Abstract
In most organisms, thioredoxin (Trx) and/or glutathione (GSH) systems are essential for redox homeostasis and deoxyribonucleotide synthesis. Platyhelminth parasites have a unique and simplified thiol-based redox system, in which the selenoprotein thioredoxin-glutathione reductase (TGR), a fusion of a glutaredoxin (Grx) domain to canonical thioredoxin reductase domains, is the sole enzyme supplying electrons to oxidized glutathione (GSSG) and Trx. This enzyme has recently been validated as a key drug target for flatworm infections. In this study, we show that TGR possesses GSH-independent deglutathionylase activity on a glutathionylated peptide. Furthermore, we demonstrate that deglutathionylation and GSSG reduction are mediated by the Grx domain by a monothiolic mechanism and that the glutathionylated TGR intermediate is resolved by selenocysteine. Deglutathionylation and GSSG reduction via Grx domain, but not Trx reduction, are inhibited at high [GSSG]/[GSH] ratios. We found that Trxs (cytosolic and mitochondrial) provide alternative pathways for deglutathionylation and GSSG reduction. These pathways are operative at high [GSSG]/[GSH] and function in a complementary manner to the Grx domain-dependent one. Despite the existence of alternative pathways, the thioredoxin reductase domains of TGR are an obligate electron route for both the Grx domain- and the Trx-dependent pathways. Overall, our results provide an explanation for the unique array of thiol-dependent redox pathways present in parasitic platyhelminths. Finally, we found that TGR is inhibited by 1-hydroxy-2-oxo-3-(N-3-methyl-aminopropyl)-3-methyl-1-triazene (NOC-7), giving further evidence for NO donation as a mechanism of action for oxadiazole N-oxide TGR inhibitors. Thus, NO donors aimed at TGR could disrupt the entire redox homeostasis of parasitic flatworms.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Multifunctional Enzymes, Parasite, Selenium, Deglutathionylation, Glutaredoxin, Glutathione, Thioredoxin, Thioredoxin Reductase
Introduction
Platyhelminths (also known as flatworms) cause severe human parasitic diseases, such as schistosomiasis, cysticercosis, and hydatid disease. Parasitic platyhelminths share a unique biochemical scenario regarding thiol-based redox pathways (1–4). These organisms possess a linked thioredoxin-glutathione system that provides reducing equivalents to both the thioredoxin (Trx)2 and the glutathione (GSH) pathways. In contrast to their mammalian hosts, platyhelminth parasites lack thioredoxin reductase (TR) and glutathione reductase (GR) and hence conventional Trx and GSH systems. The distinctive feature of the redox array of platyhelminth parasites is that reduction of Trx and GSSG is carried out by a single selenoenzyme, termed thioredoxin glutathione reductase (TGR). Thus, these organisms have an absolute reliance on TGR for redox homeostasis and DNA synthesis. Furthermore, in parasitic flatworms, mitochondrial and cytosolic TGR variants are derived from a single gene, and the encoded mature polypeptides are identical in sequence (3).
TGR is a complex flavoenzyme that contains several thiol-based redox centers and is composed of an N-terminal glutaredoxin (Grx) domain fused to canonical large TR domains (5). In the platyhelminth parasite Echinococcus granulosus (class cestoda), the Grx domain has a dithiol CPYC redox center, and the TR module contains a CX4C redox center and the conspicuous C-terminal motif GCUG, where U is the redox active amino acid selenocysteine (Sec) (1). Detailed biochemical studies with sets of mutants of human and E. granulosus TGR have shown the following: (i) both oxidized Trx and oxidized glutathione (GSSG) reduction are dependent on the redox-active C-terminal Sec residue, and (ii) reduction of GSSG, but not of oxidized Trx, requires the Grx domain (6, 7). In the case of E. granulosus and Taenia crassiceps (class cestoda) TGRs, it has also been shown that their GR activity, but not their TR activity, is inhibited at high concentrations of oxidized glutathione (GSSG) (4, 6). This phenomenon has been proposed to be due to glutathionylation of critical Cys residues (6).
Evidence obtained in vitro and in vivo is consistent with an essential role of TGR for platyhelminth parasites. RNAi of Schistosoma mansoni TGR led to parasite death in vitro (8). Auranofin, a drug that specifically targets TGR and inhibits both GSSG and Trx reduction, has been shown to kill platyhelminth parasites in vitro and partially cure S. mansoni-infected mice in vivo (6, 8, 9). The essentiality of TGR, the dissimilar biochemical scenarios between flatworm parasites and their mammalian hosts, and the need for novel drug to target flatworm infections have led to identification, by quantitative high throughput screening, of new TGR inhibitors (10). Further investigations in this field have confirmed N-oxides as new drug leads targeting TGR (11).
Despite the essential functions of TGR in flatworm parasites and its importance as a novel drug target, there are several aspects relating TGR functions, catalytic mechanism, and regulation that have not yet been addressed or are poorly understood. Open questions that have driven our investigations are as follows: (i) whether TGR has additional activities associated with the Grx domain, such as deglutathionylase activity of GSH-protein mixed disulfides (protein-S-SG) or nitrosoglutathione (GSNO) reductase activity; (ii) whether the activities mediated by the Grx domain occur via a dithiol or a monothiol mechanism; and (iii) whether there are alternative pathways for GSSG reduction that operate at high GSSG concentrations. Regarding this latter issue, it is relevant to mention that in some GR-less organisms, such as Drosophila melanogaster, GSSG reduction is carried out by Trx, which in turn is recycled by TR (12). This is also the case of Saccharomyces cerevisiae null mutants in GR, where the Trx system compensates for the lack of GR activity (13). In this study we present two major findings. First, we demonstrate that TGR can support GSH-independent deglutathionylase activity. Second, we demonstrate the existence of Trx-dependent alternative pathways for GSSG reduction and deglutathionylation downstream of TGR. In addition, we show that a NO donor inhibits TGR, providing further support to the proposed mechanism of action of oxadiazole N-oxides (11).
