Abstract
Ionizing radiation (IR) and bleomycin (BLM) are used to treat various types of cancers. Both agents generate cytotoxic double strand breaks (DSB) and abasic (apurinic/apyrimidinic (AP)) sites in DNA. The human AP endonuclease Ape1 acts on abasic or 3′-blocking DNA lesions such as those generated by IR or BLM. We examined the effect of siRNA-mediated Ape1 suppression on DNA repair and cellular resistance to IR or BLM in human B-lymphoblastoid TK6 cells and HCT116 colon tumor cells. Partial Ape1 deficiency (∼30% of normal levels) sensitized cells more dramatically to BLM than to IR cytotoxicity. In both cases, expression of the unrelated yeast AP endonuclease, Apn1, largely restored resistance. Ape1 deficiency increased DNA AP site accumulation due to IR treatment but reduced the number of DSB. In contrast, for BLM, there were more DSB under Ape1 deficiency, with little change in the accumulation of AP sites. Although the role of Ape1 in generating DSB was greater for IR, the enzyme facilitated removal of AP sites, which may mitigate the cytotoxic effects of IR. In contrast, BLM generates scattered AP sites, and the DSB have 3′-phosphoglycolate termini that require Ape1 processing. These DSB persist under Ape1 deficiency. Apoptosis induced by BLM (but not by IR) under Ape1 deficiency was partially p53-dependent, more dramatically in TK6 than HCT116 cells. Thus, Ape1 suppression or inhibition may be a more efficacious adjuvant for BLM than for IR cancer therapy, particularly for tumors with a functional p53 pathway.
Keywords: Apoptosis, Cancer Therapy, DNA Damage, DNA Repair, Radiation Inactivation
Introduction
For nonsurgical cancer therapy, tumor DNA is a frequent target for treatment with ionizing radiation (IR)2 or the radiomimetic drug bleomycin (BLM). IR generates numerous types of damaged bases and abasic sites, along with single strand breaks terminated by 3′-phosphoglycolate esters and other fragmentary products (1). When these occur in clusters affecting both DNA strands, highly cytotoxic double strand breaks (DSB) can be formed either directly from closely opposed single strand breaks or indirectly via the cleavage activity of DNA repair enzymes (2–4). BLM produces a much more restricted set of lesions, namely 4′-oxidized abasic sites and direct strand breaks with 3′-phosphoglycolates (3′-PG). These may be juxtaposed on opposite strands leading to DSB directly or following incision by repair enzymes (3–5). However, intrinsic or acquired resistance of tumors against IR or BLM is a common cause of therapeutic failure (5, 6). Several factors have been proposed to account for this resistance, including increased capacity for the tolerance or repair of DNA damage. Consequently, the inhibition of DNA repair might constitute an effective adjuvant treatment (7).
A significant fraction of DSB repair in mammalian cells is carried out by the nonhomologous end-joining pathway (8–12). The efficiency of nonhomologous end joining would be affected by the nature of the strand break termini, and the location of various lesions at or near the DSB ends. Ligation requires a nick bracketed by normal nucleotides with a 3′-OH and a 5′-phosphate, which means that the 3′-PG at oxidative breaks must first be converted to 3′-OH prior to gap filling (13). The main activity for this excision reaction in mammalian cells is Ape1, which is also the predominant mammalian apurinic/apyrimidinic (AP) endonuclease (14–16). The presence of additional lesions such as oxidized bases or AP sites near the strand breaks at a site of clustered damage would also hinder rejoining. Consistent with this view, the rejoining efficiency of DSB via nonhomologous end joining decreases with increasing structural complexity of a site of clustered lesions (9, 17).
Solo oxidized bases, AP sites, and single strand breaks are repaired primarily by base excision DNA repair (BER) pathways (18). Excision of oxidized bases by DNA glycosylases generates AP sites; oxidized abasic sites such as 2-deoxyribonolactone are also generated as direct free radical products (4, 19). In mammalian cells, Ape1 initiates AP and oxidized abasic site repair by incision on their immediate 5′ side (13, 18). Some glycosylases may also cleave the AP sites they generate (and perhaps other AP sites) by means of their associated AP lyase activities, which leave behind unsaturated abasic residues that still require excision by Ape1 (13). Dual cutting of closely opposed AP/abasic sites (within about 6 bp) generates so-called de novo DSB (17), which account for 30–50% of total radiation-induced DSB, and are major contributors to cytotoxicity (3, 4, 20). In comparison, BLM-induced lesions are simpler, with strand break termini having either directly generated 3′-PG or 5′-abasic residues that result from incision of a 4′-oxidized abasic site (5).
Inhibition of BER to enhance alkylating agent chemotherapy has shown some tantalizing results. Methoxyamine, which reacts with AP sites to prevent incision by Ape1 or AP lyases, increased tumor cell killing upon co-treatment with the potent alkylating drug temozolomide (21, 22). We have investigated the role of Ape1 in repairing IR- or BLM-generated DNA damage and the contribution of the enzyme to cell survival. We show here that, for IR, Ape1 has two opposing effects as follows: increasing cytotoxicity by converting non-DSB clustered lesions to de novo DSBs, but counteracting cytotoxicity by removing toxic AP sites and enabling BER in general. Overall, Ape1 showed a small protective effect against IR damage. In contrast, Ape1 showed a stronger protective effect against BLM toxicity, perhaps due to its role in removal of 3′-PG ends.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions
The immortalized but untransformed TK6 human B-lymphoblastoid cell line and the derived p53-deficient TK6-E6 cells were the generous gifts of Prof. J. B. Little, Harvard School of Public Health. The human colon cancer line HCT116 and its p53 knock-out derivative (HCT116 p53−/−) cells were kindly provided by Dr. Bert Vogelstein, The Johns Hopkins University Medical School. For the re-expression of p53, a vector pcDNA3-FLAG-p53 was obtained from Dr. Zhi-Min Yuan (University of Texas Health Science Center, San Antonio) and was transfected into TK6-E6 and HCT116 p53−/− cells using Lipofectamine 2000 (Invitrogen). The TK6 cells were maintained in RPMI 1640 medium with 10% horse serum and the HCT116 cells in McCoy5A cell medium supplemented with 10% fetal bovine serum, 0.1 mg/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen), at 37 °C in a 5% CO2-humidified atmosphere.
