Abstract
Increasingly lignocellulosic biomass hydrolysates are used as the feedstock for industrial fermentations. These biomass hydrolysates consist of complex mixtures of different fermentable sugars, but also contain inhibitors and salts that affect the performance of the product-generating microbes. The performance of six industrially relevant microorganisms, i.e., two bacteria (Escherichia coli and Corynebacterium glutamicum), two yeasts (Saccharomyces cerevisiae and Pichia stipitis) and two fungi (Aspergillus niger and Trichoderma reesei) were compared for their ability to utilize and grow on different feedstock hydrolysates (corn stover, wheat straw, sugar cane bagasse and willow wood). Moreover, the ability of the selected hosts to utilize waste glycerol from the biodiesel industry was evaluated. P. stipitis and A. niger were found to be the most versatile and C. glutamicum, and S. cerevisiae were shown to be the least adapted to renewable feedstocks. Clear differences in the utilization of the more abundant carbon sources in these feedstocks were observed between the different species. Moreover, in a species-specific way the production of various metabolites, in particular polyols, alcohols and organic acids was observed during fermentation. Based on the results obtained we conclude that a substrate-oriented instead of the more commonly used product oriented approach towards the selection of a microbial production host will avoid the requirement for extensive metabolic engineering. Instead of introducing multiple substrate utilization and detoxification routes to efficiently utilize lignocellulosic hydrolysates only one biosynthesis route forming the product of interest has to be engineered.
Key words: second generation feedstock, bioprocessing, filamentous fungi, industrial biotechnology
The utilization of second-generation fermentation feedstocks in industrial biotechnology is challenging in several ways. Given an efficient hydrolysis—chemical or enzymatic—of lignocellulosic biomass, the resulting hydrolysate contains mixtures of pentoses and hexoses, with high levels of other C-sources and inhibitors.1 To this end, selection of industrial production hosts was primarily based on a microorganism's product producing and not on its substrate utilization capacities. In recent years, a strong bias has been emerging towards industrial production hosts, which are either known to produce large amounts of a certain product or can easily be engineered to achieve this goal. Moreover, there is a preference for microorganisms that are well-characterized—physiologically as well as genetically—and therefore have the potential to become microbial production platforms. The most well known example of this development is illustrated by efforts to enable yeast Saccharomyces cerevisiae to utilize xylose, arabinose2 and cellulose.3
The selection of a product-generating microbe based on substrate utilization potential therefore becomes increasingly important. In a recent study,4 we used this substrate-based selection approach by comparing a selection of industrial production strains in relation to second-generation feedstock metabolism. We selected wild-type strains of two bacteria, two yeast and two filamentous fungi species to compare their native abilities and general suitability for the utilization of second-generation feedstocks. The six specific strains were selected on the basis of their industrial relevance as producers of ethanol, organic and amino acid as well as proteins.5–8
Hydrolysates—either through acid hydrolysis (AH) or through enzymatic hydrolysis (EH)—of sugar cane bagasse, wheat straw, corn stover as well as willow wood were prepared and subsequently diluted to 15 g glucose/l in the final synthetic minimal medium.4 The concentration of crude glycerol in the final synthetic minimal medium was 40 g/l. Samples of cultures were taken for all phases of the fermentations and analyzed for residual glucose, xylose, arabinose or glycerol, as well as biomass. In addition, the following compounds were also monitored: Ethanol, hydroxymethyl furfural, furfural, acetic, butyric, citric, lactic, oxalic acid. Analytical methods used for this analysis are described by Rumbold et al.4
Results are presented for each microorganism on lignocellulose in Figures 1–6. Only P. stipitis, A. niger and T. reesei were able to grow on crude glycerol (Fig. 7). No results are shown for organisms that did not grow on a particular substrate.
Figure 1.

Substrate utilization and product production performance of P. stipitis on sugar cane bagasse (AH), wheat straw (AH), corn stover (AH), glycerol, wheat straw (EH), sugar cane bagasse (EH) and willow wood (AH).
Figure 6.