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of E. granulosus Wild-type TGR, Mutant TGRs, and Wild-type Trxs
Constructs for wild-type TGR (TGRWT), a TGR mutant lacking the entire N-terminal Grx domain (Grx-less TGR), and a TGR mutant truncated at the Sec596 residue of the C-terminal redox center (truncated TGR) were previously generated (6). Constructs for wild-type mitochondrial and cytosolic Trxs (mTrx and cTrx, respectively) were also described previously (6). In this study, two TGR mutants at the N-terminal Grx domain were cloned as follows: Cys31 to Ser (TGRC31S) and Cys34 to Ser (TGRC34S) mutants. Site-directed mutagenesis was carried out by overlap extension (14). Pfu DNA polymerase (Fermentas) was used for PCRs, starting from a wild-type TGR construct template. All constructs for selenoproteins contained the Sec insertion sequence element of Escherichia coli formate dehydrogenase H at a 10-nucleotide distance from the penultimate UGASec codon to allow stop codon recoding to Sec, as described previously (6, 15). The amplified products were first cloned into pGEM-T-Easy (Promega), and the construct sequences were verified prior to subsequent subcloning into pET28a to obtain N-terminal His-tagged fusions (Novagen). For recombinant protein expression of TGRs, the corresponding constructs were used to transform E. coli BL21(DE3) cells or, in the case of selenoprotein constructs, BL21(DE3) cells previously transformed with pSUABC, a plasmid that supports high level expression of genes involved in Sec synthesis and decoding (selA, selB, and selC) (15). Expression of all recombinant proteins was carried out following the protocol described previously (16), which has been optimized for expression of selenoproteins. Essentially, induction of recombinant proteins was carried out with 100 μm isopropyl β-thiogalactopyranoside at late exponential phase (A600 = 2.4), during 24 h at 24 °C. Recombinant clones were grown in modified LB, as described previously (17), supplemented with 0.1 g/liter cysteine and 0.37 g/liter methionine (18) in the presence of kanamycin (50 μg/ml) and chloramphenicol (33 μg/ml); the latter was used only in the case of bacterial cultures harboring constructs encoding selenoproteins. At the time of induction, the culture was supplemented with 5 μm sodium selenite, 20 μg/ml riboflavin, 20 μg/ml pyridoxine, and 20 μg/ml niacin according to Ref. 17. The bacterial cultures were centrifuged, and the pellets were resuspended in modified nickel-nitrilotriacetic acid lysis buffer (300 mm NaCl, 50 mm sodium phosphate, 20 mm imidazole, pH 7.2) containing 1 mm PMSF and 1 mg/ml lysozyme and sonicated (10 pulses of 1 min with 1-min pauses). The lysates were centrifuged for 1 h at 30,000 × g, and supernatants were applied to a nickel-nitrilotriacetic acid column (Qiagen), washed with 300 mm NaCl, 50 mm sodium phosphate, 30 mm imidazole, pH 7.2, and eluted with 250 mm imidazole. The protein-containing fractions were applied to PD10 desalting columns (GE Healthcare) using phosphate-buffered saline (PBS), 150 mm potassium chloride, 50 mm sodium phosphate, pH 7.2. Fractions containing the recombinant proteins were stored at −70 °C before use. Total protein concentration and FAD content were determined spectrophotometrically at 280 (ϵ = 54.2 mm−1 cm−1) and 460 nm (ϵ = 11.3 mm−1 cm−1), respectively. The selenium content of selenoproteins was determined by atomic absorption using a plasma emission spectrometer (Jarrell-Ash 965 ICP) in the Chemical Analysis Laboratory, University of Georgia. Active selenoenzyme concentrations were calculated considering their selenium contents (selenium contents were close to 10% for all selenoproteins). The purity of the recombinant proteins was analyzed by running 10% SDS-polyacrylamide gels, under reducing conditions, and by size exclusion chromatography on a Superose 12 column (GE Healthcare).
Recombinant protein expression and purification of Trxs was performed as described previously (6). Fractions containing the recombinant proteins were stored at −70 °C prior to use. Protein concentration was determined spectrophotometrically at 280 nm (ϵ = 7.6 and 6.1 mm−1 cm−1 for cytosolic and mitochondrial Trx, respectively). The purity of the recombinant proteins was analyzed by running 15% SDS-polyacrylamide gels under reducing conditions and by size exclusion chromatography on a Superdex 75 column (GE Healthcare). The insulin reduction assay for Trx activity (19) was used as a TR-independent assay to verify these recombinant Trxs were active.
Deglutathionylation Assay
The deglutathionylase activity was evaluated using a modification of the method described previously (20). The NADPH-dependent reduction of the glutathionylated substrate peptide SQLWC(glutathione)LSN (Peptron Inc., South Korea) was followed by the decrease in absorbance at 340 nm due to NAPDH oxidation (ϵ = 6.2 mm−1 cm−1). The reaction mixtures contained 100 μm NADPH and 40 μm glutathionylated peptide.
GSSG Reduction Assay
The GR activity was assayed as the NADPH-dependent reduction of oxidized glutathione (GSSG), which is followed by the decrease in absorbance at 340 nm due to NADPH oxidation (ϵ = 6.2 mm−1 cm−1) (21). The reaction mixtures contained 100 μm NADPH and 40 μm or 1 mm GSSG.
Insulin Reduction Assay for TR Activity
The Trx-coupled assay of TR activity takes advantage of the NADPH-dependent reduction of Trx by TR, which is followed by the decrease in absorbance at 340 nm due to NADPH oxidation (ϵ = 6.2 mm−1 cm−1). In this assay, excess insulin is used as an electron sink to maintain a constant concentration of oxidized Trx (22, 23). The reaction mixtures contained 100 μm NADPH, 0.5 mg/ml insulin, and 10 μm cTrx or mTrx.
GSNO Reduction Assay
GSNO was synthesized using a modification of the method described previously (24). Fresh GSNO stock solutions were prepared by allowing equimolar concentrations of GSH (200 mm in 50 mm potassium phosphate buffer, pH 7.4, 0.1 mm diethylenetriaminepentaacetic acid) and acidified sodium nitrite (200 mm in 50 mm HCl) to react for 5 min on ice in the dark and then immediately used for the enzymatic assay. Fresh GSH and sodium nitrite solutions were prepared on the day of use. Final GSNO concentrations were calculated using an extinction coefficient of 0.77 mm−1 cm−1 at 336 nm. The GSNO reductase activity was evaluated using NADPH (100 μm) as electron donor by following the decrease in absorbance at 340 nm due to NAPDH oxidation (ϵ = 6.2 mm−1 cm−1) and GSNO reduction. Different GSNO concentrations were used, ranging from 250 μm to 1 mm.
