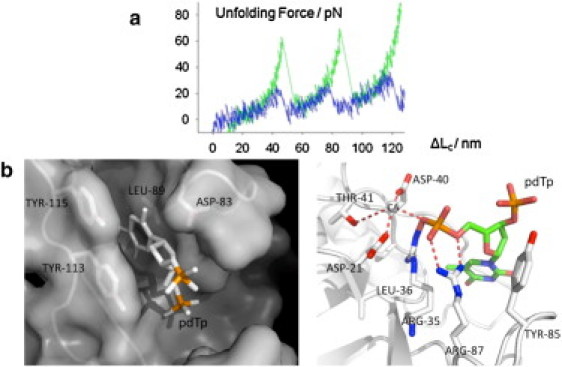
(a) A comparison of typical force-extension unfolding traces of SNase domains without (low force peak) and with (high force peak) pdTp present. (b) Detail of active site in SNase-pdTp-Ca2+ complex from PDB ID 2SNS, drawn by PyMOL (DeLano Scientific, South San Francisco, CA). The surface model shows that the thymine ring of pdTp fits well into a hydrophobic pocket formed by four major residues (left). (Solid dotted lines) Hydrogen bonds or metal interactions between pdTp and Ca2+ and the residues around the active site (right) (4).
First, the thymine ring of pdTp fits well into a hydrophobic pocket, formed by Tyr115, Tyr 113, Leu89, and Asp83.
Second, Arg35, on the one hand, forms a pair of hydrogen bonds with the 5′ phosphate, and on the other hand, forms two additional hydrogen bonds with Leu36 and with Val39.
Third, similarly, Arg87 forms a pair of hydrogen bonds with the 5′ phosphate and also interacts either with Tyr85 or Gly86 or with both.
Fourth, the 5′-phosphoryl-oxygen atom of pdTp directly interacts with the calcium ion (Fig. 4 b).
Fifth, Asp40, Val41, and Asp21 are also involved in the complex with calcium (4). Thus, calcium seems to mediate the interactions between the enzyme and the inhibitor (5,6,8,9). However, the binding of calcium alone does not seem to stabilize SNase mechanically (data not shown). In that regard the effect of calcium is different from the mechanically stabilizing effect of nickel ions which were found to directly clamp engineered bihistidine residues in GB1 protein (17,49).
Sixth, three water molecules are inserted at the interface between pdTp and the active site of the enzyme, two directly as calcium ligands, and one as a bridge between Glu43 and 5′-phosphoryl-oxygen (4).
Taken together, all these observations indicate that the pdTp, specifically the 5′-phosphate, and Ca2+ are precisely positioned at the hydrolytic site where they seem to mechanically reinforce the enzyme. It is believed that the crystal structure of SNase-pdTp-Ca2+ should approximate the actual structure of the enzyme-substrate-Ca2+ complex (4). Therefore, it is possible that the forces between the enzyme hydrolytic site and the inhibitor represent some of the interactions between the enzyme and its substrates. These forces likely participate in straining the substrate into the transition state, to facilitate the hydrolytic reaction. However, these speculations will need to be verified with experimental force spectroscopy studies of SNase in the presence of DNA substrates and will need to involve enzyme mutagenesis and also steered molecular dynamics simulations that may directly capture details of mechanical unfolding pathways of the enzyme with and without the inhibitor or the substrate.
Several single-molecular studies have already examined the effect of ligand binding on the mechanical behavior of proteins including some enzymes (40,50–58). The results are complex and indicate that some proteins are mechanically reinforced by interactions with ligands, whereas for other proteins these interactions do not alter the mechanical stability (51). Moreover, the effect of ligand binding on the mechanical stability of a given protein appears to depend on the pulling direction (51).
Because of its robust mechanical response to ligand binding demonstrated here, SNase, the classical model protein for folding studies, seems also to be a good system for future detail studies of the significance of enzyme mechanics to enzyme catalysis.
Acknowledgments
We thank the members of the Marszalek laboratory: Minkyu Kim, Mahir Rabbi, and Whasil Lee for valuable discussions. We also thank Dr. Chin-Kun Hu and Dr. Ming-Chya Wu for discussions about the protein folding mechanism. C.-C.W. thanks Dr. Chia-Seng Chang and the members of his group for training in AFM and for the possibility to use their equipment at the Academia Sinica.
This work was supported in part by grant No. GM079663 to P.E.M. from the National Institutes of Health (United States), and grant No. NSC97-2752-B-005-001-PAE from the National Science Council (Taiwan) to Y-H.H. C.-C.W.was partially supported by grant No. NSC97-2917-I-005-101 from the National Science Council (Taiwan).
Contributor Information
Yau-Heiu Hsu, Email: yhhsu@nchu.edu.tw.
Piotr E. Marszalek, Email: pemar@duke.edu.