DNA Damage Treatments
In experiments for IR-mediated DNA damage, exponentially growing cells were exposed to 1–10 Gy of x-rays at a dose rate of 0.6 Gy/min from a Philips Electronic Instruments 100 keV MG-100 x-ray machine. For BLM treatment, the exponentially growing cells were treated with the indicated concentrations for 1 h at 37 °C. For both IR and BLM treatments, the cells were then either harvested immediately after the treatment or the medium was exchanged to fresh medium, and the incubation was continued at 37 °C to allow for repair.
siRNA Retroviral Constructs
Sites in the APE1-coding sequence (5′-tgacaaagaggcagcagga-3′) and the luciferase-specific (LUC) negative control (5′-cttacgctgagtacttcga-3′) (23) were chosen as siRNA targets. For each target, a complementary pair of 64-bp oligonucleotides containing the 19-nucleotide target in both sense and antisense orientations, separated by a 9-nucleotide spacer sequence (supplemental Fig. 1), were obtained from OligoEngine (Seattle), annealed, and subcloned into the BglII and HindIII sites of the commercial retroviral siRNA vector pSUPER-retro-puro (OligoEngine). Infection and selection of the infected cell population were conducted using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the supplier's instructions. For the expression of Ape1 (human) or Apex (mouse) proteins, vectors were generated by inserting the coding sequences of APE1 and APEX into the AgeI-BamHI sites of the retroviral vector pQCXIH (Clontech). The APN1 expression vectors have been described previously (23, 24). For LUC studies, the target cells were co-infected with the retroviral expression vectors pRevTRE-Luc and pRevTet-Off (both from BD Biosciences). These expression vectors along with siRNA retrovirus constructs were co-infected into target TK6 or HCT116 cells, and 48 h later the cells were double-selected with the appropriate antibiotics.
Western Blot Analysis
Cells (107) were harvested and resuspended in 100 μl of buffer I (10 mm Tris-HCl, pH 7.8, 200 mm KCl), followed by the addition to the cell suspension of 100 μl of buffer II (10 mm Tris-HCl, pH 7.8, 600 mm KCl, 2 mm EDTA, 40% (v/v) glycerol, 0.2% (v/v) Nonidet P-40, 2 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Sigma)). The mixture was shaken at 4 °C for 40 min to promote cell lysis. The crude lysate was then centrifuged at 16,000 × g for 10 min to remove cellular debris and DNA. Protein concentrations were determined using the Bradford assay (25). After the addition of 2-fold concentrated loading buffer, the samples (each with 30 μg of total protein) were incubated at 95 °C for 1 min and resolved by SDS-PAGE. Proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell), incubated with blocking solution containing 3% powdered milk, and probed with the following appropriate antibodies: anti-Ape1 (sc-17774 Santa Cruz Biotechnology Inc., Santa Cruz, CA), a mouse anti-human Ape1 monoclonal IgG2b, diluted 1:200 in blocking solution, with incubation for 1 h at room temperature; anti-Apn1, a rabbit anti-yeast Apn1 polyclonal antibody from this laboratory (26) diluted 1:20 in blocking solution, with incubation at 4 °C overnight; or anti-β-actin (AC-15; Sigma), a mouse anti-β-cytoplasmic actin-N-terminal peptide monoclonal IgG1, which cross-reacts with actin from multiple species, including humans, diluted 1:500, with incubation for 1 h at room temperature. The protein bands detected were visualized with a fluorescent Western detection (ECF) system (Amersham Biosciences) and quantified by using the Storm 840 phosphorimager (Amersham Biosciences).
Cell Survival Assay
Cells were seeded at a density of 5 × 105 cells per 60-mm dish and exposed to a range of concentrations of DNA-damaging agents. Triplicate plates were used for each dose. The cells were washed with PBS, and fresh medium was added. Dishes were incubated for 4 days at 37 °C in a 5% CO2 incubator. Cells were trypsinized, stained with trypan blue (Sigma), and counted under microscopy. The results are expressed as the number of cells in damage-treated plates relative to control plates. As needed, we verified the trypan blue results with a colony formation assay (27) or a cell viability assay (cell counting kit-8 from Dojindo Laboratories, Gaithersburg, MD, used following the manufacturer's instructions).
Apoptosis Assays
IR- or BLM-treated and untreated TK6 cells (about 105 cells) were harvested, then immediately stained with annexin V-FITC and propidium iodide (PI), and assayed by flow cytometry following the protocol of the manufacturer (BD Biosciences, ApoAlert kits). Flow cytometric analysis was performed to monitor the green fluorescence of FITC-conjugated annexin V (530 ± 30 nm) and the red fluorescence of DNA-bound PI (630 ± 22 nm) by using a Beckman-Coulter ELITE flow cytometer (Kresge Center for Environmental Health Sciences, Harvard School of Public Health), and the data were analyzed with a CellQuest software (BD Biosciences).
Neutral Comet Assay
Double strand DNA breaks were measured by neutral microgel electrophoresis using a commercial comet assay kit (the CometAssay kit, Trevigen, Gaithersburg, MD), following the manufacturer's instructions with some modifications. To prevent cleavage at heat- and/or alkali labile-related DNA strand breaks, all procedures were performed at pH 7.4 and ≤37 °C. Briefly, exponentially growing TK6 cells were treated with BLM or IR as indicated for the individual experiments and then incubated for various times (0–5 h) at 37 °C to allow for repair. After incubation, cells were cooled immediately to 4 °C, and a 50-μl aliquot of cells (at 1 × 105 cells/ml) was added to 500 μl of 0.5% low melting agarose that had been boiled and then cooled at 37 °C for 20 min. After mixing the sample, a 50-μl aliquot was pipetted onto an area of the CometSlide. The slide was incubated at 4 °C for 10 min to accelerate gelling of the agarose disc and then transferred to pre-chilled lysis solution for 30 min at 4 °C. After lysis, the cells were first treated with ribonuclease A for 2 h and then with proteinase K for 48 h, both at 20 °C. Following enzyme digestions, slides were subjected to electrophoresis under neutral conditions (100 mm Tris, 300 mm sodium acetate, pH adjusted to 8.5 with acetic acid) in a horizontal chamber at 1 V/cm) for 10 min at room temperature. The slide was fixed in ice-cold 100% methanol for 5 min, then immersed in 100% ethanol at 20 °C for 5 min, and air-dried. For observation, samples were stained with SYBR Green (Molecular Probes, Eugene, OR), diluted 1:10,000 in 10 mm Tris-HCl, pH 7.5, 1 mm EDTA. Images of comets were visualized with a fluorescence microscope (Nikon, Tokyo, Japan). For DNA damage analysis, we used CometScore 1.5 software (TriTek Corp. Fredericksburg, VA) to compute the tail moment (TM). One hundred cells were examined from each slide. The mean tail moment reflecting DNA damage was calculated by subtracting the control value from the tail moment for the treated cells. Each experiment (triplicate samples per group) was repeated at least three times.