DTNB Reduction Assay for TR Activity
The reduction of 5,5′-dithiobis(2-dinitrobenzoic acid) (DTNB) with concomitant NADPH oxidation was determined by the increase in absorbance at 412 nm due to the formation of 5′-thionitrobenzoic acid (ϵ = 13600 m−1 cm−1) (22). The reaction mixtures contained 100 μm NADPH and 5 mm DTNB.
Assay for TGR Inhibition by NO Donors
The effect of NOC-7 (1-hydroxy-2-oxo-3-(N-3-methylaminopropyl)-3-methyl-1-triazene; Dojindo Laboratories, Japan) on the TR activity of TGR was evaluated using the DTNB reduction assay. A fresh 50 mm NOC-7 stock solution in 0.01 n NaOH was prepared. Reaction mixtures containing 1 nm TGRWT, 100 μm NADPH, and different NOC-7 concentrations were preincubated for 5 min at room temperature. The reaction was started by the addition of DTNB.
All enzymatic assays were carried out in 50 mm potassium phosphate buffer, pH 7.0, containing 1 mm EDTA, at 25 °C, and 1-ml reaction volume, using a PG-T70+ (PG Instruments, UK) spectrophotometer. Analyses of the kinetic data were performed using the ORIGIN software (OriginLab).
Molecular Modeling and Conservation Analysis
Multiple sequence alignments were carried out with the standalone ClustalW (version 2.0.3) program. Visualization of alignments was done with JalView (version 2.3); details on the coloring schemes used are given in the figure legends. For any given multiple alignment, the conservation for each aligned position was evaluated, as described previously (25), and performed with JalView 2.0.3. The position-specific conservation values obtained were then exported, processed for visualization purposes (i.e. normalized for the highest), and finally plotted to the model of E. granulosus Trx, with in-house Python version 2.5 scripts. The output of the procedure is illustrated in supplemental Fig. S2. Homology modeling of E. granulosus Trx was made with Swiss-Model, using the structure with Protein Data Bank code 2OE1 as a template. The substrate, GSSG, was downloaded from the Protein Data Bank repository. The SP4 force field and the AMMP-mom method were employed to assign charges to GSSG and to minimize the structure. The substrate was analyzed in the intercalation site using a docking box of 20 × 20 × 20 Å centered on the approximate center of mass of the active site CXXC motif of E. granulosus Trx. Docking analysis were performed with the program ArgusLab, employing the GAdock docking engine, a genetic algorithm search technique, with the following parameters: population size 250, maximum generations 100,000, mutation rate 0.02, grid resolution 0.2 Å, and flexible ligand mode (other parameters were kept with default values).
In Vitro Culture of Larval Worms
50,000 protoscoleces, obtained from asceptical puncture of a single hydatid cyst from bovine lung, were washed several times with PBS and then incubated at 37 °C, 5% CO2, in DMEM supplemented with antibiotics and 20 mm HEPES, pH 6.8. Cultured protoscoleces were treated with NOC-7 and NOC-12 (1-hydroxy-2-oxo-3-(N-ethyl-2-aminoethyl)-3-ethyl-1-triazene; Dojindo Laboratories, Japan) at concentrations ranging from 50 μm to 5 mm or with the vehicle (0.01 n NaOH). Protoscoleces were observed under the microscope, and viability was assessed by exclusion of the vital dye eosin over 4 days.
RESULTS
TGR Can Support GSH-independent Deglutathionylation of Peptide-S-SG Mixed Disulfides via Its Grx Domain
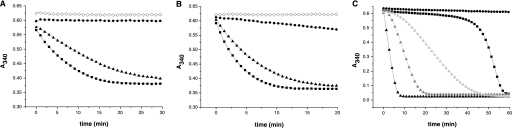
We assayed deglutathionylase activity using a modification of a recently described method that uses the glutathionylated peptide SQLWC(glutathione)LSN as a substrate (see “Experimental Procedures”). We found that recombinant TGR possesses deglutathionylase activity. In contrast, a TGR mutant truncated at the penultimate Sec residue of the C-terminal redox center, and a Grx-less TGR do not display deglutathionylase activity, indicating that both the C-terminal and the Grx redox centers are required for this catalysis (Fig. 1). These results clearly show that TGR can support GSH-independent deglutathionylation of peptide-S-SG mixed disulfides.
FIGURE 1.
Deglutathionylation of glutathionylated peptide by TGR. The deglutathionylase activities of wild-type TGR (TGRWT) (black box), Grx-less TGR (gray right arrow), and truncated TGR (gray left arrow) were evaluated using a glutathionylated peptide (peptide-S-SG) and NADPH as substrates, following the decrease in absorbance at 340 nm due to NADPH oxidation. The full time courses obtained are shown. The assay was carried out at 100 μm NADPH, 40 μm peptide-S-SG, and 2 nm enzyme. Reaction mixtures without enzyme (white diamond), and with TGRWT and the parental nonglutathionylated peptide (gray diamond) were used as controls. The overall NADPH oxidation corresponds to the consumption of 40 μm peptide-S-SG, which is the limiting substrate.
Deglutathionylase Activity, like Glutathione Reductase Activity, Is Inhibited at High [GSSG]/[GSH]
We have previously shown that the GR activity of TGR was inhibited at high [GSSG]/[GSH] ratios, exhibiting a hysteretic behavior (a lag phase in GSSG reduction), whereas the TR activity was not regulated (6). Thus, we analyzed whether deglutathionylase activity, which is also dependent on the Grx domain, could be affected by the [GSSG]/[GSH] ratio. Indeed, we found that deglutathionylation was inhibited at high (1 mm) but not at low (40 μm) GSSG concentrations (Fig. 2). Under conditions of no hysteresis, TGR reduces GSSG more efficiently than the peptide-S-SG mixed disulfide.
FIGURE 2.