References
- 1.Schechter A.N., Chen R.F., Anfinsen C.B. Kinetics of folding of staphylococcal nuclease. Science. 1970;167:886–887. doi: 10.1126/science.167.3919.886. [DOI] [PubMed] [Google Scholar]
- 2.Chen H.M., You J.L., Tsong T.Y. Kinetic analysis of the acid and the alkaline unfolded states of staphylococcal nuclease. J. Mol. Biol. 1991;220:771–778. doi: 10.1016/0022-2836(91)90116-n. [DOI] [PubMed] [Google Scholar]
- 3.Heins J.N., Suriano J.R., Anfinsen C.B. Characterization of a nuclease produced by Staphylococcus aureus. J. Biol. Chem. 1967;242:1016–1020. [PubMed] [Google Scholar]
- 4.Cotton F.A., Hazen E.E., Jr., Legg M.J. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3′,5′-bisphosphate-calcium ion complex at 1.5-Å resolution. Proc. Natl. Acad. Sci. USA. 1979;76:2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker P.W., Hazen E.E., Jr., Cotton F.A. Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. I. Isolation; physical and enzymatic properties. Mol. Cell. Biochem. 1978;22:67–77. doi: 10.1007/BF00496235. [DOI] [PubMed] [Google Scholar]
- 6.Tucker P.W., Hazen E.E., Jr., Cotton F.A. Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. II. Solution studies of the nucleotide binding site and the effects of nucleotide binding. Mol. Cell. Biochem. 1979;23:3–16. doi: 10.1007/BF00226675. [DOI] [PubMed] [Google Scholar]
- 7.Anfinsen C.B., Cuatrecasas P., Taniuchi H. Academic Press; New York: 1971. The Enzymes. [Google Scholar]
- 8.Cuatrecasas P., Wilchek M., Anfinsen C.B. The action of staphylococcal nuclease on synthetic substrates. Biochemistry. 1969;8:2277–2284. doi: 10.1021/bi00834a007. [DOI] [PubMed] [Google Scholar]
- 9.Dunn B.M., DiBello C., Anfinsen C.B. The pH dependence of the steady state kinetic parameters for staphylococcal nuclease-catalyzed hydrolysis of deoxythymidine-3′-phosphate-5′-p-nitrophenylphosphate in H2O and D2O. J. Biol. Chem. 1973;248:4769–4774. [PubMed] [Google Scholar]
- 10.Mikulski A.J., Sulkowski E., Laskowski M., Sr. Susceptibility of dinucleotides bearing either 3′- or 5′-monophosphate to micrococcal nuclease. J. Biol. Chem. 1969;244:6559–6565. [PubMed] [Google Scholar]
- 11.Sulkowski E., Laskowski M.S. Hydrolysis of ribo- and deoxyribo-dinucleotides by micrococcal nuclease. Biochim. Biophys. Acta. 1970;217:538–540. doi: 10.1016/0005-2787(70)90553-8. [DOI] [PubMed] [Google Scholar]
- 12.Sugawara T., Kuwajima K., Sugai S. Folding of staphylococcal nuclease A studied by equilibrium and kinetic circular dichroism spectra. Biochemistry. 1991;30:2698–2706. doi: 10.1021/bi00224a018. [DOI] [PubMed] [Google Scholar]
- 13.Ishii T., Murayama Y., Sano M. Probing force-induced unfolding intermediates of a single staphylococcal nuclease molecule and the effect of ligand binding. Biochem. Biophys. Res. Commun. 2008;375:586–591. doi: 10.1016/j.bbrc.2008.08.073. [DOI] [PubMed] [Google Scholar]
- 14.Taniuchi H., Morávek L., Anfinsen C.B. Ligand-induced resistance of staphylococcal nuclease and nuclease-T to proteolysis by subtilisin, α-chymotrypsin, and thermolysin. J. Biol. Chem. 1969;244:4600–4606. [PubMed] [Google Scholar]
- 15.Cao Y., Li H. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nat. Mater. 2007;6:109–114. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- 16.Cao Y., Li H. How do chemical denaturants affect the mechanical folding and unfolding of proteins? J. Mol. Biol. 2008;375:316–324. doi: 10.1016/j.jmb.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Cao Y., Yoo T., Li H. Single molecule force spectroscopy reveals engineered metal chelation is a general approach to enhance mechanical stability of proteins. Proc. Natl. Acad. Sci. USA. 2008;105:11152–11157. doi: 10.1073/pnas.0803446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clausen-Schaumann H., Seitz M., Gaub H.E. Force spectroscopy with single bio-molecules. Curr. Opin. Chem. Biol. 2000;4:524–530. doi: 10.1016/s1367-5931(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 20.Dietz H., Berkemeier F., Rief M. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA. 2006;103:12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietz H., Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez J.M., Li H.B. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 23.Lee G., Abdi K., Marszalek P.E. Nanospring behavior of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 24.Marszalek P.E., Lu H., Fernandez J.M. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 25.Oberhauser A.F., Carrión-Vázquez M. Mechanical biochemistry of proteins one molecule at a time. J. Biol. Chem. 2008;283:6617–6621. doi: 10.1074/jbc.R700050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberhauser A.F., Marszalek P.E., Fernandez J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 27.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 28.Steward A., Toca-Herrera J.L., Clarke J. Versatile cloning system for construction of multimeric proteins for use in atomic force microscopy. Protein Sci. 2002;11:2179–2183. doi: 10.1110/ps.0212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serquera D., Lee W., Itzhaki L.S. Mechanical unfolding of an ankyrin repeat protein. Biophys. J. 2010;98:1294–1301. doi: 10.1016/j.bpj.2009.12.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Oberhauser A.F., Fernandez J.M. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. USA. 2000;97:6527–6531. doi: 10.1073/pnas.120048697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams P.M., Fowler S.B., Clarke J. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446–449. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