Abasic Site Assay
Genomic DNA was isolated from 2 × 106 cells using Qiagen blood and cell culture DNA kits. Samples of 1 μg of DNA were subjected to an AP site quantification assay based on the aldehyde-reactive probe (ARP kit, Kamiya Biomedical Co., Seattle) according to the manufacturer's instructions. Data are expressed as number of AP sites per 105 nucleotides. In some experiments, to correct for artifactual AP sites formed during the assay, methoxyamine (Sigma) was introduced before the DNA extraction as described previously (24).
Statistical Analysis
Statistical significance between means was determined by using the standard t test and graphed with the mean ± S.E.
RESULTS
Ape1 Deficiency Sensitizes More to BLM than to IR
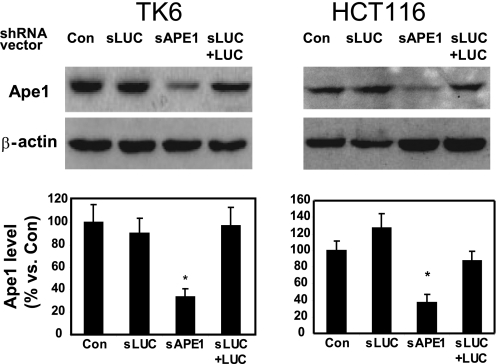
To examine the cellular role of Ape1 in IR- or BLM-induced damage, we used several specific short hairpin RNA (shRNA) molecules expressed from a retroviral vector (pSUPER) to suppress cellular Ape1 levels in human TK6 (immortalized but otherwise normal lymphocytes) (28) and the HCT116 colon tumor line. Because Ape1 is essential even at the cell level (24, 29), we carefully performed shRNA titration experiments (supplemental Fig. 2) and selected an amount of shRNA that partially reduced Ape1 levels to ∼30% of normal (Fig. 1). This partial depletion did not significantly change overall cell proliferation rates or spontaneous apoptosis compared with Ape1-proficient control cells (data not shown). We included two additional controls as follows: shRNA against the nonendogenous luciferase gene (sLUC), and co-expression of sLUC and the target LUC gene to activate RNA interference pathways (Fig. 1).
FIGURE 1.
Ape1 suppression in shRNA-treated cells. TK6 or HCT116 cells were infected with the indicated retroviral vectors, and after 2 days, puromycin (1.5 μg/ml) was added to the culture medium for 5–7 days for selection. Abbreviations used are as follows: Con, uninfected; sLUC, luciferase-specific shRNA vector; sAPE1, APE1-specific shRNA vectors; sLUC+LUC, sLUC, co-expressed with LUC expression retroviral vector. After an additional 3 days of selection, cell samples were subjected to immunoblotting with an Ape1-specific antibody, which was quantified by phosphorimaging and normalized to immunoblotting for β-actin (bar graphs). The upper panels show a representative immunoblot (see “Experimental Procedures” for details) and the lower panels the quantification from three independent experiments. Standard deviations are shown, and the asterisk indicates significant difference from control with p < 0.05.
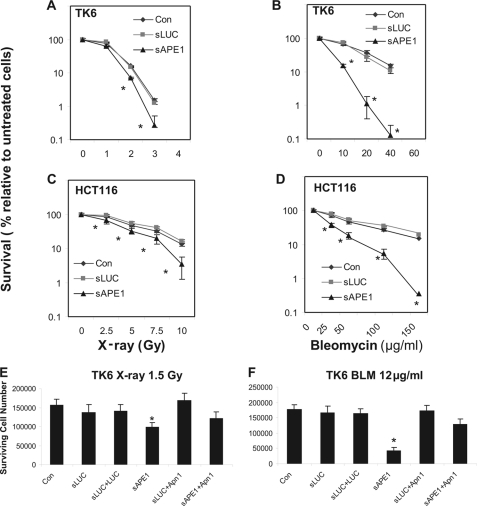
The shRNA-treated and control TK6 and HCT116 cells were exposed to increasing doses of IR or BLM, and the cell survival rates were determined. Both TK6 and HCT116 cells treated with Ape1 shRNA (sAPE1) displayed increased sensitivity to either x-ray or BLM treatment compared with non-shRNA-treated control cells (Fig. 2, A–D), although the relative increase in BLM toxicity was greater than that seen for x-rays. The shRNA control sLUC cells retained normal resistance to both x-rays and BLM (Fig. 2, A–D). Activating RNAi pathways by expression of sLUC and the LUC target gene, without depleting Ape1, did not sensitize TK6 cells to either IR or BLM (Fig. 2, E and F). The TK6 cells appeared to be more greatly sensitized to BLM by Ape1 depletion than were HCT116 cells, although the modest sensitization to x-rays was similar in both lines. The cytotoxicity experiments were routinely analyzed by a cell-counting assay (see details under “Experimental Procedures”) and verified using a colony-forming assay (supplemental Fig. 3).
FIGURE 2.
Cytotoxicity induced by IR or BLM in Ape1-deficient TK6 and HCT116 cells. Cells were untreated (Con) or infected with retroviral siRNA expression vectors for luciferase (sLUC) or APE1 (sAPE1). Additionally, in E and F, the yeast Apn1 protein was co-expressed with sLUC (sLUC+Apn1) or sAPE1 (sAPE1+Apn1) and luciferase was co-expressed with sLUC (sLUC+LUC) as a control for the activation of RNAi pathways. A and B, TK6 cells were treated with increasing doses of x-rays (100 keV, 1–4 Gy) or BLM (10–60 μg/ml, 60 min). C and D, HCT116 cells treated with 2.5–10 Gy x-rays or 25–150 μg/ml BLM for 60 min. E and F, TK6 cells treated with 1.5 Gy x-rays, or 12 μg/ml BLM for 60 min. The cells were washed twice, and the incubation continued for 4 days. One thousand cells per sample were then reseeded in 60-mm dishes or a 96-well plate, and the incubation was continued in fresh medium for 96 h. The cell number was determined by trypan blue cell counting or a commercial cell counting kit (cell counting kit-8). The data were quantified from three independent experiments. Standard deviations are shown, and asterisk indicates significant difference from control with p < 0.05.