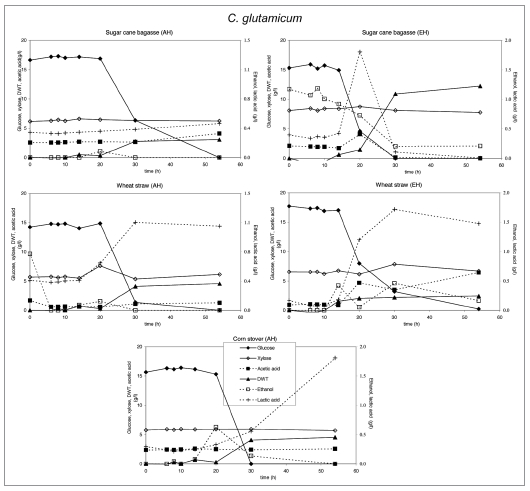
Substrate utilization and product production performance of C. glutamicum on sugar cane bagasse (AH), wheat straw (AH), corn stover (AH), wheat straw (EH), sugar cane bagasse (EH).
Figure 7.
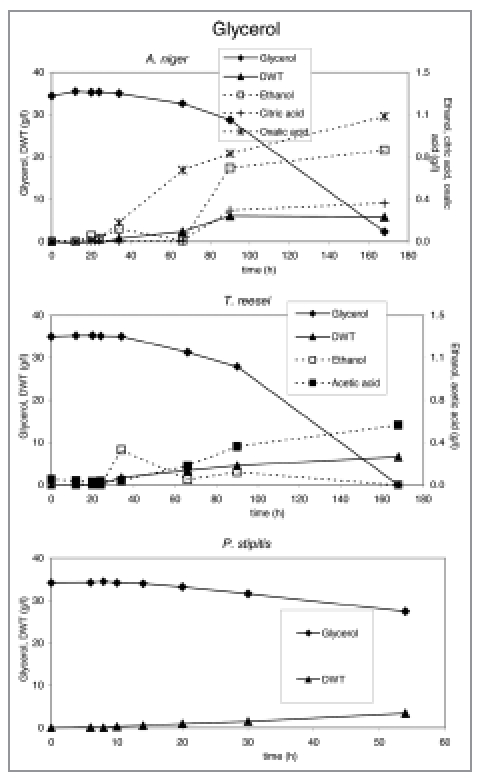
Substrate utilization and product production performance of P. stipitis, A. niger and T. reesei on crude glycerol.
As expected all strains consumed glucose efficiently. The xylose concentration in the feedstocks was between 5 and 9 g/l and its consumption was observed with A. niger, T. reesei, P. stipitis and E. coli, the latter with exceptions.
Crude glycerol at a level of 35 g/l in the medium was utilized by all strains except E. coli and C. glutamicum. S. cerevisiae. P. stipitis grew slowly on glycerol and none of the products were detected (Fig. 7). Interestingly, P. stipitis (Fig. 4) and S. cerevisiae (Fig. 3) produced up to 2 g/l glycerol on wheat straw (EH). T. reesei (Fig. 6) and A. niger (Fig. 5) produced up 1 and 1.5 g/l glycerol, respectively on corn stover (AH). All fungi produced lower concentrations of glycerol on the other lignocellulosic feedstocks. Bacteria did not produce glycerol on lignocellulose.
Figure 4.
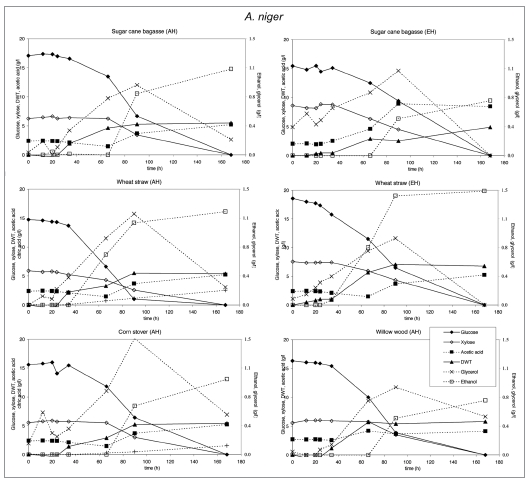
Substrate utilization and product production performance of A. niger on sugar cane bagasse (AH), wheat straw (AH), corn stover (AH), glycerol, wheat straw (EH), sugar cane bagasse (EH) and willow wood (AH).
Figure 3.
