Abstract
Mutations in the human TGFBI gene encoding TGFBIp have been linked to protein deposits in the cornea leading to visual impairment. The protein consists of an N-terminal Cys-rich EMI domain and four consecutive fasciclin 1 (FAS1) domains. We have compared the stabilities of wild-type (WT) human TGFBIp and six mutants known to produce phenotypically distinct deposits in the cornea. Amino acid substitutions in the first FAS1 (FAS1-1) domain (R124H, R124L, and R124C) did not alter the stability. However, substitutions within the fourth FAS1 (FAS1-4) domain (A546T, R555Q, and R555W) affected the overall stability of intact TGFBIp revealing the following stability ranking R555W>WT>R555Q>A546T. Significantly, the stability ranking of the isolated FAS1-4 domains mirrored the behavior of the intact protein. In addition, it was linked to the aggregation propensity as the least stable mutant (A546T) forms amyloid fibrils while the more stable variants generate non-amyloid amorphous deposits in vivo. Significantly, the data suggested that both an increase and a decrease in the stability of FAS1-4 may unleash a disease mechanism. In contrast, amino acid substitutions in FAS1-1 did not affect the stability of the intact TGFBIp suggesting that molecular the mechanism of disease differs depending on the FAS1 domain carrying the mutation.
Keywords: Amyloid, Extracellular Matrix, Extracellular Matrix Proteins, Protein Folding, Protein Structure
Introduction
Protein misfolding is associated with human disorders, collectively known as “protein deposition diseases” (1) or “protein misfolding diseases” (2). More than 40 of these disorders have been described, including neurodegenerative diseases such as Parkinson disease, Alzheimer disease, and Creutzfeldt-Jacob disease as well as various types of systemic amyloidoses (3). Protein aggregation is generally presumed to be initiated by global or partial unfolding, resulting in exposure of hydrophobic moieties from the protein core (4). In addition, conformational states resulting from thermal fluctuation in native, globular proteins have recently been proposed to be intermediates in protein aggregation (1) suggesting that even minor changes in protein stability may influence the aggregation propensity. Single amino acid substitutions have been shown to enhance these effects in proteins such as human lysozyme (5), human superoxide dismutase 1 (6), and immunoglobulin (7).
Transforming growth factor-β-induced protein (TGFBIp)4 is a 683-amino acid residue extracellular protein expressed in various human tissues including the cornea (8–14). TGFBIp has been found, by sequence identity, to consist of an N-terminal Cys-rich EMI domain (15) and four consecutive fasciclin 1 (FAS1) domains. Furthermore, authentic TGFBIp molecules purified from human and pig corneas exist as monomers with no apparent post-translational modifications (16). TGFBIp has been shown to interact in vitro with numerous macromolecules including several types of integrins (17, 18), collagens, and proteoglycans (19, 20), indicating that TGFBIp exerts a role in mediating contact between cells and the extracellular matrix.
More than 30 different mutations in the TGFBI gene have been reported to result in corneal protein deposits (21). These TGFBI-associated corneal dystrophies are a heterogeneous group of hereditary, bilateral diseases characterized by a progressive accumulation of abnormal protein deposits in the cornea (22). This study addresses the stability changes in TGFBIp brought about by amino acid substitutions associated with different types of corneal dystrophies. Specifically, we investigated the effects of mutations in the first FAS1 domain (FAS1-1) including R124H, R124L, R124C, respectively giving rise to granular corneal dystrophy (GCD) type II, GCD type III, and lattice corneal dystrophy (LCD) type I (21), and the fourth FAS1 domain (FAS1-4) including R555W, R555Q, and A546T resulting in GCD type I, Thiel-Behnke corneal dystrophy, and LCD type IIIA, respectively (21).
The analyses of intact WT and mutant TGFBIp revealed that amino acid substitutions in the FAS1-4 domain (A546T and R555W) affected the unfolding in different ways, making A546T less stable and R555W more stable than WT, while R555Q appeared to retain a slightly lower stability than the WT. This behavior was mirrored by the FAS1-4 domain, analyzed separately. Because all the mutations are known to cause disease, this suggests that both an increase and a decrease in stability may cause protein aggregation. In contrast, amino acid substitutions within the FAS1-1 domain including R124H, R124L, R124C, did not appear to affect the overall stability. This suggests that the molecular mechanism of disease differs depending on the domain containing the amino acid substitution.
EXPERIMENTAL PROCEDURES
Materials
Chemicals were purchased from Sigma-Aldrich. The ChampionTM pET SUMO Expression System was purchased from Invitrogen. Mammalian HEK293T cells were obtained from American Type Cell Culture, LGC Standards.
Cloning, Expression, and Purification of WT and Mutant TGFBIp
WT TGFBIp mutants were expressed and purified as previously described (23). Briefly, the WT TGFBI gene was subcloned from a human TGFBI (BIGH3) cDNA clone (Invitrogen) into the pCMV-SPORT6 vector. The genes encoding the R124H, R124L, R124C, A546T, R555Q, and R555W mutants of human TGFBIp were constructed by introducing the point mutations in a QuikChange reaction (Stratagene) using the cloned WT TGFBI gene as template (Table 1). Mutations were confirmed by sequencing both strands of the constructs. Expressions of full-length WT and mutant TGFBIp were conducted in the human cell line HEK293T after transfection using the calcium phosphate precipitation method (24).
TABLE 1.