Ape1 is a multifunctional protein with activities in DNA repair and transcriptional control (29, 30), so in principle some of the observed effects of Ape1 depletion could be due to such nonrepair functions. To control for this possibility, we co-expressed the Saccharomyces cerevisiae Apn1 protein together with APE1-specific shRNA. Although Apn1 shares repair activities with Ape1 (13), the yeast protein is not a homolog of Ape1, and it lacks other known Ape1 nonrepair functions (13, 30). We have previously shown that Apn1 can replace Ape1 repair functions (24). For both IR and BLM challenges, co-expression of Apn1 with sAPE1 in TK6 cells rescued cell survival to near that observed for the non-shRNA-treated or sLUC controls (Fig. 2, E and F), whereas co-expression of Apn1 with sLUC did not alter cellular resistance. Thus, IR or BLM sensitization by depleting Ape1 in human cells was due to insufficient Ape1-dependent DNA repair. At the same time, the normal level of Ape1 does not seem to be the limiting factor for repair of lethal IR or BLM damage in repair-proficient cells (because normal resistance was not further enhanced by expression of Apn1).
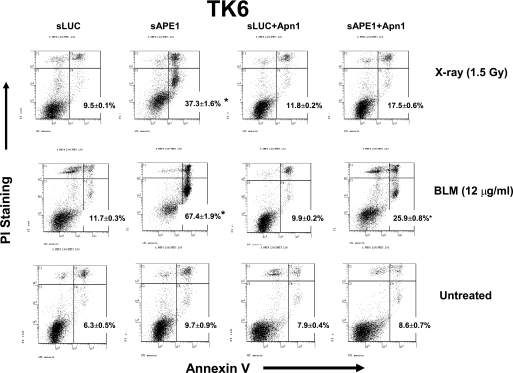
The increased cell death in response to either IR or BLM in Ape1-depleted cells was accompanied by the appearance of morphologic characteristics of apoptosis, such as cytoplasmic shrinkage, and condensed chromatin with an intact nuclear membrane (data not shown). To confirm the findings from cell survival tests, we examined IR- and BLM-induced apoptosis by flow cytometry. Apoptosis was measured by the annexin-V/propidium iodide double-staining method (31). As shown in Fig. 3 and Table 1, 24 h after treatment with 1.5 Gy of x-rays, the fraction of early apoptotic (annexin V-positive and propidium iodide-negative) cells was sharply increased in Ape1-deficient TK6 (sAPE1; from 9.7 to 37%). In contrast, x-ray-induced apoptosis in the control sLUC cells was minimal (increased to 9.5% from 6.3% in mock-transfected cells). For BLM, the apoptotic increase due to depletion of Ape1 was even more dramatic; a 1-h treatment with 12 μg/ml BLM yielded 67% apoptosis with sAPE1, compared with 11.7% with sLUC. Again, co-expression of Apn1 with sAPE1 largely reversed the apoptotic yield (from 37 to 17.5% for IR and from 67 to 26% for BLM). The consistency of the apoptosis data and with the cytotoxicity measurements reinforces the conclusion that repair function(s) of Ape1 are an important factor against IR- and BLM-induced cell killing, with perhaps a greater role against the latter.
FIGURE 3.
Apoptosis induced by IR or BLM in Ape1-deficient TK6 cells. TK6 cells were untreated (Con) or infected with retroviral shRNA expression vectors for luciferase (sLUC), APE1 (sAPE1), or co-expressed with Apn1 (sLUC+Apn1 and sAPE1+Apn1). The cells were treated with x-rays (1.5 Gy) or BLM (12 μg/ml) for 60 min. The cells were washed twice, and the incubation was continued in fresh medium for 24 h. Apoptosis was determined by the annexin V-FITC and propidium iodide assay. The panel shows a representative experiment, and the data were quantified from three independent experiments. The number in each panel indicates the percentage of apoptotic (annexin V-positive) cells and standard deviations. Asterisk indicates significant difference from the untreated control with p < 0.05. The PI staining indicates cell membrane leaking, a hallmark of early necrosis, whereas annexin V-positive indicates early apoptosis. The PI (+)/annexin V (−) (upper left quadrant in each panel) corresponds to necrosis; PI(−)/annexin V (+) (lower right quadrant) indicates apoptosis; PI(+)/annexin V (+) (upper right quadrant) indicates late cell death that could be due to either necrosis or apoptosis; PI(−)/annexin V (−) (lower left quadrant) corresponds to normal viable cells.
TABLE 1.
Annexin V-propidium iodide assay for DNA damage-induced cell death+
See text for details of cell treatment and analysis. An example of a flow cytometry experiment is shown in Fig. 3. Anx indicates annexin.