Inhibition of TGR deglutathionylase activity at high GSSG concentrations. Full time courses for NADPH oxidation (A340 decrease) at different concentrations of peptide-S-SG and/or GSSG are shown. The comparison of 40 μm peptide-S-SG (gray triangle) with 40 μm peptide-S-SG and 1 mm GSSG (gray box) time courses indicates that deglutathionylation is inhibited at high GSSG concentrations. The time courses for NADPH oxidation using 40 μm GSSG (dark gray circle), 1 mm GSSG (light gray circle), and 40 μm peptide-S-SG and 40 μm GSSG (black box) are included as references. All assays were carried out at 100 μm NADPH and 2 nm wild-type TGR. Note that in each case the overall NADPH oxidation (A340 decrease) corresponds to the total consumption of the limiting substrate(s).
Deglutathionylase and Glutathione Reductase Activities of TGR Proceed via a Grx Monothiol Mechanism
The redox active site of the Grx domain of TGR contains the 31CPYC34 redox center. To investigate whether the Grx domain-dependent activities require one or both Cys residues (i.e. if the mechanism is monothiolic or dithiolic), we generated Cys to Ser mutants in the attacking (nucleophilic) Cys31 and in the putative resolving Cys34 (TGRC31S and TGRC34S, respectively) and assayed their deglutathionylase and GR activities. The TGRC31S mutant had negligible deglutathionylase and GR activities when compared with wild-type TGR (Fig. 3, A and B). In contrast, both the deglutathionylase and GR activities of the TGRC34S mutant were only slightly lower, indicating that Cys34 does not significantly contribute to these activities (Fig. 3, A and B). These results indicate that the mechanism is primarily monothiolic. The fact that the truncated TGR mutant lacking Sec does not possess activity indicates that the Cys31 glutathionylated intermediate is reduced by the Sec residue of the C-terminal redox center. Unexpectedly, the typical hysteretic behavior of GR activity for the wild-type enzyme was significantly attenuated in the TGRC34S mutant (Fig. 3C). This implicates the Cys34 residue in the observed hysteretic phenomenon. It also reveals that the molecular basis underlying hysteresis is more complex than what has been anticipated (6).
FIGURE 3.
Deglutathionylase and GR activity of TGR mutants in the Grx domain. The deglutathionylase (A) and GR (B) activities of TGRWT (black box), TGRC34S (black triangle), and TGRC31S (black circle) were compared at 40 μm peptide-S-SG or GSSG, 100 μm NADPH, and 2 nm enzyme. Reaction mixtures without enzyme (white diamond) were also assayed for comparison. Full time courses for NADPH oxidation (A340 decrease) are shown. C, GR activities of TGRWT (black box), TGRC34S (black triangle), and TGRC31S (black circle) were compared at hysteresis conditions for the wild-type form as follows: 2 nm (low enzyme concentration) and 1 mm GSSG (high GSSG concentration). For TGRC34S, additional enzyme concentrations were assayed: 0.5 nm (light gray triangle) and 1 nm (dark gray triangle). In all cases full time courses for NADPH oxidation (A340 decrease) are shown. Note that in each case the overall NADPH oxidation (A340 decrease) corresponds to the total consumption of the limiting substrate. The results show that deglutathionylation and GSSG reduction are primarily monothiolic (A and B) and that TGRC34S displays a significantly attenuated hysteretic behavior at high GSSG concentration (C).
Cytosolic and Mitochondrial Trxs Can Reduce GSSG at High [GSSG]/[GSH]
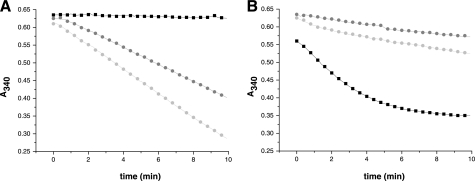
Because TGR cannot reduce GSSG at high GSSG concentrations, we addressed whether there are alternative pathways for GSSG reduction operative under these conditions. Thus, we tested whether E. granulosus Trxs can effectively reduce GSSG under conditions at which the Grx domain of TGR is inhibited (i.e. under hysteresis). For this purpose, we used the TGRC31S mutant (able to reduce Trxs, but devoid of GR activity), in combination with cytosolic and mitochondrial Trxs at 1 mm GSSG. Both Trxs were able to reduce GSSG in the presence of NADPH (Fig. 4A). However, at low GSSG concentrations, reduction of GSSG by Trxs was not as efficient as at high GSSG concentrations. Besides, GR activity at low substrate concentration was more efficient through the Grx domain of TGR than via Trxs (Fig. 4B). These results demonstrate the following: (i) Trxs can accept reducing equivalents from the TR domains of TGR and shuttle them to GSSG, and (ii) the existence of two alternative and complementary pathways for GSSG reduction in both mitochondria and cytosol; although the route via the Grx domain of TGR is operative at low GSSG concentrations, the one via Trxs is operative at high GSSG concentrations.
FIGURE 4.
Alternative pathways for GSSG reduction. The GR activity of TGRWT was compared with that of Trxs under different assay conditions. A, GR activities of TGRWT (black box), cTrx (dark gray circle), and mTrx (light gray circle) at 1 mm GSSG. B, GR activity of TGRWT (black box), cTrx (dark gray circle), and mTrx (light gray circle) at 40 μm GSSG. TGRC31S, devoid of GR activity, was used to regenerate the reduced forms of cTrx and mTrx. Time courses for NADPH oxidation (A340 decrease) are shown. The reaction mixtures contained 100 μm NADPH, 2 nm TGRWT, or TGRC31S and 10 μm cTrx or mTrx. The results show that both Trxs efficiently reduce GSSG at high concentrations.
Cytosolic and Mitochondrial Trxs Can Deglutathionylate Peptide-S-SG Mixed Disulfides
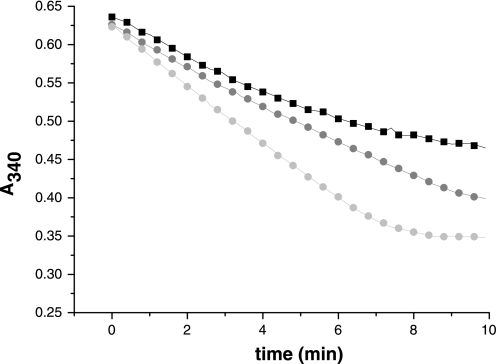
We next addressed whether E. granulosus Trxs provide an additional route for reduction of mixed disulfides as well. For this purpose, we used TGRC31S mutant (able to reduce Trxs but devoid of deglutathionylase activity) in combination with cytosolic and mitochondrial Trxs. The results are presented in Fig. 5. Both Trxs were able to reduce the peptide-S-SG mixed disulfide in the presence of NADPH, revealing the existence of pathways for deglutathionylation that are not dependent on the Grx domain of TGR. These Trx-dependent routes for deglutathionylation would be particularly relevant at high GSSG concentrations, because TGR is unable to deglutathionylate mixed disulfides under these conditions (see Fig. 2). Given that Trxs offer a rescue pathway for GSSG reduction (see Fig. 4A) and that they also reduce glutathionylated peptides, our results indicate that, in all likelihood, Trxs also constitute a rescue pathway for deglutathionylation at high GSSG concentrations.