- 32.Rabbi M., Marszalek P.E. Probing polysaccharide and protein mechanics by AFM. In: Selvin Paul R., Ha Taekjip., editors. Single Molecule Techniques: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. pp. 371–394. [Google Scholar]
- 33.Marszalek P.E., Oberhauser A.F., Fernandez J.M. Polysaccharide elasticity governed by chair-boat transitions of the glucopyranose ring. Nature. 1998;396:661–664. doi: 10.1038/25322. [DOI] [PubMed] [Google Scholar]
- 34.Florin E.L., Rief M., Gaub H.E. Sensing specific molecular-interactions with the atomic-force microscope. Biosens. Bioelectron. 1995;10:895–901. [Google Scholar]
- 35.Bustamante C., Marko J.F., Smith S. Entropic elasticity of λ-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 36.Carra J.H., Privalov P.L. Thermodynamics of denaturation of staphylococcal nuclease mutants: an intermediate state in protein folding. FASEB J. 1996;10:67–74. doi: 10.1096/fasebj.10.1.8566550. [DOI] [PubMed] [Google Scholar]
- 37.Dill K.A., Shortle D. Denatured states of proteins. Annu. Rev. Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 38.Taniuchi H., Anfinsen C.B. An experimental approach to the study of the folding of staphylococcal nuclease. J. Biol. Chem. 1969;244:3864–3875. [PubMed] [Google Scholar]
- 39.Oberhauser A.F., Badilla-Fernandez C., Fernandez J.M. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 40.Junker J.P., Hell K., Rief M. Influence of substrate binding on the mechanical stability of mouse dihydrofolate reductase. Biophys. J. 2005;89:L46–L48. doi: 10.1529/biophysj.105.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law R., Carl P., Discher D.E. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys. J. 2003;84:533–544. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rief M., Pascual J., Gaub H.E. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 43.Randles L.G., Rounsevell R.W.S., Clarke J. Spectrin domains lose cooperativity in forced unfolding. Biophys. J. 2007;92:571–577. doi: 10.1529/biophysj.106.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M., Abdi K., Marszalek P.E. Fast and forceful refolding of stretched α-helical solenoid proteins. Biophys. J. 2010;98:3086–3092. doi: 10.1016/j.bpj.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L., Wetzel S., Fernandez J.M. Stepwise unfolding of ankyrin repeats in a single protein revealed by atomic force microscopy. Biophys. J. 2006;90:L30–L32. doi: 10.1529/biophysj.105.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu H., Schulten K. The key event in force-induced unfolding of Titin's immunoglobulin domains. Biophys. J. 2000;79:51–65. doi: 10.1016/S0006-3495(00)76273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isralewitz B., Gao M., Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 48.Lu H., Schulten K. Steered molecular dynamics simulations of force-induced protein domain unfolding. Proteins. 1999;35:453–463. [PubMed] [Google Scholar]
- 49.Cao Y., Er K.S., Li H. A force-spectroscopy-based single-molecule metal-binding assay. ChemPhysChem. 2009;10:1450–1454. doi: 10.1002/cphc.200900333. [DOI] [PubMed] [Google Scholar]
- 50.Junker J.P., Ziegler F., Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 51.Bertz M., Rief M. Ligand binding mechanics of maltose binding protein. J. Mol. Biol. 2009;393:1097–1105. doi: 10.1016/j.jmb.2009.08.066. [DOI] [PubMed] [Google Scholar]
- 52.Wilcox A.J., Choy J., Matouschek A. Effect of protein structure on mitochondrial import. Proc. Natl. Acad. Sci. USA. 2005;102:15435–15440. doi: 10.1073/pnas.0507324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao Y., Balamurali M.M., Li H. A functional single-molecule binding assay via force spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:15677–15681. doi: 10.1073/pnas.0705367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hann E., Kirkpatrick N., Brockwell D.J. The effect of protein complexation on the mechanical stability of Im9. Biophys. J. 2007;92:L79–L81. doi: 10.1529/biophysj.106.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao Y., Yoo T., Li H. Protein-protein interaction regulates proteins’ mechanical stability. J. Mol. Biol. 2008;378:1132–1141. doi: 10.1016/j.jmb.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 56.Rief M., Junker J.P., Neupert W. Response to the Comment by Ainavarapu et al. Biophys. J. 2006;91:2011–2012. [Google Scholar]
- 57.Ainavarapu S.R., Li L., Fernandez J.M. Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys. J. 2005;89:3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ainavarapu S.R., Li L., Fernandez J.M. Fingerprinting DHFR in single-molecule AFM studies. Biophys. J. 2006;91:2009–2012. doi: 10.1529/biophysj.106.085126. [DOI] [PMC free article] [PubMed] [Google Scholar]