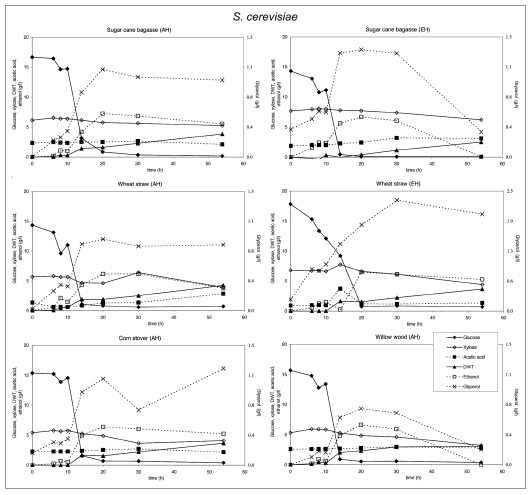
Substrate utilization and product production performance of S. cerevisiae on sugar cane bagasse (AH), wheat straw (AH), corn straw (AH), sugar cane bagasse (EH), wheat straw (EH) and willow wood (AH).
Figure 5.
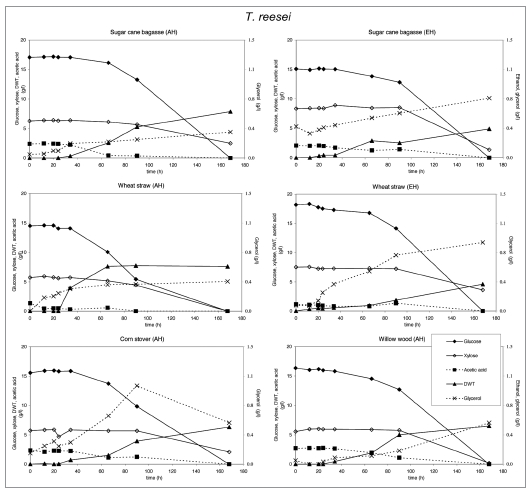
Substrate utilization and product production performance of T. reesei on sugar cane bagasse (AH), wheat straw (AH), corn stover (AH), glycerol, wheat straw (EH), sugar cane bagasse (EH) and willow wood (AH).
As was shown in earlier experiments4 acid hydrolysis of raw lignocellulose-containing substrates leads to the formation by-products like 5-hydroxymethyl-2-furaldehyde (HMF) and 2-furaldehyde (furfural). These compounds act as inhibitors in fermentation processes for all host organisms included in our study.4
In order to analyze whether these inhibitory compounds may be used as substrate, shake flask experiments were carried out with A. niger using furfural or HMF as the sole carbon sources. No significant growth was observed suggesting the absence of an efficient catabolic pathway for these compounds in Aspergillus. To determine if these compounds are at all metabolized, samples derived from a controlled fermentation of A. niger using acid hydrolysed corn stover were analyzed for HMF and furfural and their respective acid and alcohol derivatives were measured as described by Wierckx et al.9 (Fig. 8). As shown, HMF and furfural as present in the acid-treated cornstover were quickly converted into their alcohol derivatives. Subsequently conversion to the acid derivatives occurred. These latter compounds were not utilized further. From these results we concluded that A. niger was able to rapidly detoxify these compounds, similar as is described for a fungal strain selected for bioabatement.10
Figure 8.
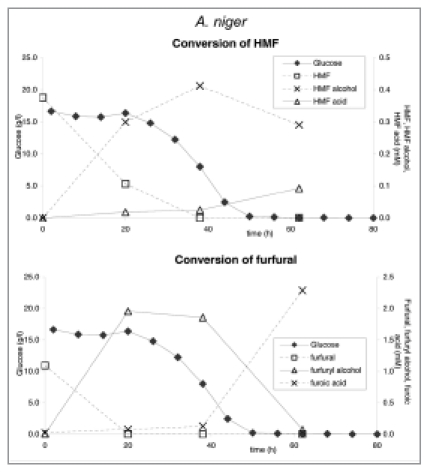
Conversion of HMF and furfural in hydrolyzed corn stover fermentations of A. niger strain N402 at pH 4.5.
In general, yeasts and bacteria grow faster than fungi, but xylose is not utilized by S. cerevisiae and C. glutamicum. A. niger, T. reesei and P. stipitis start fermenting xylose after glucose is more or less consumed. E. coli does not utilize xylose from acid hydrolyzed wheat straw and enzymatically hydrolyzed sugar cane bagasse.
The production and level of metabolites as well as biomass did not vary strongly on different biomass feedstocks, except for glycerol. P. stipitis and S. cerevisiae mainly produced ethanol and glycerol, P. stipitis produced more biomass than the other species.
E. coli produced acetic and lactic acid as well as some ethanol, whereas biomass levels were low in those cases were xylose was not consumed. C. glutamicum produced lactic acid and low biomass levels, except on enzymatically hydrolyzed sugar cane bagasse, where significantly more biomass was produced after consumption of lactic acid. Growth inhibition in both bacteria was caused by the rapid acidification of the medium after glucose consumption.