FIGURE 5.
Alternative pathways for deglutathionylation. The deglutathionylase activity of TGRWT (black box) was compared with that of cTrx (dark gray circle) and mTrx (light gray circle) at 40 μm peptide-S-SG. TGRC31S, devoid of deglutathionylase activity, was used to regenerate the reduced forms of cTrx and mTrx. Time courses for NADPH oxidation (A340 decrease) are shown. The reaction mixtures contained 100 μm NADPH, 2 nm TGRWT, or TGRC31S and 10 μm cTrx or mTrx. Note that the overall NADPH oxidation (A340 decrease) corresponds to the consumption of 40 μm peptide-S-SG, which is the limiting substrate.
Docking and Sequence Analysis of Trxs Suggests That Mitochondrial and Cytosolic Trxs Reduce GSSG in a Similar Manner
Some cytosolic Trxs are known to reduce GSSG (e.g. those from D. melanogaster, Plasmodium falciparum, S. cerevisiae, and Homo sapiens), but no information is available on mitochondrial isoforms, except for E. granulosus (this study). We performed a multiple sequence analysis, including several cytosolic and mitochondrial Trxs from different species. Conserved residues, including those known to be Trx signatures, were identified in all proteins, and overall, no obvious differences between mitochondrial and cytosolic Trxs could be observed (supplemental Fig. S1). Furthermore, when looking at the structural level, we found that the most consistently conserved region is around the active site (supplemental Fig. S2A). This region plays a crucial role in the recognition and reduction of protein disulfide substrates. GSSG and glutathionylated peptides are probably recognized and reduced in a similar manner to other thioredoxin disulfide substrates. Preliminary docking analysis conducted on cytosolic E. granulosus Trx supports this hypothesis, i.e. an energetically favored docking site for GSSG is superimposable with the highly conserved active site region (compare supplemental Fig. S2, A and B). This, together with the conservation of most active site features (i.e. primary, secondary, and tertiary structure) among mitochondrial and cytosolic Trxs, would suggest these oxidoreductases share the ability to reduce GSSG. Furthermore, a recent study demonstrated that only a few residues of S. cerevisiae Trx (Protein Data Bank code 3f3r) are involved in GSH stabilization, and only one of them (Met72, numbering of code 3f3r) is involved in H-bond interactions with GSH. Mutations in this position introducing positively charged residues increase the stability of the glutathionylated Trx complex, lowering the turnover rate for GSSG reduction (26). Interestingly, this position is well conserved in both cytosolic and mitochondrial Trxs (Met, in most cases, or similarly sized hydrophobic residues, see supplemental Fig. S1). The other conserved positions are located in the structural core of the protein and far from the reaction center (supplemental Fig. S2A), where they are involved in structural stabilization of the Trx fold (27).
Linked Trx-GSH System Does Not Efficiently Reduce Nitrosoglutathione
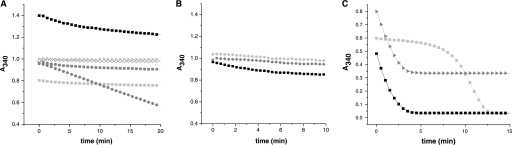
Because GSNO has also been shown to be a substrate for the Trx system (28, 29), we investigated whether the E. granulosus linked Trx-GSH system could play a role in nitrosothiol reduction. We found that TGR catalyzed the NADPH-dependent GSNO reduction but not efficiently; a moderate activity was only observed at high substrate and enzyme concentrations (Fig. 6A). This activity was dependent on the Grx domain. Similarly to what was observed for TGR, neither cytosolic nor mitochondrial Trxs supported efficient reduction of GSNO (Fig. 6B). GSNO has also been reported to be an inhibitor of GR (30). We analyzed whether TGR is inhibited by GSNO and found that the GR activity of TGR (an activity that requires all redox centers to be functionally active) was not affected by this compound (Fig. 6C). Likewise, Trx activity was not inhibited by GSNO (data not shown).
FIGURE 6.
GSNO reduction by TGR and Trxs. A, the ability of TGRWT and Grx-less TGR to reduce GSNO was tested using NADPH as an electron donor and following NADPH oxidation (A340 decrease). For TGRWT, different GSNO and enzyme concentrations were assayed as follows: 2 nm TGRWT and 250 μm (light gray box), 500 μm (dark gray box), and 1 mm (black box) GSNO concentrations, and 40 nm TGRWT and 500 μm GSNO (dark gray circle). For Grx-less TGR, 2 nm enzyme and 500 μm GSNO were used (light gray right triangle). A reaction mixture without enzyme and containing 500 μm GSNO was run for comparison (white diamond). The initial NADPH concentration was 100 μm in all cases. Note that as both reduced NADPH and GSNO contribute to A340, initial absorbance values vary according to GSNO concentrations. A marginal GSNO reduction activity is observed only at high TGRWT. B, TGRWT at 2 nm (black box), and Grx-less TGR (2 nm) plus 10 μm cTrx (dark gray circle) or 10 μm mTrx (light gray circle) were compared for GSNO reduction at 500 μm GSNO and 100 μm NADPH. No significant activity was found for either Trx. C, the effect of GSNO addition on the GR activity of TGRWT was studied by comparing the time courses for NADPH oxidation (A340 decrease) by wild-type TGR (5 nm) with GSSG 100 μm in the absence (black box) or presence (dark gray right triangle) of 500 μm GSNO. The time course of TGRWT (5 nm) at 1 mm GSSG is included as a control of an inhibited TGR (light gray box). The results indicated that at the conditions assayed TGR is not inhibited by GSNO. Note that the differences in initial absorbance values for the reaction mixtures containing different GSNO concentrations are due to the contribution of GSNO to A340.
