Abstract
Ectomycorrhiza (ECM) is a mutualistic association between fungi and the roots of the vast majority of trees. These include numerous ecologically and economically relevant species and the participating fungal symbionts are predominantly filamentous basidiomycetes. In natural ecosystems the plant nutrient uptake from soil takes place via the extraradical mycelia of these ECM mycosimbionts as a trade for plant photosyntates. The symbiotic phase in the life cycle of ECM basidiomycetes is the dikaryotic hyphae. Therefore, studies on symbiotic relevant gene functions require the inactivation of both gene copies in these dikaryotic fungi. RNA silencing is a eukaryotic sequence homology-dependent degradation of target RNAs which is believed to have evolved as a protection mechanism against invading nucleic acids. In different eukaryotic organisms, including fungi, the RNA silencing pathway can be artificially triggered to target and degrade gene transcripts of interest, resulting in gene knock-down. Most importantly, RNA silencing can act at the cytosolic level affecting mRNAs originating from several gene copies and different nuclei thus offering an efficient means of altering gene expression in dikaryotic organisms. Therefore, the pHg/pSILBAγ silencing vector was constructed for efficient RNA silencing triggering in the model mycorrhizal fungus Laccaria bicolor. This cloning vector carries the Agaricus bisporus gpdII-promoter, two multiple cloning sites separated by a L. bicolor nitrate reductase intron and the Aspergillus nidulans trpC terminator. pSILBAγ allows an easy two-step PCR-cloning of hairpin sequences to be expressed in basidiomycetes. With one further cloning step into pHg, a pCAMBIA1300-based binary vector carrying a hygromycin resistance cassette, makes the pHg/pSILBAγ plasmid compatible with Agrobacterium-mediated transformation. The pHg/pSILBAγ-system results in predominantly single integrations of RNA silencing triggering T-DNAs in the fungal genome and the integration sites of the transgenes can be resolved by plasmid rescue. Besides the optimized use in L. bicolor, general consideration was taken to build a vector system with maximum compatibility with other homobasidiomycetes and different transformation techniques.
Key words: RNA silencing, RNAi, gene knock-down, basidiomycetes, laccaria, mycorrhiza, ectomycorhiza, symbiosis
The ectomycorrhizal (ECM) symbiosis is an ancient mutualistic association between fungi, mostly homobasidiomycetes, and the roots of the vast majority of tree species of boreal and temperate zones. These include economically important trees such as pines, spruces, birches and poplars. In natural ecosystems the plant nutrient uptake (nitrogen, phosphorus and several micronutrients) from soil occurs via the extraradical mycelia of these symbiotic fungi as an exchange for plant photosyntates. This association also improves water acquisition, heavy metal tolerance and pathogen resistance of the host trees.
Despite the high ecological and economic importance of ECM the current comprehension of host-mycosymbiont recognition, establishment of dual-organs and their functions is still rather poor. During the last ten years the exploitation of novel molecular technologies such as EST-libraries, and cDNA micro- and macro arrays has dramatically increased our understanding of the genetic regulation underlying the ECM interaction. Most importantly, the decision of the Department of Energy Joint Genome Institute (JGI) to sequence the genomes of micobionts of the first genome-sequenced tree species, poplar, took mycorrhizal research to the genomic era. As a result, the full genomic sequence of the basidiomycete ECM fungus Laccaria bicolor monokaryotic strain S238N-H82 was resolved in collaboration between JGI and the Laccaria Genome Consortium (http://genome.jgi-psf.org/Lacbi1/Lacbi1.home.html). The Laccaria genome sequence1 is currently being used for genome-wide expression profiles at different stages of the ECM development. This approach is generating new and highly valuable data on symbiotic plant-fungal interactions. However, the genomic ECM research faces serious technical obstacles. Further studies for resolving the biological relevance of the identified symbiosis regulated Laccaria genes depend on the availability of reverse genetic tools functional in this fungus. These tools include, firstly, an efficient genetic transformation technique and, secondly, a methodology for targeted modification of gene expression in dikaryotic mycelium, the phase of the fungal lifecycle engaged in the symbiotic interaction.