Primers used for generating TGFBI mutations
| Mutant | Primer sequencea | Changes introduced |
|---|---|---|
| R124H | CTCAGCTGTACACGGACCACACGGAGAAGCTGAGG | G418 to A |
| R124L | CTCAGCTGTACACGGACCTCACGGAGAAGCTGAGG | G418 to T |
| R124C | CTCAGCTGTACACGGACTGCACGGAGAAGCTGAGG | C417 to T |
| A546T | TTGCTCCCACAAATGAAACCTTCCGAGCCCTGCCA | G1683 to A |
| R555W | CCTGCCACCAAGAGAATGGAGCAGACTCTTGGGAG | C1710 to T |
| R555Q | CCTGCCACCAAGAGAACAGAGCAGACTCTTGGGAG | G1711 to A |
a Only the sequence of the forward primer is shown. The reverse mutation primer is complementary to the forward primer. Changes introduced are shown in bold italics. Nucleotide numbering is according to entry NM_000358.
Cell culture media from transfected HEK293T cells were harvested over a period of 3 days, dialyzed against 20 mm Tris-HCl, pH 7.4 (buffer A) and separated by heparin affinity chromatography (5-ml HiTrap Heparin HP, GE Healthcare). Proteins were eluted from the column using a linear gradient from buffer A to buffer B (20 mm Tris-HCl, 1 m NaCl, pH 7.4). TGFBIp-containing fractions were pooled, dialyzed, and further purified by anion-exchange chromatography (1-ml HiTrap Q column, GE Healthcare). Proteins were separated by a linear gradient of buffer A to buffer B, and TGFBIp-containing fractions were identified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
Cloning and Expression of WT and Mutant FAS1-4
The WT and mutant FAS1-4 domains (residues 502–657) were constructed from the corresponding TGFBI clones using the following primers: 5′-gcaggtatggggactgtcatggatgtcct-3′ (forward) and 5′-tcatgcttgtttgaagatctcaagcgca-3′ (reverse), which add two additional amino acids (Ala-Gly) at the N terminus of FAS1-4 domain compared with the native TGFBIp amino acid sequence (SwissProt accession number Q15582) and include the authentic C terminus of corneal TGFBIp (16). PCR products were subsequently cloned into the ChampionTM pET SUMO Protein Expression System (Invitrogen) according to the manufacturer's instructions.
Expression of FAS1-4 variants was performed in the Escherichia coli strain BL21(DE3), which was grown in lysogeny broth (LB) medium supplemented with the antibiotic kanamycin (50 μg ml−1) at 37 °C. FAS1-4 expression was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at an absorbance (A) at 600 nm of 0.7–0.8. Cells were harvested by centrifugation at 6,000 × g when no further growth was detected, and stored at −20 °C until used.
Purification of FAS1-4 Domains
Harvested cells from the expression of the four FAS1-4 variants were resuspended in 50 mm Tris-HCl, 60 mm KCl, 7 mm β-mercaptoethanol, 10% glycerol, 100 μm PMSF, pH 7.6 (Start buffer), and sonicated for 6 × 30 s at 10 μm amplitude while kept on ice. The samples were then centrifuged for 30 min at 33,000 × g and filtered using a 0.22-μm filter before applied on a nickel-chelating column (5 ml HiTrap Chelating HP equilibrated in 0.1 m NiCl2 and Start buffer). After washing the column successively with 50 mm Tris-HCl, 7 mm MgCl2, 0.5 m NaCl, 5 mm β-mercaptoethanol, 10% glycerol, pH 7.6 (Wash buffer) containing 20 mm and 35 mm imidazole, specifically bound protein was eluted with 50 mm Tris-HCl, 40 mm KCl, 5 mm β-mercaptoethanol, 10% glycerol, 200 mm imidazole, pH 7.6 (elution buffer). Purity of the protein sample was assessed using SDS-PAGE. The purified N-terminally His-tagged SUMO-FAS1-4 fusion proteins were then dialyzed against 50 mm Tris-HCl, 95 mm NaCl, pH 7.6, and the SUMO moiety was cleaved off by adding 0.2% Nonidet P-40, 5 mm DTT and SUMO protease to a final concentration of 4 units ml−1 and incubated at 30 °C for 2–3 h. The resulting samples were dialyzed against 20 mm sodium phosphate, 15 mm NaCl, pH 7.4 and passed over a nickel-chelating column and the pure FAS1-4 proteins were collected in the flow through.
Polyacrylamide Gel Electrophoresis
Proteins were separated by SDS-PAGE in 5–15% polyacrylamide gels (25). Samples were boiled for 5 min in the presence of 30 mm dithiothreitol (DTT) and 1% SDS prior to electrophoresis.
Transverse Urea Gradient (TUG) Gel Electrophoresis
A gradient mixer was used to cast (10 cm × 10 cm × 1.5 mm) 7% polyacrylamide TUG gels containing a linear 0–8 m urea gradient using the glycine/2-amino-2-methyl-1,3-propanediol/HCl buffer system described previously (25). The gels were rotated 90°, and samples containing ∼10 μg of full-length TGFBIp (WT and mutants) or 150 μg of FAS1-4 protein (WT or mutants) were loaded evenly across the top of the gel (26). Refolding TUG gels were performed by unfolding the samples in 8 m urea for 60 min at 23 °C prior to applying the samples to the gels. Electrophoresis was performed at 23 °C, and proteins were visualized by silver staining or Coomassie Brilliant Blue. Quantitative interpretations of the equilibrium unfolding transitions ([urea]1/2) were estimated as described elsewhere (27).