| Treatment | Transfection | Normal PI(−)Anx(−) | Necrosis PI(+)Anx(−) | Apoptosis PI(−)Anx(+) | Late cell death PI(+)Anx(+) | |
|---|---|---|---|---|---|---|
| % | % | % | % | |||
| 1 | Untreated | sLUC | 76.7 ± 1.6 | 6.7 ± 0.7 | 6.3 ± 0.5 | 10.3 ± 0.4 |
| 2 | Untreated | sAPE1 | 72.7 ± 1.9 | 6.4 ± 0.3 | 9.7 ± 0.9 | 11.2 ± 0.7 |
| 3 | Untreated | sLUC+Apn1 | 71 ± 1.1 | 13.2 ± 0.7 | 7.9 ± 0.4 | 7.9 ± 0.4 |
| 4 | Untreated | sAPE1+Apn1 | 75.4 ± 1.1 | 7.2 ± 0.3 | 8.6 ± 0.7 | 8.8 ± 0.5 |
| 5 | X-ray (1.5 Gy) | sLUC | 56.8 ± 2.0 | 13.4 ± 0.2 | 9.5 ± 0.1 | 20.1 ± 0.6 |
| 6 | X-ray (1.5 Gy) | sAPE1 | 35.4 ± 1.2 | 7.2 ± 0.8 | 37.3 ± 1.6 | 20.1 ± 0.6 |
| 7 | X-ray (1.5 Gy) | sLUC+Apn1 | 70.2 ± 2.1 | 5.3 ± 0.2 | 11.8 ± 0.2 | 12.7 ± 0.4 |
| 8 | X-ray (1.5 Gy) | sAPE1+Apn1 | 61.9 ± 1.7 | 6.1 ± 0.4 | 17.5 ± 0.6 | 14.5 ± 0.6 |
| 9 | BLM 12 μg/ml | sLUC | 60.3 ± 1.2 | 15.7 ± 0.3 | 11.7 ± 0.3 | 12.3 ± 0.3 |
| 10 | BLM 12 μg/ml | sAPE1 | 14 ± 1.1 | 7.3 ± 0.5 | 67.4 ± 1.9 | 11.3 ± 0.5 |
| 11 | BLM 12 μg/ml | sLUC+Apn1 | 72.1 ± 1.9 | 5.3 ± 0.1 | 9.9 ± 0.2 | 12.7 ± 0.9 |
| 12 | BLM 12 μg/ml | sAPE1+Apn1 | 53.5 ± 1.4 | 6.1 ± 0.3 | 25.9 ± 0.8 | 14.5 ± 0.6 |
Ape1 Role in IR or BLM-induced DSB
DSB are thought to be the predominant cytotoxic lesions formed by IR or BLM. We therefore examined the role of Ape1 in DSB repair following treatment with these agents. A modified neutral comet assay was used to quantify cellular DSB (32). The TM, which is estimated as the “comet” tail DNA intensity and migration distance (= % DNA in tail × tail length), was used to quantify total cellular DSBs (32).
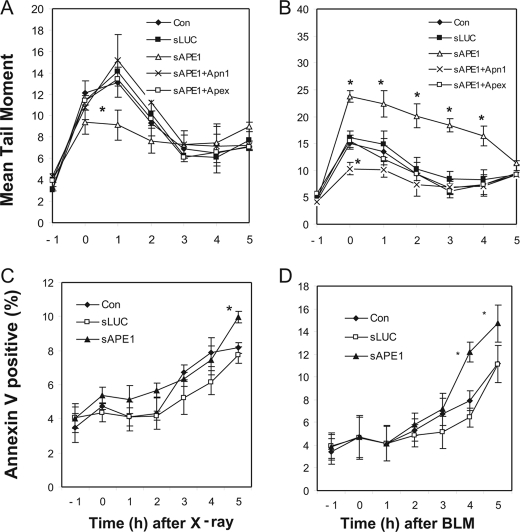
We compared sLUC and sAPE1 treatments with control cells in a kinetic study of DSB formation and repair in TK6 cells. Initial IR exposures were performed under conditions unfavorable for DNA repair (cells chilled on ice), which resulted in overwhelming (>35%) death in sAPE1-treated cells within 5 h after DNA damage (data not shown). That observation may indicate severe repair deficiency in Ape1-deficient cells. In comparison, for IR exposures at room temperature and at 37 °C for BLM, the spontaneous apoptosis rate was ≤5% in APE1-deficient cells (Fig. 4, C and D). These conditions were used for the subsequent experiments.
FIGURE 4.
DSB formation by IR and BLM and subsequent cellular processing. TK6 cells were untreated (Con) or infected with retroviral shRNA expression vectors for luciferase (sLUC), APE1 (sAPE1), or sAPE1 co-expressed with Apn1 (sAPE1+Apn1), or Apex (sAPE1+Apex). A and C, cells treated with x-rays (1.5 Gy). B and D, cells were treated with 12 μg/ml BLM for 60 min and then washed incubated in fresh medium for the indicated times. DSB were quantified as the tail moment in a neutral comet assay (A and B). Apoptosis was determined by the annexin V-FITC/PI assay (C and D). The data were quantified from three independent experiments. Standard deviations are shown, and asterisk indicates significant difference from control with p < 0.05.
Before treatment (−1 h), the basal TM levels were comparable in all the tested groups (Fig. 4, A and B). The cells were exposed to IR or BLM, with the time at the end of the treatment set as 0 h in the kinetic study. As expected, immediately following 2.5 Gy IR treatment, the TM of the no-shRNA or sLUC-treated controls were rapidly increased (2–3-fold relative to the basal level at 0 h). Interestingly, for Ape1-proficient cells, the TM rose continually to a peak at 1 h after treatment, indicating the post-IR formation of DSBs. By 3 h post-IR, the TM had decreased to <30% of its peak value and stayed at this level through at least 5 h (Fig. 4A). For Ape1-deficient cells, the post-damage DSB increase (0–1 h) was not observed (Fig. 4A). The post-damage DSB could be caused by delayed damage after exposure or by the generation of de novo DSB during repair of clustered lesions. To address these possibilities, the cells were co-transfected with sAPE1 and with expression vectors for Apn1 or the mouse Apex protein (whose mRNA is immune to siRNA against human APE1, due to six mismatched bases present in the target sequence; data not shown), and post-damage DSB peaks reappeared in both cases (Fig. 4A). Thus, the post-damage DSB are associated with repair nuclease activity, consistent with lesion processing generating the extra DSB post-IR.
For BLM damage (Fig. 4B), the dynamics of DSB changes were strikingly different from what was seen with IR. The TM of the no-shRNA control cells increased during the 1-h BLM exposure to its peak (about 3-fold over untreated cells) and then monotonically declined. There was no post-damage DSB peak in either Ape1-deficient or Ape1-proficient cells. However, the initial DSB damage (at 0 h) was more severe in Ape1-deficient cells (sAPE1) than in the no-shRNA or sLUC controls. The TM of Ape1-deficient cells remained significantly higher than for the controls (control or sLUC) during 0–4 h after BLM damage, with the slower recovery finally reducing the DSB levels in all cases to about the same level after 5 h. As for the IR treatment, the expression of either Apex or Apn1 in sAPE1-treated cells completely restored the DSB to the control levels (Fig. 4B), which indicates that the DSB changes were caused by insufficient Ape1 repair. Strikingly, Apn1 expression in Ape1-proficient cells reduced the TM even below control levels, which Apex did not do (Fig. 4B, 0 h). Such an effect would be consistent with the more active 3′-processing activity of Apn1 (33) compared with mammalian Ape1 family proteins (13).