Homobasidiomycetes are rather recalcitrant to genetic modifications. This is partly due to the fact that traditional fungal transformation methods require formation and regeneration of protoplast, steps which are especially challenging with this group of filamentous fungi. On the other hand, targeted inactivation of genes is difficult to accomplish due to very low homologous recombination (HR) rates in these species. Also the dikaryotic nature of the symbiotic mycelium creates a special challenge for generating gene-null mutants of ECM fungi. To overcome these obstacles we have established a high throughput Agrobacterium-mediated transformation method (AMT) based on hygromycin B resistance for Laccaria dikaryotic and monokaryotic strains.2 Moreover, Laccaria possesses a complete set of genes known to be needed for RNA silencing in eukaryotic cells. We have demonstrated that the post-transcriptional RNA silencing pathway is functional in L. bicolor and that it can be triggered via AMT. Targeted gene knockdown in dikaryotic mycelium was also shown to produce functional phenotypes altered in the symbiotic capacity confirming that RNA silencing is a powerful novel way to study symbiosis-regulated genes.3 These studies have now initiated the RNA silencing era in mycorrhizal research, a field that has been hindered by the lack of proper genetic tools.
However, the efficient use of RNA silencing for studying Laccaria depends on access to a universal RNA silencing/transformation vector compatible with AMT. Such a tool was, until now, not available to basidiomycete research. Therefore, our aim was to develop a multiuse cloning vector adapted to both gene expression and RNA silencing studies in the fungus. Besides the optimized use in Laccaria, general consideration was taken to build a vector system with maximum compatibility with other homobasidiomycetes and different transformation methodologies as well. The requirements for such vector system were: (a) an easy to clone hpRNA expression cassette under a fungal promoter widely recognized by homobasidiomycetes and with an intronic spacer for RNA silencing studies, (b) the possibility to use the same RNA silencing cassette also for transgene cloning and gene expression studies, (c) a hygromycin B resistance cassette under a widely recognized fungal promoter for transformant selection, (d) compatibility with AMT, (e) a plasmid rescue-motif for easy transgene integration site recovery and analysis, (f) the possibility to use the silencing triggering/gene expression cassette without this plasmid rescue component, (g) the possibility to launch simultaneous silencing of two target genes from two separate silencing cassettes, and (h) the possible compatibility of the system with other plasmid-based transformation methods.
The silencing/transformation binary vector system pHg/pSILBAγ fulfilling these requirements was designed and its functionality was evaluated in L. bicolor by successful silencing of nitrate reductase (Lbnr) (protein ID 254066) in the dikaryotic strain S238N and inositol-1,4,5, triphosphate 5-phosphatase gene (protein ID 306121) in the monokaryotic strain S238N-H82. Two other plasmid variants (pSILBA and pSILBAa) their intronic sequences and the silencing cassette orientations were also tested for their silencing triggering capacity on Lbnr-target gene. The performance of pSILBAγ in producing strongly silenced strains was found superior and this effect was traced back to methylation-free promoter sequences in the given construct most likely allowing the maximum silencing trigger production from it.4
The pHg/pSILBAγ-vector system is based on separating the construction of the RNA silencing trigger cassette in pSILBAγ plasmid (SIL: silencing), BA: basidiomycetes and the fungal selection cassette for hygromycin B resistance in the Agrobacterium binary vector pHg (Fig. 1). This separation was done in order to allow easy cloning steps and manipulation of the smaller size plasmid pSILBAγ and to maintain the maximum number of unique restriction sites for inverted repeated sequence cloning around the intronic sequence of Lbnr. Joining these two plasmids together creates a silencing/AMT transformation vector of predicted T-DNA organization with a plasmid rescue motif for T-DNA RB-rescue (with BamHI independently of the sequences cloned by each researcher) under ampicilline resistance in E. coli. The rescued T-DNA-gDNA junctions can be sequenced with universal primers M13/pUC-reverse (−26) 17mer or with T3 promoter/pUC (Fig. 2).