Limited Proteolysis of TGFBIp using Trypsin
Recombinant WT TGFBIp was incubated at 37 °C and titrated with increasing amounts of trypsin. Proteolysis was inhibited after 90 min with 1 mm (final concentration) phenylmethylsulphonyl fluoride (PMSF), for 30 min at 23 °C. Samples were subsequently boiled in reducing SDS sample buffer and separated using SDS-PAGE (28).
Edman Degradation
Proteolytic fragments of TGFBIp were separated by SDS-PAGE as described above. For this analysis, the stacking gel was allowed to polymerize overnight prior to electrophoresis and samples were heated for 3 min at 80 °C only. After electrophoresis proteins were transferred to a polyvinylidene diflouride membrane (Immobilon-P, Millipore) in 10 mm CAPS, 10% (v/v) methanol, pH 11 as described previously (29). Samples were analyzed by automated Edman degradation using an Applied Biosystems PROCISETM 494 HT sequencer with on-line phenylthiohydantoin analysis by HPLC (Applied Biosystems Model 120A).
Nanoelectrospray Mass Spectrometry
Protein bands were excised and digested with trypsin (30). The resulting peptides were analyzed by nano-liquid chromatography (LC)-MS/MS using an EASY-nLC (Proxeon Biosystems) system connected in-line to a Q-TOF Ultima API (Micromass/Waters Corp.) mass spectrometer. The data were processed using Masslynx 4.0 (Micromass) and the generated peak list files were used to query the Swiss-Prot database using the Mascot program (31). The searches were performed with propionamide as fixed modification, peptide mass, and fragment mass tolerances offset to 0.2 Da, and no missed tryptic cleavage sites were allowed.
Far-UV Circular Dichroism (CD) Spectroscopy
Far-UV CD wavelength spectra and temperature scans were recorded on a Jasco J-810 spectropolarimeter (Jasco Spectroscopic Co. Ltd., Hachioji City, Japan) equipped with a JASCO PTC-348WI temperature control unit. Unless specifically mentioned, the buffer was 20 mm sodium phosphate, pH 7.4 and 15 mm NaCl. Thermal scans were performed with a scan rate of 90 °C h−1. All scans were done in triplicate. Unless specifically indicated, proteins were used at a concentration of 10 μm as determined using the Bradford method. For urea denaturation unfolding experiments, samples were incubated in 0–8 m urea and allowed to equilibrate for 2 h before recording and analyzing the spectra using ellipticity at 222 nm as described (32) to obtain the midpoint concentration of denaturation [urea]50%. Urea denaturation refolding experiments were performed by first denaturing the protein in 8 m urea for 2 h, followed by diluting the solution to 0.5–8 m urea, equilibration for 2 h, and carrying out the analysis as for the unfolding experiments.
Fibrillation Assay
FAS1-4 protein (0.4 mg ml−1) in 20 mm NaCl, 20 mm sodium phosphate, pH 7.0 was mixed with 40 μm thioflavin T (ThT) in a 96-well clear-bottomed plate (Nunc) and incubated at 37 °C in a Genios Pro (Tecan, Männesdorf, Switzerland) plate reader with excitation at 450 nm and emission at 485 nm. Data collection proceeded for 150 h, with 3 min intervals of agitation, commencing every 5 min. Samples for electron microscopy were prepared by incubating FAS1-4 protein (0.5 mg ml−1) in 137 mm NaCl, 20 mm sodium phosphate, pH 7.4 in a 96-well clear-bottomed plate (Nunc) for 8 days at 37 °C including a 10 s double orbital shaking every 45 min.
Electron Microscopy
Protein aliquots of 5 μl were placed on 400-mesh carbon-coated, glow-discharged grids for 30 s followed by washing with distilled water and staining with phosphotungstic acid (pH 6.8). Samples were visualized using a Jeol 1010 transmission electron microscope.
RESULTS
TGFBIp WT and Mutants Display Different Susceptibilities to Chemical Denaturation
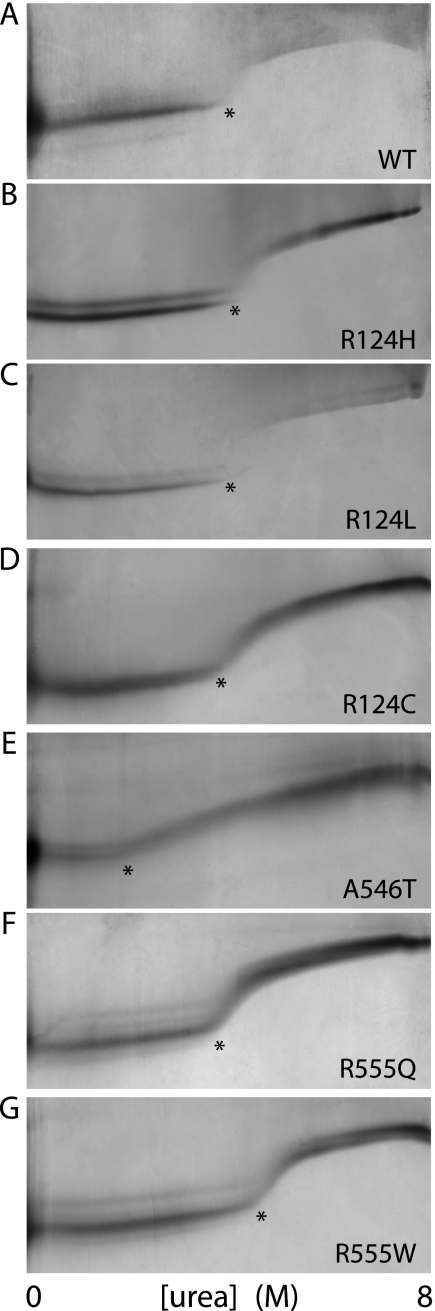
To investigate if naturally occurring amino acid substitutions affected the stability of recombinant TGFBIp, WT TGFBIp and a selection of mutants including R124H, R124L, R124C, A546T, R555Q, and R555W were analyzed by TUG gel electrophoresis (representative gels are shown in Fig. 1). In TUG gel electrophoresis the protein migrates as a single band spanning the gel as it encounters a gradient of 0–8 m urea perpendicular to the direction of migration. The unfolded protein will be conformationally more extended than in the native state and will therefore migrate more slowly. The resulting data showed the unfolding of all TGFBIp variants, as a smooth transition from folded to unfolded protein as the urea concentrations increased (Fig. 1) (33). Given that TGFBIp is a multi-domain protein, it is likely that the domains unfold independently of each other. But individual transitions are often difficult to distinguish, particularly if they occur in similar denaturant concentration ranges. Therefore we used the midpoint urea denaturation concentration ([urea]1/2) for comparative purposes as an empirical indication of the overall stability of TGFBIp. The data revealed that amino acid substitutions within the FAS1-1 domain (R124H, R124L, R124C) failed to significantly affect the stability (Table 2). In contrast, substitutions A546T or R555W, both found in the FAS1-4 domain, gave rise to pronounced changes in [urea]1/2 compared with WT TGFBIp, respectively reducing and increasing the stability of TGFBIp.