Extending the repair incubation until 8 h did not allow complete elimination of DSB for any of the samples tested (data not shown). It has been reported that, during early apoptosis, the nucleus releases large DNA fragments that could contribute to the comet tail (34). To investigate whether apoptosis was a confounding factor, we used annexin V/propidium iodide double staining. Small increases in the number of apoptotic cells were detected at 3 h post-treatment for most of the samples (Fig. 4, C and D). Therefore, a fraction of the comet DNA may come from apoptotic cells 3 h or later after treatment with the IR or BLM. However, most of the post-IR DSB formation seemed to occur within the 1st h after the initial damage, so that apoptosis should not be a factor. In BLM-damaged cells, the eventual apoptosis observed at 4–5 h post-treatment was significantly higher with Ape1 deficiency compared with the controls (Fig. 4D). However, the extra DSB in Ape1-deficient cells were observed immediately after BLM treatment, after which they declined (Fig. 4B). This observation would indicate that the defective repair of BLM-induced DSB in Ape1-deficient cells potentiates the later apoptotic death of these cells.
Ape1 Role in Repair of Abasic Sites Induced by Radiation and BLM
IR and BLM both also produce another type of DNA damage, abasic sites, both as directly free radical products and indirectly by the action of DNA glycosylases on damaged bases. The latter are hydrolytic AP sites, although the former (direct) products include various oxidized abasic residues. Both IR and BLM produce large quantities of AP sites, 10–20-fold the number of DSB (24, 35, 36), such that AP sites are also a significant source of cytotoxicity. In mammalian cells, Ape1 protein is the major repair enzyme to initiate the removal of AP sites. We therefore evaluated the formation of abasic sites in TK6 cells treated with IR or BLM.
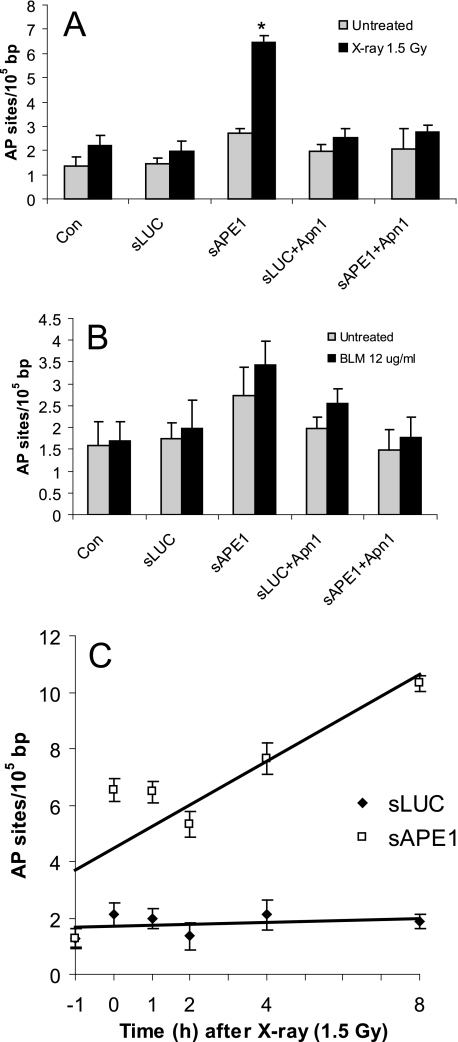
As shown in Fig. 5A, 1 h after exposure to IR, the residual number of abasic sites was the same in two control samples (Con and sLUC cells) compared with unirradiated cells, reflecting efficient repair. As expected for sAPE1-treated cells, residual IR-induced AP sites were increased (2.4-fold) compared with the controls. For BLM treatment, no difference in the level of abasic sites was found among the Ape1-proficient cells samples. For Ape1-deficient cells, the basal level of abasic sites was slightly higher than in the Ape1-proficient samples, and little additional change was observed due to BLM damage. Ectopic expression of Apn1 in the Ape1-deficient cells diminished the abasic site level to that for the Ape1-proficient cells (Fig. 5A). These results in conjunction with the previous cytotoxicity studies (Fig. 2E) support the hypothesis that abasic sites have a major cell-killing potential in IR damage.
FIGURE 5.
Formation of abasic sites in Ape1-deficient cells. TK6 cells were untreated (Con) or infected with retroviral shRNA expression vectors for luciferase (sLUC) and APE1 (sAPE1) or co-expressed with Apn1 (sLUC+Apn1 and sAPE1+Apn1). The cells were treated with 1.5 Gy x-rays (A) or BLM at 12 μg/ml for 60 min (B). At 1 h after finishing the treatment, the cells were subjected to DNA extraction, and the levels of abasic sites were determined using an aldehyde-reactive probe. C, kinetics of abasic site levels following IR (1.5 Gy x-rays). The data were quantified from three independent experiments. Standard deviations are shown, and asterisk indicates significant difference from untreated control with p < 0.05. The data were used to generate the least squares lines shown in the figure.
The persistence of abasic sites in Ape1-deficient cells indicates a mechanism for their modest hypersensitivity to IR (Figs. 2A and 4C). It is possible that Ape1 has opposing effects in IR-induced cell killing, one involving increased cell killing via de novo DSB generation and the other a protective role, by removing toxic IR-induced AP sites. Overall, the Ape1 protective effect appears to be slightly higher for TK6 cells. In addition, we also examined the effect of Ape1 on the kinetics of AP site levels. Samples were obtained from cells 1 h before the treatment and 0 h (immediately post-IR) and 2–8 h after x-irradiation. This study (Fig. 5C) showed increased abasic sites in Ape1-deficient cells (more than 3-fold over Ape1-proficient cells) that were apparent immediately after IR and persisted up to 2 h later. Because the latter increase (4–8 h post-IR) was coincident with the rise of apoptosis (Fig. 4C), some of the observed AP sites may be a consequence of apoptosis.