Figure 1.
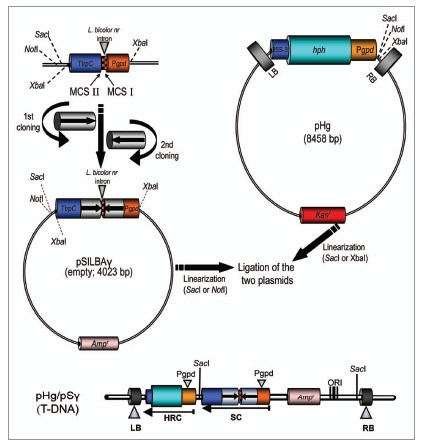
The pHg/pSILBAγ vector system. The ihpRNA expression cassette is constructed in pSILBAγ under A. bisporus gpdII promoter by two directed cloning steps. Laccaria Lbnr intron forms the spacer for the inverted repeats. The hygromycin B resistance cassette is located in the T-DNA of the binary vector pHg. Joining of pSILBAγ and pHg creates the AMT/silencing/rescue-vector with a predicted T-DNA structure.
Figure 2.
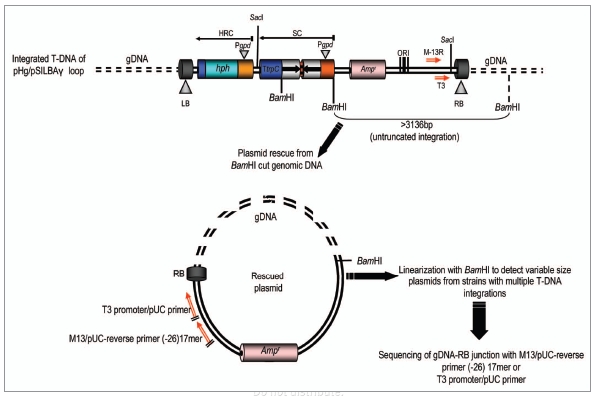
Plasmid rescue of genomic T-DNA integration sites from pHg/pSILBAγ-transformed Laccaria strains by RB rescue with BamHI l and ampicilline selection in E. coli. Untruncated integrations generate rescue plasmids of a minimal size of ∼3.1 kb and linearizable with BamHI. Fungal genomic sequences can be resolved by sequencing with the universal primers T3 promoter/pUC or M13/pUC-reverse (−26) 17 mer. HRC, hygromycin B resistance cassette; SC, silencing cassette.
The PgpdII from Agaricus bisporus5 is widely recognized in homobasidiomycetes. This promoter, besides A. bisporus and L. bicolor, has been shown to be functional at least in Suillus bovinus, Hebeloma cylindsrosporum, Coprinus cinereus, Hypholoma sublateritium and Moniliophthora perniciosa.6–10 Since the tryptophan synthetase terminator of Aspergillus nidulans (TtrpC) is present in the cloning cassette of pSILBA., it is suited not only for ihpRNA triggering but also for transgene expression studies in basidiomycetes with the possibility to incorporate an intronic sequence before or after the gene coding region of interest (Fig. 3C). An intronic sequence is often needed for efficient transgene expression in homobasidiomycetes.8,11–14
Figure 3.