FIGURE 1.
TUG gels of full-length TGFBIp WT and mutants. Chemical denaturation of TGFBIp WT and mutants was analyzed by TUG gel electrophoresis. Native recombinant WT (A), R124H (B), R124L (C), R124C (D), A546T (E), R555Q (F), and R555W (G) TGFBIp were analyzed by TUG gel electrophoresis. The starting points of unfolding are marked by asterisks.
TABLE 2.
Stabilities of intact TGFBIp variants
| Mutant | [Urea]1/2 | Δ[Urea]1/2a |
|---|---|---|
| m | m | |
| WT | 4.1 | − |
| R124H | 4.2 | +0.1 |
| R124L | 4.3 | +0.2 |
| R124C | 4.0 | −0.1 |
| A546T | 3.3 | −0.8 |
| R555Q | 4.2 | +0.1 |
| R555W | 4.6 | +0.5 |
a Compared to WT TGFBIp (mutant minus WT). The midpoints of the transitions were calculated from TUG gels as described in “Experimental Procedures.”
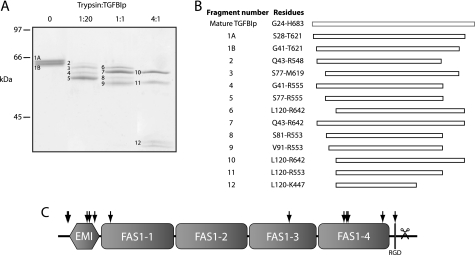
The FAS1-4 Domain Is Susceptible to Trypsin Proteolysis
The conformational stability of WT TGFBIp was further tested by limited trypsin proteolysis (Fig. 2). The results revealed a tryptic fragment resistant to proteolysis even at a trypsin:TGFBIp ratio of 4:1 (w/w) (Fig. 2A). Peptide mass fingerprinting and Edman degradation of the resulting fragments showed that the fragment was composed of most of the first three FAS1 domains (residues 120–447), indicating that this region is particularly rigid (Fig. 2, B and C). This was in contrast to both the EMI domain and the FAS1-4 domain, which were both cleaved at multiple sites. These results supported that the FAS1-4 domain is more flexible than the remaining FAS1 domains encompassing the bulk of the TGFBIp molecule.
FIGURE 2.
TGFBIp contains a trypsin-resistant core. Limited proteolysis of TGFBIp by trypsin. A, SDS-PAGE of trypsin digests. Trypsin to TGFBIp ratios are indicated at the top. B, LC-MS/MS analyses of analyzed fragments. C, schematic representation showing approximate trypsin cleavage sites in TGFBIp (arrows) based on peptide mass fingerprinting. The scissors indicate the C-terminal truncation reported in TGFBIp purified from human corneas (16). A large part of TGFBIp is resistant to trypsin proteolysis.
Chemical Denaturation of FAS1-4 Variants using TUG Gel Electrophoresis
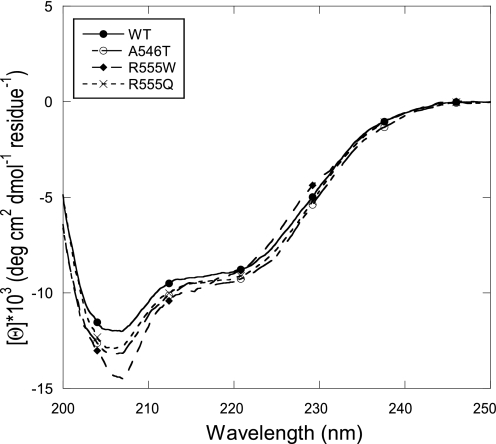
Amino acid substitutions in the FAS1-4 domain appeared to have the most pronounced effect on the TGFBIp stability, thus this domain was analyzed further. The stability effects introduced by amino acid substitutions in the FAS1-4 domain were examined using purified recombinant FAS1-4 (residues 502–657) WT, and the mutants A546T, R555Q, and R555W. To confirm proper folding of the purified protein, the secondary structure compositions of the purified FAS1-4 domains were evaluated by far-UV CD spectroscopy. The spectra of the FAS1-4 variants all showed a major negative band at 208–210 nm and a minor negative band at 222 nm, characteristic of α+β structures (34) (Fig. 3). There are some differences among the four proteins in the absolute magnitude of the far-UV CD spectrum, but we attribute this to errors in the determination of protein concentration and to contributions from the single Trp residue only present in R555W. Spectral deconvolution using the K2d algorithm (35) suggested 25% α-helicity and 19% β-strand for all the four variants tested.