p53 Is Involved in BLM-induced Cell Death in Ape1-deficient Cells
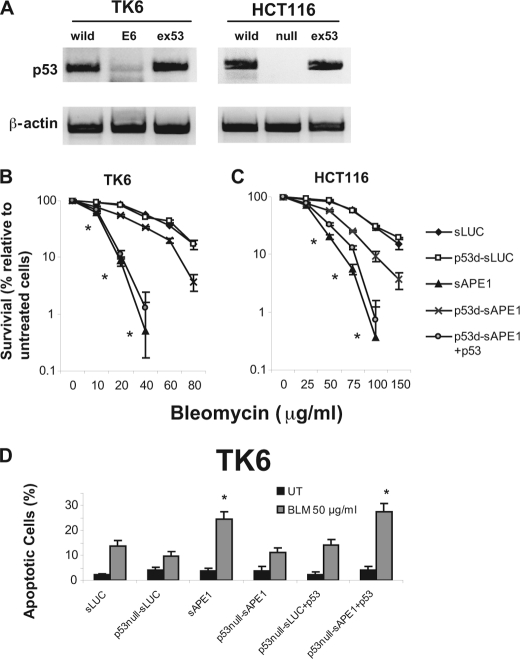
The p53 protein is an important mediator of cellular DNA damage responses. The TK6 and HCT116 cell lines used in our studies both preserve normal p53 signaling pathways (37, 38). To determine whether p53 plays a role in DNA damage-induced cell death, we compared these cells to p53-deficient cells derived directly from them; TK6-E6 expresses human papillomavirus-16 E6 protein, which causes depletion of endogenous p53 by proteolysis (39), and HCT116-p53−/− cells have had both p53 alleles disrupted in cell culture (38). The reduced p53 levels were confirmed by immunoblotting (Fig. 6A). TK6-E6 and HCT116-p53−/− cells were subjected to sAPE1 treatment reducing the Ape1 level to ∼30% of normal (data not shown). Because previous cytotoxicity studies (Fig. 2, A and B) showed that Ape1 deficiency had a more pronounced effect toward BLM damage than for IR, we examined only BLM toxicity in these experiments. When Ape1 was expressed at normal levels, survival assays (conducted as described for Fig. 2, B and D) indicated that the p53 status made no difference in either TK6 (Fig. 6B) or HCT116 cells (Fig. 6C). As already shown in Fig. 2, Ape1 suppression in both p53-proficient cell types strongly sensitized them to BLM. However, p53 suppression in TK6-E6 (Fig. 6B) or elimination of p53 in HCT116-p53−/− (Fig. 6C) significantly alleviated the BLM hypersensitivity. Importantly, restoring p53 activity in p53-deficient cells by transfection with the vector pcDNA3-FLAG-p53 (Fig. 6A) restored the BLM hypersensitivity in both cell lines (Fig. 6, B and C), demonstrating a specific effect of p53. The cell counting results were verified in colony-forming assays (supplemental Fig. 4).
FIGURE 6.
Effect of p53 deficiency on BLM sensitivity in TK6 and HT116 cells. A, level of p53 protein was examined by immunoblotting in TK6 cells (transfected with vector only (WT) or with E6 protein (E6) or E6 with ectopic expression of p53 (ex53)) and in HCT116 cells (WT; homozygous p53 deficiency (null); null with ectopic expression of p53 (ex53)). The upper panels show p53, and the lower panels show β-actin as a loading control. B and C, BLM sensitivity of cells treated with control siRNA-LUC (sLUC) or APE1-specific siRNA (sAPE1) as a function of p53 status (p53d, p53-deficient). BLM treatment was as described for Fig. 2. D, BLM-induced apoptosis in TK6 cells as a function of p53 status. Cells were infected with the control (sLUC) or APE1-specific siRNA (sAPE1), challenged with BLM (30 μg/ml, 1 h), or untreated (UT) and 24 h later assayed for apoptosis by annexin V-FITC and propidium iodide staining. The data were quantified from three independent experiments. Standard deviations are shown, and asterisk indicates significant difference from sLUC control with p < 0.05.
Because of the relative sensitivity of TK6 cells to Ape1 depletion during exposure to BLM, we also compared the effect of p53 status on the BLM-induced apoptosis rate in cells using the annexin V/propidium iodide double-staining method. BLM-induced apoptosis in TK6-E6 was only about 30% that observed for the p53-proficient TK6 counterpart (Fig. 6D), consistent with the survival assays. Again, re-expression of p53 in TK6-E6 cells restored normal BLM-induced apoptosis (Fig. 6D). We conclude that Ape1-proficient cells efficiently eliminate bleomycin-induced DNA lesions that would otherwise trigger p53-dependent apoptosis. Conversely, p53-deficient human cells would be less dependent on their Ape1 status for BLM resistance.
DISCUSSION
Radiation is a widely used therapy for treating various types of cancers, and it remains one of most effective modalities. An antibiotic isolated from Streptomyces verticillus, BLM also acts as an effective antitumor agent (5). In combination with cisplatin and etoposide, BLM is 90% curative for testicular cancer (40). These two anti-cancer agents (IR and BLM) share a similar tumor-killing mechanism: the production of large quantities of difficult-to-repair DSB lesions, which can be highly toxic to growing and dividing cells (41). Those generated by BLM and IR include oxidative DNA lesions (1, 5), which complicate their handling in cells. These agents are less effective for many other cancers, which may have a good initial response but eventually develop resistance. The basis of this developed resistance has been elusive, with recent proposals related to cancer stem cells. Cancer stem cells are ascribed with the capacity to initiate and sustain the growth of heterogeneous tumors through self-renewal and differentiation (42). The underlying resistance mechanisms could include lower rates of proliferation, more efficient DNA damage response and repair, increased drug efflux activity, defects in apoptotic pathways, and lower cellular free radical levels (43). As novel DNA repair pathways have not been reported for cancer stem cells (44), the inhibition of known DNA repair pathways may be a useful approach to enhance treatment efficacy (7).