NO Donors Inhibit TGR
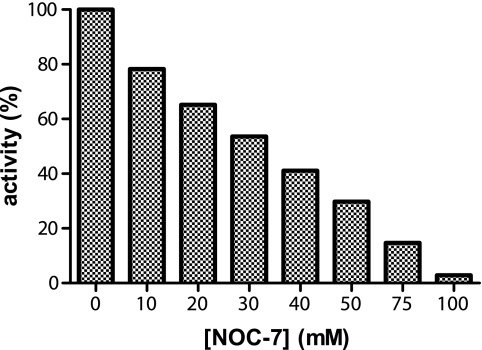
It has been hypothesized that oxadiazole 2-oxides, the recently identified TGR inhibitors and drug leaders against schistosomiasis, may exert its inhibitory effect through NO release (11). In this study, we assayed the effect of NO donors on the TR activity of E. granulosus TGR. NOC-7 was used as a NO donor (half-life 10 min at 22 °C) in a range of concentrations between 10 and 100 μm in the enzymatic assay. A linear dose-dependent inhibition was observed, and complete inactivation was achieved at 100 μm NOC-7 (Fig. 7).
FIGURE 7.
Effect of NO donors on TR activity of TGR. The effect of NOC-7 on the TR activity of TGRWT was evaluated using the DTNB reduction assay. The assay was carried out at 5 mm DTNB, 100 μm NADPH, and 1 nm TGRWT. The corresponding reaction mixtures without DTNB were preincubated for 5 min at room temperature, and the reaction was started by the addition of DTNB. The plot of the residual activity, calculated from the initial velocities under each condition (mean of duplicate experiments) versus NOC-7 concentration, is shown.
Parasites in Vitro Tolerate High Concentrations of NO
After observing an inhibitory effect of NO on TR activity of TGR, we assayed the effect of NO donors on E. granulosus larval worms in vitro. Nonoates were used in a concentration range from 50 μm to 5 mm (NOC-7 as a fast donor and NOC-12 as a slow donor) (see “Experimental Procedures”). No effect was observed with NOC-7 or NOC-12 up to 1 mm concentration. With NOC-7, a significant percentage (30%) of larval worms were killed at 2.5 mm and 100% at 5 mm. Because NO can passively diffuse through membranes, the results indicate that larval worms can tolerate high NO concentrations.
DISCUSSION
In parasitic flatworms, TGR is the key enzyme for reduction of both GSSG and Trx. TGR has also been shown to reduce mixed disulfides using 2-hydroxyethyl mercaptan-glutathione (HED assay) as an artificial substrate. Here, we show that TGR is also able to reduce physiological glutathionylated substrates. This deglutathionylase activity, like GR activity, depends on the Grx domain. The mechanism for both Grx domain-dependent activities of TGR is monothiolic, as observed for the deglutathionylation activity in conventional Grxs. Fig. 8 schematically depicts the proposed mechanism for deglutathionylation and GSSG reduction in TGR. This implies the formation of a glutathionylated intermediate involving the attacking Cys31 and glutathione. In contrast to the mechanism observed for conventional Grxs in which the glutathionylated intermediate is resolved by GSH, in TGR this intermediate can be resolved by Sec. This intramolecular and GSH-independent mechanism avoids the need for GSH diffusion and, at the same time, allows the parasite to work under a broader range of conditions. For example, this would confer an advantage if the GSH/GSSG redox potential becomes too low to allow reduction of the glutathionylated intermediate, such as in oxidative stress situations. In addition, in compartments and localized areas of the cell where GSH concentrations may be low, TGR would support GSH-independent deglutathionylation. These results highlight an advantage of the Grx-TR domain fusion provided by TGR over conventional Grxs.
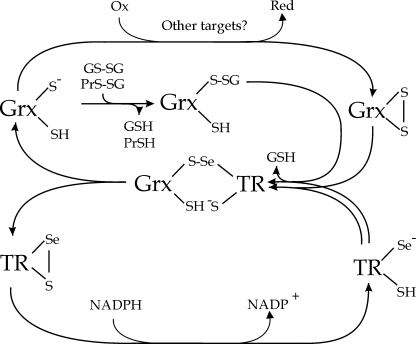
FIGURE 8.
TGR deglutathionylase and glutathione reductase activities occur by a GSH-independent monothiol mechanism. The attacking Cys residue (Cys31) of the Grx redox active center of TGR initiates the mechanism, attacking the GSSG or PrS-SG disulfide, forming glutathionylated Grx intermediate, and releasing GSH or PrSH. The intermediate is resolved by the nucleophilic attack of the selenolate at the C-terminal redox active center of the TR domains of TGR, releasing GSH, and forming a selenenylsulfide intermediate between both redox centers; then, the resolving Cys (Cys34) of the C-terminal redox center attacks this selenenylsulfide forming a C-terminal selenenylsulfide, releasing the Grx domain in its reduced form. Finally, the C-terminal selenenylsulfide is reduced by NADPH in a similar manner as in conventional TRs.
Given that the mechanism for these activities is monothiolic and that the Cys34 mutant displays an attenuated hysteretic behavior, the need for conserving both cysteines in the Grx domain is intriguing. This may reflect either an important role of Cys34 in other functions, such as protein disulfide reduction, and/or selective pressure to avoid an unpaired cysteine thereby protecting Cys31 from oxidative damage. Similar arguments have been proposed for other Grxs with paired cysteines at their active sites, even if the second cysteine has been shown to be detrimental for the Grx activity when compared with the Ser mutant (20).
Both Grx-dependent activities (GR and deglutathionylase) are inhibited at high [GSSG]/[GSH] ratios. In this study, we show the existence of alternative pathways for GSSG reduction and deglutathionylation involving Trxs and the TR domains of TGR. Interestingly, these Trx-dependent pathways are operative at high [GSSG]/[GSH] ratios and would therefore function as a backup. Furthermore, the pathways depending on the Grx domain of TGR and the ones through Trxs have markedly different efficiencies for GSSG reduction, depending on [GSSG]. The Trx-dependent pathways work optimally at high [GSSG] but are inefficient at low [GSSG], and vice versa for the Grx domain-dependent ones. Taken together, these results indicate that alternative and complementary pathways for deglutathionylation and GSSG reduction operate in this parasite, depending on [GSSG]/[GSH] ratios, as depicted in Fig. 9. Alternative pathways via Trxs have been described as backup routes for GSSG reduction in some organisms (12, 13, 31). Our results and those obtained for S. cerevisiae Trxs (32) support the idea that Trxs are also relevant as backup for Grxs deglutathionylase activity. It is important to note that, even though alternative routes for GSSG reduction and deglutathionylation exist, the TR domains are the common node to all of them, reinforcing the concept that the whole redox wiring relies on selenium.