FIGURE 3.
Far-UV CD spectroscopy of purified FAS1-4 variants. Far-UV circular dichroism spectra of the FAS1-4 variants WT, A546T, R555Q, and R555W. All FAS1-4 variants have similar CD spectra indicating that their ratios of secondary structure elements are identical. The minimum at ∼210 nm together with the slight dip at ∼222 nm are characteristic of α+β structures.
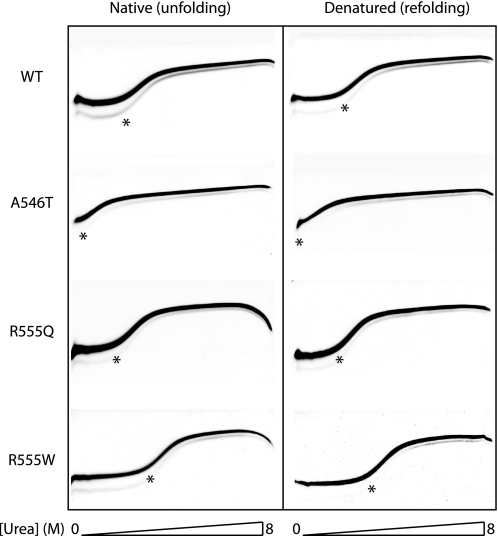
As for the full-length TGFBIp variants, the stabilities of the isolated FAS1-4 domains were examined using TUG gel electrophoresis (Fig. 4). The unfolding midpoints in the 0–8 m urea gradient were measured to 1.0 m (A546T), 2.5 m (R555Q), 2.7 m (WT), and 3.6 m (R555W) (Table 3). Importantly, the same midpoints of denaturation were obtained when the proteins were loaded onto the TUG gels in a denatured state (8 m urea). This underlines the reversibility of the unfolding transition. In addition, the data revealed that the isolated FAS1-4 domains retained the same stability profile as intact TGFBIp, where A546T was less stable than WT or R555Q and R555Q was more stable.
FIGURE 4.
Unfolding and refolding TUG gels of FAS1-4 variants. Chemical denaturation of FAS1-4 variants was analyzed by TUG gel electrophoresis for WT, A546T, R555Q, and R555W FAS1-4. Proteins were loaded on the TUG gels in either native buffer (left panel) or 8 m urea (right panel). The gels show rapid two-state unfolding kinetics for all FAS1-4 variants. The A546T variant is significantly less stable than the WT domain. The stability of mutant R555Q is slightly less than that of WT. Mutant R555W displays an increased stability compared with the WT FAS1-4 domain. The 0–8 m urea gradient is shown below the TUG gels.
TABLE 3.
Stability of FAS1-4 variants
Transition midpoints were calculated using the program KaleidaGraph by fitting curves to the data points.
| [Urea]1/2 (TUGunfolding) | [Urea]1/2 (TUGrefolding) | [Urea]50% | |
|---|---|---|---|
| m | m | m | |
| WT | 2.7 | 2.7 | 3.30 ± 0.059 |
| A546T | 1.0 | 1.1 | 2.01 ± 0.09 |
| R555Q | 2.5 | 2.5 | 3.12 ± 0.041 |
| R555W | 3.6 | 3.7 | 4.00 ± 0.056 |
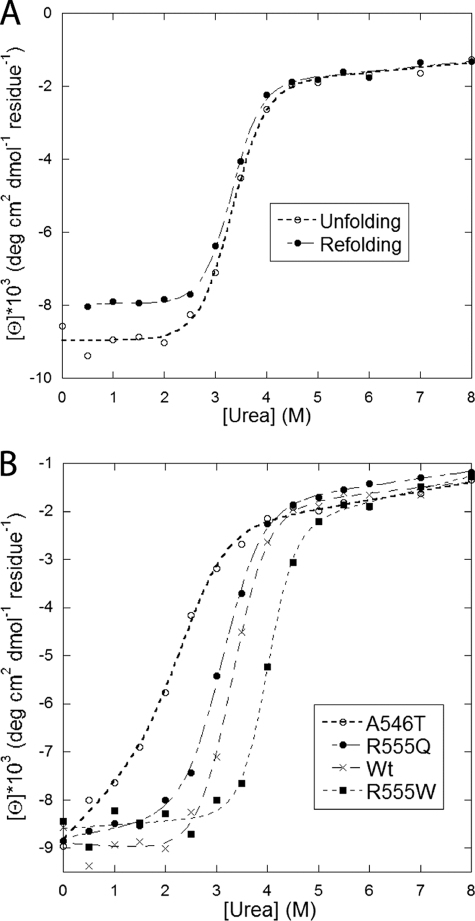
Chemical Denaturation and Refolding of FAS1-4 Variants using Urea
The stability of the FAS1-4 variants were further assessed in a more quantitative fashion using urea-induced chemical denaturation detected by far-UV CD spectroscopy (Fig. 5). To monitor the unfolding process during denaturation, the ellipticity was measured at 222 nm, where the most dramatic signal changes during the transition from folded state to random coil (denatured) state was expected. In agreement with the TUG data, about 90% of WT FAS1-4 refolded displaying the same midpoint of transition as was observed for for the native protein (Fig. 5A). The molar urea concentrations required to induce a 50% denaturation ([urea]50%) were measured to 2.01 ± 0.09 m, 3.30 ± 0.06 m, 3.12 ± 0.04 m, and 4.00 ± 0.06 m for the A546T, WT, R555Q, and R555W, respectively (Table 3 and Fig. 5B). These results support the conclusions reached in the TUG gel experiments, illustrating the same stability ranking R555W>WT>R555Q>A546T.