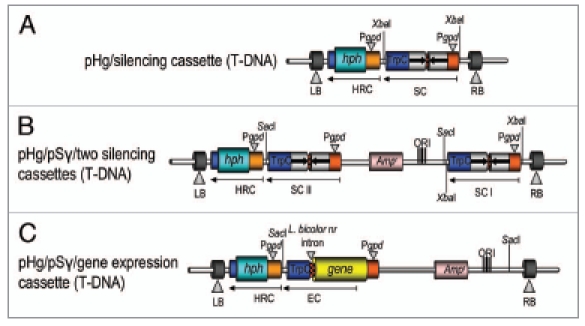
Different uses of the pHg/pSILBAγ vector system. The figure represents the T-DNAs of binary vectors for (A) ihpRNA triggering without the plasmid rescue motif; (B) simultaneous silencing of two different genes or boosting ihpRNA triggering of a single target; (C) the use in transgene expression studies with an intronic sequence included into the expression cassette. HRC, hygromycin B resistance cassette; SC, silencing cassette; EC, expression cassette.
For RNA silencing the PCR-amplified target gene sequence repeats are directionally cloned into unique restriction sequences in the multiple cloning sites (MCS) I and II around the Laccaria intronic spacer in pSILBAγ (Fig. 1). Afterwards, the plasmid is linearized with SacI (or if a SacI site is present in the inverted repeated sequences, NotI can be used instead). The binary vector pHg has a hygromycin B resistance cassette under the same A. bisporus gpdII promoter. This plasmid also carries a unique SacI site in its T-DNA for linearization. The pSILBAγ with the inverted repeated sequence can be cloned as a complete plasmid into the T-DNA of pHg and the joined plasmids are selected under ampicilline and kanamycin in E. coli. Due to certain incompatibility between pHg and pSILBA plasmids only one ligation orientation is viable producing T-DNAs with a predicted structure (Fig. 1). Constructing a RNA silencing/AMT vector can be accomplished by using a minimum of three PCR primers (one blunt end and two sticky end sites) and three ligation reactions. The pSILBA-plasmids tolerate well inverted repeated arms of 700 bp while repeats of over 900 bp were noticed to generate some structural instability and are thus not recommended.
Alternatively, if the rescue motif of the pSILBA backbone is not desired, the ihpRNA expression cassette (or a transgene expression cassette) can be excised from pSILBAγ with XbaI and cloned into the unique XbaI site available in the pHg T-DNA (Fig. 3A). Also the introduction of two separate RNAi/gene expression cassettes of interests into pHg is possible by first cloning the XbaI-liberated cassette and afterwards the other cassette as a full pSILBAγ-plasmid using the SacI site of pHg (Fig. 3B). The RNA silencing efficiency is linked to the relative abundance of both the target mRNA and the silencing trigger in cells and silencing of rare transcripts can thus be expected to be more difficult than the abundant ones. Therefore this double silencing cassette approach could also be used for boosting the silencing triggering of a single target gene. The simultaneous double cassette RNAi triggering of two separate target genes or boosting silencing of a single gene have not been tested yet in Laccaria. However, the T-DNA of pHg seems to tolerate the presence of two inverted repeated sequence cassettes suggesting that such constructs would not cause technical problems in AMT of the fungus.
Currently the pHg/pSILBAγ vector system is successfully being used in launching RNA silencing of several mycorrhiza-regulated Laccaria genes. This tool, in combination with genomics and genome-wide transcriptomics, and with cell biology and biochemistry/molecular biology methods, will certainly allow the study of symbiosis-regulated fungal genes and reveal their true functions in ectomycorrhiza.
Acknowledgements
The Laccaria genome sequence data were produced by the US Department of Energy Joint Genome Institute www.jgi.doe.gov. This work was supported by Universidad Nacional de Quilmes, CONICET and Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT).