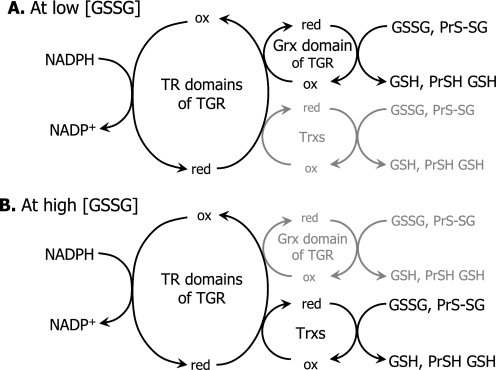
FIGURE 9.
Alternative and complementary pathways for glutathione reductase and deglutathionylase activities in E. granulosus. A, at low GSSG concentrations, proteinS-SG mixed disulfides (PrS-SG) and GSSG are efficiently reduced by TGR, via its “in-built” Grx domain that receives electrons from the TR domains. B, at high GSSG concentrations, the Grx domain is inhibited, and cytosolic and mitochondrial Trxs provide a rescue pathway for GSSG and PrS-SG mixed disulfides reduction.
To our knowledge, mitochondrial Trxs have not been reported as capable of reducing GSSG. This may simply reflect that GSSG reduction by mitochondrial Trxs has not been studied yet. The comparison of mitochondrial and cytosolic Trxs carried out in this study suggests that this may well be the case. Most likely, the ability of Trxs to reduce GSSG and glutathionylated peptides would be due to its general protein-disulfide reductase activity, rather than to specific recognition of the glutathione moiety as in Grxs (33, 34). Recently, the structure of S. mansoni TGR in complex with GSH was solved by x-ray crystallography (35). This study found that GSH was bound to TGR at the Grx domain. The GSH pocket is composed of three segments of the polypeptide chain, each capable of binding noncovalently one of the three residues of GSH. Amino acids Asp84–Gln86 (numbering corresponds to S. mansoni TGR) contour the γ-glutamyl moiety, amino acids Thr71 and Val72 orient the sulfur atom toward the Grx domain active site, and amino acids Lys25 and Gln60 bind the glycine moiety. These are key residues that define the GSH-binding groove in Grxs (33, 36–39) and are also conserved in TGRs (supplemental Fig. S3); however, some differences are also observed, which may confer subtle biochemical differences.
Besides glutathionylation, thiols are subject to modification by nitrosylation, affecting protein function (28). Trxs have been described as one of the thiol denitrosylases, and at the same time they have been shown to be inhibited by nitrosylation. We could only detect a marginal GSNO reductase activity by TGR-dependent pathways, but neither TGR nor Trxs were inhibited by GSNO.
TGR has been shown to be a promising drug target for flatworm infections. Recently, it has been proposed that oxadiazole 2-oxides, identified as potent inhibitors of TGR, exert their action through TGR-triggered NO release (11). Therefore, we studied the effect of well known NO donors on TGR and found that it was susceptible to inhibition by NO donor concentrations in the upper micromolar range (100 μm). Effective oxadiazole 2-oxides operate in the low micromolar range (10 μm) (10). Considering that the drug scaffold may allow significant NO concentrations to be achieved on TGR, our results are consistent with the proposed inhibition mechanism. As for S. mansoni, E. granulosus in vitro-cultured larval worms were resistant to NO donor concentrations up to 1 mm. This suggests that, although parasites are well shielded against NO, NO releasing pro-drugs that can reach sufficient intracellular concentrations and are able to fit into the TGR active site can lead to parasite death. In addition, our results highlight that oxadiazole 2-oxides may serve as drug leads not only for schistosomiasis but also for other flatworm infections.
Supplementary Material
Acknowledgment
We greatly appreciate the gift of pSUABC from Dr. Elias Arnér (Karolinska Institutet).
This study was supported, in whole or in part, by National Institutes of Health FIRCA Grant 1R03TW008588-01 (to G. S.) and Grant GM065204 (to V. N. G.). This study was also supported by Grants PDT 63-105 from the Uruguayan Research Council (to G. S.), and PEDECIBA/ANII (Uruguay) and Comisión Sectorial de Investigación Científica, UdelaR (Uruguay) postgraduate fellowships (to M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Trx
- thioredoxin
- DTNB
- 5,5′-dithiobis (2-dinitrobenzoic acid)
- GR
- glutathione reductase
- Grx
- glutaredoxin
- NOC-7
- 1-hydroxy-2-oxo-3-(N-3-methyl-aminopropyl)-3-methyl-1-triazene
- NOC-12
- 1-hydroxy-2-oxo-3-(N-ethyl-2-aminoethyl)-3-ethyl-1-triazene
- peptide-S-SG
- glutathionylated peptide
- Sec
- selenocysteine
- TGR
- thioredoxin glutathione reductase
- TGRWT
- wild-type TGR
- TR
- thioredoxin reductase
- mTrx
- mitochondrial Trx
- cTrx
- cytosolic Trx.