FIGURE 5.
Denaturation of FAS1-4 variants in urea monitored by the ellipticity at 222 nm. A, WT FAS1-4 unfolding and refolding data (i.e. the protein was transferred from respectively 0 and 8 m urea to different concentrations of urea). The superimposition of the two curves shows that unfolding is completely reversible. B, unfolding data for WT FAS1-4 and the three mutants A546T, R555Q, and R555W. Data were fitted to a two-state unfolding scheme as described (32), and midpoint denaturation concentrations are summarized in Table 3.
Aggregation of FAS1-4 Mutants Follow the Same Pattern as in Vivo Deposits
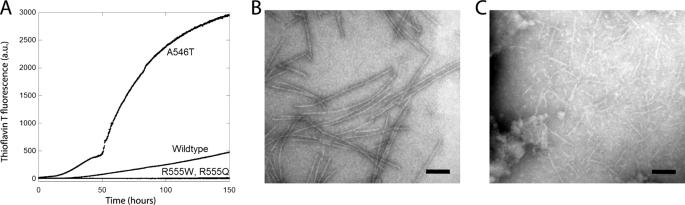
Finally, we investigate how the amino acid substitutions affected the ability of FAS1-4 to form amyloid deposits under physiological salt and pH conditions. The wild-type FAS1-4 and the three mutants were incubated at 37 °C and amyloid formation was followed by a ThT assay (Fig. 6). Based on the onset of the ThT signal the propensity of A546T to form amyloid fibrils was significantly increased as compared with the wild-type FAS1-4. In addition, the two mutants, R555W and R555Q, showed no significant rise in fluorescence at all (Fig. 6A). This is consistent with the observation that TGFBIp containing the A546T amino acid substitutions forms amyloid deposits, while R555W and R555Q form amorphous (i.e. non-amyloid) deposits in vivo (21). The formation of amyloid fibril by A546T and to some degree WT FAS1-4 was confirmed by electron microscopy (Fig. 6, B and C, respectively). During the same conditions, both the R555W and the R555Q mutants failed to form visible aggregates.
FIGURE 6.
Aggregation of WT and mutant FAS1-4. A, ThT fluorescence for WT and mutant FAS1-4. The A546T mutant displays a 6-fold higher ThT signal than WT FAS1-4, after 150 h. R555Q and R555W display no significant increase in fluorescence over the monitored time period, suggesting no fibrillation. B, electron micrograph of A546T FAS1-4 after 8 days at 37 °C. C, electron micrograph of WT FAS1-4 after 8 days at 37 °C. Scale bars in panels B and C denote 100 nm.
DISCUSSION
In this study we have investigated the stability of WT TGFBIp, three different mutants of the FAS1-1 domain (R124H, R124L, R124C) and three FAS1-4 mutants (A546T, R555Q, and R555W). These mutants were chosen to represent different phenotypic manifestations observed in patients suffering from TGFBI-associated corneal dystrophy.
The TGFBIp variants were initially analyzed by TUG gel electrophoresis, revealing that the overall stability of intact TGFBIp was affected only by amino acid substitutions within the FAS1-4 domain, whereas changes in the FAS1-1 failed to change the stability. Limited trypsin proteolysis of WT TGFBIp supported that the FAS1-4 domain was more labile than the bulk of the molecule containing the FAS1-1. Because mutations in the FAS1-4 domain, in contrast to the FAS1-1 mutations, appeared to affect the stability of TGFBIp, the FAS1-4 domain was further studied in isolation.
TUG gel electrophoresis and CD spectroscopy of the isolated FAS1-4, WT, A546T, R555Q, and R555W variants, provided a quantitative view of the unfolding process. Both TUG gel electrophoresis and chemical denaturation followed by CD spectroscopy supported the following stability ranking of the four FAS1-4 variants; R555W>WT>R555Q>A546T. This underlines that single amino acid substitutions might have a dramatic effect on the overall stability of the FAS1-4. The same stability ranking of intact TGFBIp (WT and mutants) and the isolated FAS1-4 domains suggested that FAS1-4 is a feasible model for understanding the molecular mechanism governing the stability of TGFBIp. Electron micrographs of FAS1-4 samples incubated for 1 week with agitation showed a formation of amyloid structures for the A564T mutant, as expected from the ThT assay. The WT FAS1-4 was also found to form fibrillar (or protofibrillar) structures, although to a much lesser extend. This observation points toward an inherent propensity of WT FAS1-4 to form amyloid structures. The R555W and R555Q did not generate aggregates, implying that other factors may be required for the formation of the amorphous aggregation observed in vivo.