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/12385
References
- 1.Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 2.Kemppainen M, Circosta A, Tagu D, Martin F, Pardo AG. Agrobacterium-mediated transformation of the ectomycorrhizal symbiont Laccaria bicolor S238N. Mycorrhiza. 2005;16:19–22. doi: 10.1007/s00572-005-0008-7. [DOI] [PubMed] [Google Scholar]
- 3.Kemppainen M, Duplessis S, Martin F, Pardo AG. RNA silencing in the model mycorrhizal fungus Laccaria bicolor. Gene knock-down of nitrate reductase results in inhibition of symbiosis with Populus. Environ Microbiol. 2009;11:1878–1896. doi: 10.1111/j.1462-2920.2009.01912.x. [DOI] [PubMed] [Google Scholar]
- 4.Kemppainen MJ, Pardo AG. pHg/pSILBAγ vector system for efficient gene silencing in homobasidiomycetes: optimization of ihpRNA-triggering in the mycorrhizal fungus Laccaria bicolor. Microb Biotechnol. 2010;3:178–200. doi: 10.1111/j.1751-7915.2009.00122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Stone M, Schlagnhaufer C, Romaine CP. A fruiting body tissue method for efficient Agrobacterium-mediated transformation of Agaricus bisporus. Appl Environ Microbiol. 2000;66:4510–4513. doi: 10.1128/aem.66.10.4510-4513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanif M, Pardo AG, Gorfer M, Raudaskoski M. T-DNA transfer and integration in the ectomycorrhizal fungus Suillus bovinus using hygromycin B as a selectable marker. Curr Genet. 2002;41:183–188. doi: 10.1007/s00294-002-0297-8. [DOI] [PubMed] [Google Scholar]
- 7.Combier JP, Melayah D, Raffier C, Gay G, Marmeisse R. Agrobacterium tumefaciens-mediated transformation as a tool for insertional matagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. FEMS Micorbiol Lett. 2003;220:141–148. doi: 10.1016/S0378-1097(03)00089-2. [DOI] [PubMed] [Google Scholar]
- 8.Burns C, Gregory KE, Kirby M, Cheung MK, Riquelme M, Elliott TJ, et al. Efficient GFP expression in the mushroom Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol. 2005;42:191–199. doi: 10.1016/j.fgb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Godio R, Fouces R, Guniña E, Martin J. Agrobacterium tumefaciens-mediated transformation of an antitumor clavaric acid-producing basidiomycete Hypholoma sublateritium. Curr Genet. 2004;45:287–94. doi: 10.1007/s00294-004-0533-5. [DOI] [PubMed] [Google Scholar]
- 10.Fagundes Lopes FJ, Marisa Vieira de Queiroz M, Oliveira Lima J, Oliveira Silva VA, Fernandes de Araújo E. Restriction enzyme improves the efficiency of genetic transformations in Moniliophthora perniciosa, the causal agent of witches broom disease in Theobroma cacao. Braz Arch Biol Technol. 2008;51:27–34. [Google Scholar]
- 11.Lugones LG, Scholtmeijer K, Klootwijk R, Wessels JGH. Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol Microbiol. 1999;32:681–689. doi: 10.1046/j.1365-2958.1999.01373.x. [DOI] [PubMed] [Google Scholar]
- 12.Ma B, Mayweld MB, Gold MH. The green fluorescent protein gene functions as a reporter of gene expression in Phanerochaete chrysosporium. Appl Environ Microbiol. 2001;67:948–955. doi: 10.1128/AEM.67.2.948-955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholtmeijer K, Wösten HAB, Springer J, Wessels JGH. Effect of introns and AT-rich sequences on expression of the bacterial hygromycin B resistance gene in the basidiomycete Schizophyllum commune. Appl Environ Microbiol. 2001;67:481–483. doi: 10.1128/AEM.67.1.481-483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki T, Okajima Y, Kawashima H, Tsukamoto A, Sugiera J, Shishido K. Intron-dependent accumulation of mRNA in Coriolus hirsutus of lignin peroxidase gene the product of which is involved in conversion/degradation of polychlorinated aromatic hydrocarbons. Biosci Biotechnol Biochem. 2006;70:1293–1299. doi: 10.1271/bbb.50471. [DOI] [PubMed] [Google Scholar]