T. reesei produced low levels of glycerol. However on glycerol it produced acetic acid. A. niger produced the same amount of biomass on all feedstocks and consumed most of the glycerol formed after depletion of glucose. Acetic acid and ethanol was produced on lignocellulosic feedstocks, citric acid only on acid hydrolyzed corn stover and wheat straw.
All industrial production hosts were able to grow on hydrolysates of agricultural waste or waste glycerol, with a few exceptions. In general, the method of feedstock pre-treatment has only little effect on the performance of the microbial production host.
Second-generation feedstocks have been identified as the most important feedstocks for the production of bulk chemicals in industrial biotechnology. Our present approach was to evaluate microorganisms for their suitability for industrial biotechnology using second-generation feedstocks. If an industrial production strain can not or only partly utilize a substrate, or is strongly inhibited by by-products that result from the pretreatment process, the effort of engineering such an organism to adapt to this kind of substrate is more complex. We therefore recommend concentrating metabolic engineering efforts on already adapted organism in order to improve the product production capacities.
Figure 2.
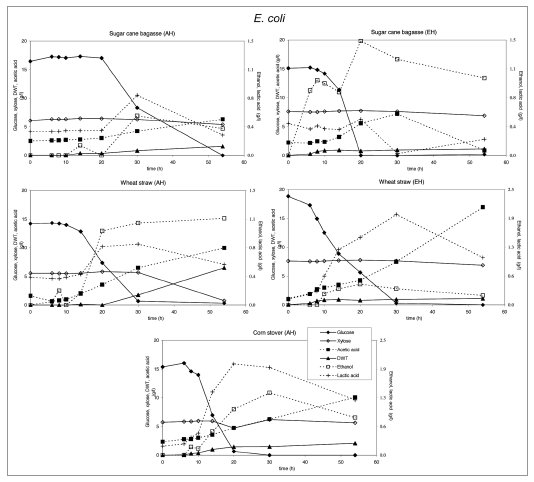
Substrate utilization and product production performance of E. coli on sugar cane bagasse (AH), wheat straw (AH), corn straw (AH), sugar cane bagasse (EH), wheat straw (EH).
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/12389
References
- 1.Palmqvist E, Hahn H. Fermentation of lignocellulosic hydrolysates I: Inhibition and detoxification. Bioresour Technol. 2000;74:17–24. [Google Scholar]
- 2.Jeffries TW. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol. 2006;17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Van Rensburg P, Van Zyl WH, Pretorius IS. Engineering yeast for efficient cellulose degradation. Yeast. 1998;14:67–76. doi: 10.1002/(SICI)1097-0061(19980115)14:1<67::AID-YEA200>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 4.Rumbold K, Van Buijsen HJJ, Overkamp KM, Van Groenestijn JW, Punt PJ, Van Der Werf MJ. Microbial production host selection for converting second-generation feedstocks into bioproducts. Microb Cell Fact. 2009;8:64. doi: 10.1186/1475-2859-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vertès AA, Inui M, Yukawa H. Technological options for biological fuel ethanol. J Mol Microbiol Biotechnol. 2008;15:16–30. doi: 10.1159/000111989. [DOI] [PubMed] [Google Scholar]
- 6.Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006;9:268–274. doi: 10.1016/j.mib.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Kubicek CP, Rohr M. Citric acid fermentation. Crit Rev Biotechnol. 1986;3:331–373. [Google Scholar]
- 8.Punt PJ, Van Biezen N, Conesa A, Albers A, Mangnus J, Van Den Hondel C. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 2002;20:200–206. doi: 10.1016/s0167-7799(02)01933-9. [DOI] [PubMed] [Google Scholar]
- 9.Wierckx N, Koopman F, Bandounas l, De Winde JH, Ruijssenaars HJ. Isolation and characterization of Cupriavidus basilensis HMF14 for biological removal of inhibitors from lignocellulosic hydrolysate. Microb Biotechnol. 2010 doi: 10.1111/j.1751-7915.2009.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols NN, Sharma LN, Mowery RA, Chambliss CK, Van Walsum GP, Dien BS, et al. Fungal metabolism of fermentation inhibitors present in corn stover dilute acid hydrolysate. Enz Microb Technol. 2008;42:624–630. [Google Scholar]