The Drosophila FAS1 (Protein Data Bank ID: 1O70) suggests that Arg-555 in human FAS1-4 is exposed on the surface (36). However, introducing a Trp residue at that position has the potential to favor interactions with the hydrophobic core, stabilizing the domain, as observed in this study. According to the CD analysis, this is achieved without introducing major changes in the secondary structure (Fig. 3B). However, it is unexpected that stabilizing amino acid substitutions promote aggregation (3). Here a positively charged hydrophilic (Arg) is replaced by a hydrophobic amino acid residue (Trp), which is likely to increase the propensity of the protein to associate, potentially lowering the solubility, simply by reducing the level of electrostatic repulsion. A similar effect, though with a less aggressive outcome, is expected by replacing a positively charged Arg with the uncharged but polar Gln residue. Alternatively, Arg-555 may be involved in an inter-domain salt bridge that stabilizes the intact TGFBIp molecule. These suggestions are in accordance with in vivo observations of R555W and R555Q showing amorphous aggregates, composed of intact TGFBIp. In contrast, the A546T mutant forms amyloid structures, characterized by the loss of native structure and formation of cross-β sheets (37). An increased level of intact R555W TGFBIp has been observed in corneal deposits in vivo (38) supporting that the protein is resistant to degradation and that an increase in protein stability may indeed have pathological consequences.
The A546T amino acid substitution was found to destabilize intact TGFBIp as well as the FAS1-4 domain alone. The FAS1 crystal structure suggests that most pathogenic mutations in FAS1-4 introduce bulky or polar groups at the hydrophobic core of the domain. This is unfavorable and likely to cause perturbation of the FAS4 structure. We propose that these substitutions may expose intrinsic fibrillation prone regions of TGFBIp (e.g. as reported by Yuan et al. (39)), causing amyloid deposits to form. Amyloids are resistant to proteolysis and indeed, other studies have shown that fragments of TGFBIp accumulated in corneas of patients with amyloid deposits (40–43).
In conclusion, this work shows that only amino acid substitutions in FAS1-4 provoke significant changes in the stability of TGFBIp. These changes were found to be destabilizing for the amyloid-associated A546T mutation and stabilizing for the granular corneal dystrophy mutation R555W. Our results suggest that different aggregation mechanisms are involved during protein deposition in the TGFBI related corneal dystrophies. Significantly, the FAS1-4 mutant A546T was found to form amyloid structures in vitro in accordance with its in vivo phenotype. Furthermore, the similar behavior of intact TGFBIp and the isolated FAS1-4 domain suggests that the FAS1-4 domain may be used as a model system to understand the FAS1-4-derived corneal dystrophies.
Acknowledgment
We thank Anne Gylling for expert technical assistance.
This work was supported by the National Eye Institute (R01 EY012712) and the Danish National Research Foundation (inSPIN).
- TGFBIp
- transforming growth factor-β-induced protein
- TGFBI gene
- transforming growth factor-β induced gene
- FAS1
- fasciclin 1
- FAS1-4
- FAS1 domain 4
- EMI
- EMILIN-1
- GCD
- granular corneal dystrophy
- LCD
- lattice corneal dystrophy
- SUMO
- small ubiquitin-like modifier
- TUG
- transverse urea gradient.
REFERENCES
- 1. Chiti F., Dobson C. M. (2009) Nat. Chem. Biol. 5, 15–22 [DOI] [PubMed] [Google Scholar]
- 2. Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 3. Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 4. Dobson C. M. (1999) Trends Biochem. Sci. 24, 329–332 [DOI] [PubMed] [Google Scholar]
- 5. Booth D. R., Sunde M., Bellotti V., Robinson C. V., Hutchinson W. L., Fraser P. E., Hawkins P. N., Dobson C. M., Radford S. E., Blake C. C., Pepys M. B. (1997) Nature 385, 787–793 [DOI] [PubMed] [Google Scholar]
- 6. Stathopulos P. B., Rumfeldt J. A., Scholz G. A., Irani R. A., Frey H. E., Hallewell R. A., Lepock J. R., Meiering E. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7021–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raffen R., Dieckman L. J., Szpunar M., Wunschl C., Pokkuluri P. R., Dave P., Wilkins Stevens P., Cai X., Schiffer M., Stevens F. J. (1999) Protein Sci. 8, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carson D. D., Lagow E., Thathiah A., Al-Shami R., Farach-Carson M. C., Vernon M., Yuan L., Fritz M. A., Lessey B. (2002) Mol. Hum. Reprod. 8, 871–879 [DOI] [PubMed] [Google Scholar]
- 9. Ferguson J. W., Thoma B. S., Mikesh M. F., Kramer R. H., Bennett K. L., Purchio A., Bellard B. J., LeBaron R. G. (2003) Cell Tissue Res. 313, 93–105 [DOI] [PubMed] [Google Scholar]