REFERENCES
- 1. Agorio A., Chalar C., Cardozo S., Salinas G. (2003) J. Biol. Chem. 278, 12920–12928 [DOI] [PubMed] [Google Scholar]
- 2. Alger H. M., Williams D. L. (2002) Mol. Biochem. Parasitol. 121, 129–139 [DOI] [PubMed] [Google Scholar]
- 3. Otero L., Bonilla M., Protasio A. V., Fernández C., Gladyshev V. N., Salinas G. (2010) BMC Genomics 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rendón J. L., del Arenal I. P., Guevara-Flores A., Uribe A., Plancarte A., Mendoza-Hernández G. (2004) Mol. Biochem. Parasitol. 133, 61–69 [DOI] [PubMed] [Google Scholar]
- 5. Sun Q. A., Kirnarsky L., Sherman S., Gladyshev V. N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3673–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonilla M., Denicola A., Novoselov S. V., Turanov A. A., Protasio A., Izmendi D., Gladyshev V. N., Salinas G. (2008) J. Biol. Chem. 283, 17898–17907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun Q. A., Su D., Novoselov S. V., Carlson B. A., Hatfield D. L., Gladyshev V. N. (2005) Biochemistry 44, 14528–14537 [DOI] [PubMed] [Google Scholar]
- 8. Kuntz A. N., Davioud-Charvet E., Sayed A. A., Califf L. L., Dessolin J., Arnér E. S., Williams D. L. (2007) PLoS Med. 4, e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martínez-González J. J., Guevara-Flores A., Alvarez G., Rendón-Gómez J. L., Del Arenal I. P. (2010) Parasitol. Res. 107, 227–231 [DOI] [PubMed] [Google Scholar]
- 10. Simeonov A., Jadhav A., Sayed A. A., Wang Y., Nelson M. E., Thomas C. J., Inglese J., Williams D. L., Austin C. P. (2008) PLoS Negl. Trop. Dis. 2, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sayed A. A., Simeonov A., Thomas C. J., Inglese J., Austin C. P., Williams D. L. (2008) Nat. Med. 14, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanzok S. M., Fechner A., Bauer H., Ulschmid J. K., Müller H. M., Botella-Munoz J., Schneuwly S., Schirmer R., Becker K. (2001) Science 291, 643–646 [DOI] [PubMed] [Google Scholar]
- 13. Tan S. X., Greetham D., Raeth S., Grant C. M., Dawes I. W., Perrone G. G. (2010) J. Biol. Chem. 285, 6118–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 15. Arnér E. S., Sarioglu H., Lottspeich F., Holmgren A., Böck A. (1999) J. Mol. Biol. 292, 1003–1016 [DOI] [PubMed] [Google Scholar]
- 16. Rengby O., Johansson L., Carlson L. A., Serini E., Vlamis-Gardikas A., Kårsnäs P., Arnér E. S. (2004) Appl. Environ. Microbiol. 70, 5159–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bar-Noy S., Gorlatov S. N., Stadtman T. C. (2001) Free Radic. Biol. Med. 30, 51–61 [DOI] [PubMed] [Google Scholar]
- 18. Müller S., Heider J., Böck A. (1997) Arch. Microbiol. 168, 421–427 [DOI] [PubMed] [Google Scholar]
- 19. Holmgren A. (1979) J. Biol. Chem. 254, 9627–9632 [PubMed] [Google Scholar]
- 20. Peltoniemi M. J., Karala A. R., Jurvansuu J. K., Kinnula V. L., Ruddock L. W. (2006) J. Biol. Chem. 281, 33107–33114 [DOI] [PubMed] [Google Scholar]
- 21. Carlberg I., Mannervik B. (1985) Methods Enzymol. 113, 484–490 [DOI] [PubMed] [Google Scholar]
- 22. Arnér E. S., Zhong L., Holmgren A. (1999) Methods Enzymol. 300, 226–239 [DOI] [PubMed] [Google Scholar]
- 23. Luthman M., Holmgren A. (1982) Biochemistry 21, 6628–6633 [DOI] [PubMed] [Google Scholar]
- 24. Giustarini D., Milzani A., Aldini G., Carini M., Rossi R., Dalle-Donne I. (2005) Antioxid. Redox. Signal. 7, 930–939 [DOI] [PubMed] [Google Scholar]
- 25. Livingstone C. D., Barton G. J. (1993) Comput. Appl. Biosci. 9, 745–756 [DOI] [PubMed] [Google Scholar]
- 26. Bao R., Zhang Y., Lou X., Zhou C. Z., Chen Y. (2009) Biochim. Biophys. Acta 1794, 1218–1223 [DOI] [PubMed] [Google Scholar]
- 27. Forman-Kay J. D., Clore G. M., Wingfield P. T., Gronenborn A. M. (1991) Biochemistry 30, 2685–2698 [DOI] [PubMed] [Google Scholar]
- 28. Benhar M., Forrester M. T., Stamler J. S. (2009) Nat. Rev. Mol. Cell Biol. 10, 721–732 [DOI] [PubMed] [Google Scholar]
- 29. Nikitovic D., Holmgren A. (1996) J. Biol. Chem. 271, 19180–19185 [DOI] [PubMed] [Google Scholar]
- 30. Becker K., Savvides S. N., Keese M., Schirmer R. H., Karplus P. A. (1998) Nat. Struct. Biol. 5, 267–271 [DOI] [PubMed] [Google Scholar]
- 31. Kanzok S. M., Schirmer R. H., Turbachova I., Iozef R., Becker K. (2000) J. Biol. Chem. 275, 40180–40186 [DOI] [PubMed] [Google Scholar]
- 32. Greetham D., Vickerstaff J., Shenton D., Perrone G. G., Dawes I. W., Grant C. M. (2010) BMC Biochem. 11, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nordstrand K., Sandström A., Aslund F., Holmgren A., Otting G., Berndt K. D. (2000) J. Mol. Biol. 303, 423–432 [DOI] [PubMed] [Google Scholar]
- 34. Fernandes A. P., Holmgren A. (2004) Antioxid. Redox. Signal. 6, 63–74 [DOI] [PubMed] [Google Scholar]
- 35. Angelucci F., Dimastrogiovanni D., Boumis G., Brunori M., Miele A. E., Saccoccia F., Bellelli A. (2010) J. Biol. Chem. 285, 32557–32567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xia T. H., Bushweller J. H., Sodano P., Billeter M., Björnberg O., Holmgren A., Wüthrich K. (1992) Protein Sci. 1, 310–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia B., Vlamis-Gardikas A., Holmgren A., Wright P. E., Dyson H. J. (2001) J. Mol. Biol. 310, 907–918 [DOI] [PubMed] [Google Scholar]
- 38. Discola K. F., de Oliveira M. A., Rosa Cussiol J. R., Monteiro G., Bárcena J. A., Porras P., Padilla C. A., Guimarães B. G., Netto L. E. (2009) J. Mol. Biol. 385, 889–901 [DOI] [PubMed] [Google Scholar]
- 39. Foloppe N., Sagemark J., Nordstrand K., Berndt K. D., Nilsson L. (2001) J. Mol. Biol. 310, 449–470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.