- 10. Kitahama S., Gibson M. A., Hatzinikolas G., Hay S., Kuliwaba J. L., Evdokiou A., Atkins G. J., Findlay D. M. (2000) Bone. 27, 61–67 [DOI] [PubMed] [Google Scholar]
- 11. LeBaron R. G., Bezverkov K. I., Zimber M. P., Pavelec R., Skonier J., Purchio A. F. (1995) J Invest. Dermatol. 104, 844–849 [DOI] [PubMed] [Google Scholar]
- 12. Lee S. H., Bae J. S., Park S. H., Lee B. H., Park R. W., Choi J. Y., Park J. Y., Ha S. W., Kim Y. L., Kwon T. H., Kim I. S. (2003) Kidney Int. 64, 1012–1021 [DOI] [PubMed] [Google Scholar]
- 13. Ohno S., Doi T., Fujimoto K., Ijuin C., Tanaka N., Tanimoto K., Honda K., Nakahara M., Kato Y., Tanne K. (2002) J Dent. Res. 81, 822–825 [DOI] [PubMed] [Google Scholar]
- 14. Rawe I. M., Zhan Q., Burrows R., Bennett K., Cintron C. (1997) Invest. Ophthalmol. Vis. Sci. 38, 893–900 [PubMed] [Google Scholar]
- 15. Doliana R., Bot S., Bonaldo P., Colombatti A. (2000) FEBS Lett. 484, 164–168 [DOI] [PubMed] [Google Scholar]
- 16. Andersen R. B., Karring H., Møller-Pedersen T., Valnickova Z., Thøgersen I. B., Hedegaard C. J., Kristensen T., Klintworth G. K., Enghild J. J. (2004) Biochemistry 43, 16374–16384 [DOI] [PubMed] [Google Scholar]
- 17. Bae J. S., Lee S. H., Kim J. E., Choi J. Y., Park R. W., Yong Park J., Park H. S., Sohn Y. S., Lee D. S., Bae Lee E., Kim I. S. (2002) Biochem. Biophys. Res. Commun. 294, 940–948 [DOI] [PubMed] [Google Scholar]
- 18. Kim J. E., Kim S. J., Lee B. H., Park R. W., Kim K. S., Kim I. S. (2000) J. Biol. Chem. 275, 30907–30915 [DOI] [PubMed] [Google Scholar]
- 19. Reinboth B., Thomas J., Hanssen E., Gibson M. A. (2006) J. Biol. Chem. 281, 7816–7824 [DOI] [PubMed] [Google Scholar]
- 20. Hashimoto K., Noshiro M., Ohno S., Kawamoto T., Satakeda H., Akagawa Y., Nakashima K., Okimura A., Ishida H., Okamoto T., Pan H., Shen M., Yan W., Kato Y. (1997) Biochim. Biophys. Acta. 1355, 303–314 [DOI] [PubMed] [Google Scholar]
- 21. Kannabiran C., Klintworth G. K. (2006) Hum. Mutat. 27, 615–625 [DOI] [PubMed] [Google Scholar]
- 22. Klintworth G. K. (2003) Front Biosci. 8, d687–713 [DOI] [PubMed] [Google Scholar]
- 23. Runager K., García-Castellanos R., Valnickova Z., Kristensen T., Nielsen N. C., Klintworth G. K., Gomis-Rüth F. X., Enghild J. J. (2009) Acta. Crystallogr. Sect F Struct. Biol. Cryst. Commun. 65, 299–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luthman H., Magnusson G. (1983) Nucleic. Acids Res. 11, 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bury A. F. (1981) J. Chromatogr. 213, 491–500 [Google Scholar]
- 26. Goldenberg D. P. (1989) in Protein Structure: A Practical Approach (Creighton T. E. ed) pp. 225–250, Oxford University Press [Google Scholar]
- 27. Hollecker M., Creighton T. E. (1982) Biochim. Biophys. Acta. 701, 395–404 [DOI] [PubMed] [Google Scholar]
- 28. Rubenstein D. S., Enghild J. J., Pizzo S. V. (1991) J. Biol. Chem. 266, 11252–11261 [PubMed] [Google Scholar]
- 29. Matsudaira P. (1987) J. Biol. Chem. 262, 10035–10038 [PubMed] [Google Scholar]
- 30. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 31. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 32. Mogensen J. E., Ipsen H., Holm J., Otzen D. E. (2004) Biochemistry 43, 3357–3367 [DOI] [PubMed] [Google Scholar]
- 33. Goldenberg D. P., Creighton T. E. (1984) Anal. Biochem. 138, 1–18 [DOI] [PubMed] [Google Scholar]
- 34. Manavalan P. J. W. (1985) Proc. Int. Symp. Biomol. Struct. Interactions, Suppl. J. Biosci. 8, 141–149 [Google Scholar]
- 35. Andrade M. A., Chacón P., Merelo J. J., Morán F. (1993) Protein Eng 6, 383–390 [DOI] [PubMed] [Google Scholar]
- 36. Clout N. J., Tisi D., Hohenester E. (2003) Structure 11, 197–203 [DOI] [PubMed] [Google Scholar]
- 37. Klintworth G. K. (2009) Orphanet. J. Rare. Dis. 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klintworth G. K., Valnickova Z., Enghild J. J. (1998) Am. J. Pathol. 152, 743–748 [PMC free article] [PubMed] [Google Scholar]
- 39. Yuan C., Berscheit H. L., Huang A. J. (2007) FEBS Lett. 581, 241–247 [DOI] [PubMed] [Google Scholar]
- 40. Takács L., Boross P., Tözser J., Módis L., Jr., Tóth G., Berta A. (1998) Exp. Eye. Res. 66, 739–745 [DOI] [PubMed] [Google Scholar]
- 41. Konishi M., Yamada M., Nakamura Y., Mashima Y. (2000) Curr. Eye. Res. 21, 891–896 [DOI] [PubMed] [Google Scholar]
- 42. Korvatska E., Henry H., Mashima Y., Yamada M., Bachmann C., Munier F. L., Schorderet D. F. (2000) J. Biol. Chem. 275, 11465–11469 [DOI] [PubMed] [Google Scholar]
- 43. Stix B., Leber M., Bingemer P., Gross C., Rüschoff J., Fändrich M., Schorderet D. F., Vorwerk C. K., Zacharias M., Roessner A., Röcken C. (2005) Invest. Ophthalmol. Vis. Sci. 46, 1133–1139 [DOI] [PubMed] [Google Scholar]