Our cytotoxicity results showed that Ape1 deficiency sensitized cells more greatly toward BLM than to IR, which is consistent with previous reports using chemical inhibitors or ectopic Ape1 expression (45, 46). However, the underlying mechanisms remained unclear. For IR, a dose of 1 Gy produces (per 106 bp) about 2 DSB, 32 single strand breaks, 29 oxidized purines, 25 abasic sites, and 10 oxidized pyrimidines (36, 47). Approximately 15% of the modified bases, abasic sites, and single strand breaks occur in clusters of damage (17). Compared with isolated single lesions, BER is less effective in repairing clustered DNA damage (48). A major problem is at the DNA ligase step; in contrast, abasic site cleavage by Ape1 was less affected. When an AP site was positioned 1–5 residues 5′ to an 8-oxo-G in the complementary strand, incision at 8-oxo-G by DNA glycosylases was slowed, but AP site incision was not impaired. For two AP sites positioned on opposing strands, both AP sites were incised, giving rise to a DSB (48). An earlier study of Ape1 incision showed polar effects at clustered lesions, and for pairs of synthetic abasic sites at various places in opposite strands, there was more interference when they were positioned 5′ with respect to one another than for the 3′ arrangement (49). A recent study compared normal, under-, or overexpression of human NTH1 or OGG1 in TK6 cells and found that the production of de novo DSB correlates positively with the level of either glycosylase (3). This study also showed that glycosylase overexpression may even enhance cell killing as a consequence of excessive glycosylase activity. Our data suggest that Ape1 cleavage activity might contribute at least 30% of de novo DSB following x-irradiation (Fig. 4A, 1-h time point). Consistent with this interpretation for human cells, Ape1-dependent generation of DSB in transfected DNA-containing lesions closely opposed on opposite strands has been reported for mouse fibroblasts (50). Thus, for IR, Ape1 activity could actually enhance some aspects of IR toxicity.
However, the contribution of Ape1 to the repair of IR and BLM damage is more complex. Isolated base lesions, abasic sites, and DNA strand breaks with 3′-PG termini can interfere with replication and transcription, and they all require Ape1 for efficient processing (13, 24, 51, 52). Ape1 is normally an abundant cellular protein (13), but when this enzyme is limiting, abasic sites may be processed in other ways. An alternative pathway has been proposed involving dual β-elimination/δ-elimination by the NEIL1 glycosylase and removal of the resulting 3′-phosphate by polynucleotide kinase (53). However, other DNA glycosylases with associated AP lyase activity, such as NTH1 protein, would cleave AP sites to yield 3′-terminal unsaturated products of deoxyribose (13), and Ape1 is again required for the efficient excision of these residues. Consequently, some problems in Ape1-deficient cells might arise from the products of AP lyase activity. Moreover, about half of direct DSB induced by IR contain 3′-PG (54), which requires Ape1 for excision (14, 15, 55, 56). Thus, Ape1 may have opposing effects on IR toxicity due to the different roles the enzyme plays in DNA repair, which would limit the effect on cell survival of depleting Ape1.
BLM generates in DNA a small subset of the types of lesions caused by IR. It forms 3′-PG and 4′-oxidized abasic sites that may be clustered, and the prevalence of 3′-PG at DSB termini is a major obstacle for repair and cell survival (5). However, BLM does not produce oxidative base damage, and removing 3′-PG is thought to be a limiting step for strand break repair, because the 3′-PG diesterase activity of Ape1 is about 100-fold slower than its incision of AP sites or the BLM-induced C4′-oxidized residues (57). This somewhat narrower role of Ape1 in repair of BLM damage is consistent with the greater effect on toxicity of suppressing the enzyme and with the persistence of unrepaired DSB in Ape1-deficient cells. Other enzymes have recently been identified that are capable of removing 3′-PG at DSB lesions as follows: tyrosyl-DNA phosphodiesterase (58) and Artemis (59). However, the repair of BLM-induced 3′-PG in cell extracts is strongly dependent on Ape1 (14). It is possible that some of the complex DSB generated by BLM and some other agents (5) require these or other proteins for their repair, and this may be further modulated by other DSB-recognizing proteins such as Ku and DNA-dependent protein kinase (58).
To enhance the effectiveness of DNA-damaging anti-cancer agents, a number of DNA repair inhibitors have been developed that target DNA-dependent protein kinase (DSB repair by nonhomologous end joining) as follows: O6-methylguanine-DNA methyltransferase, poly(ADP-ribose) polymerase-1, and Ape1 (60). Our data indicate that these, perhaps especially the Ape1 inhibitors, should be applied with caution, and their effectiveness in co-treatment will vary with the tumor type and the primary treatment agent. Beyond the greater role played by Ape1 in resistance to BLM compared with IR, the status of the cellular p53 signaling pathway is an important consideration. Based on our data, achieving success in tumor cell killing by BLM may require a functional p53 DNA damage-signaling pathway in the target cells, because p53-deficient cells were far less sensitive to Ape1 depletion for BLM toxicity.
Supplementary Material
Acknowledgments
We are grateful to Jonathan Wing for help with the experiments; to Drs. J. B. Little and B. Vogelstein for the TK6 and HCT116 cell lines, respectively; to Dr. Zhi-Min Yuan for the p53 expression vector; to Amy Imrich for help with the flow cytometry experiments; to the members of the Demple laboratory for helpful discussions, and to Dr. R. A. O. Bennett for a critical reading of the manuscript and help finalizing the figures.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM040000 (to B. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- IR
- ionizing radiation
- AP
- apurinic/apyrimidinic
- PG
- phosphoglycolate
- BLM
- bleomycin
- DSB
- double strand break
- LUC
- luciferase
- sLUC
- luciferase-specific
- TM
- tail moment
- Gy
- gray
- PI
- propidium iodide
- BER
- base excision DNA repair.
REFERENCES
- 1. Von Sonntag C. (1987) The Chemical Basis of Radiation Biology, Taylor & Francis Ltd., London [Google Scholar]
- 2. Milligan J. R., Aguilera J. A., Paglinawan R. A., Nguyen K. J., Ward J. F. (2002) Int. J. Radiat. Biol. 78, 733–741 [DOI] [PubMed] [Google Scholar]
- 3. Yang N., Chaudhry M. A., Wallace S. S. (2006) DNA Repair 5, 43–51 [DOI] [PubMed] [Google Scholar]
- 4. Sutherland B. M., Bennett P. V., Sutherland J. C., Laval J. (2002) Radiat. Res. 157, 611–616 [DOI] [PubMed] [Google Scholar]
- 5. Chen J., Stubbe J. (2005) Nat. Rev. Cancer 5, 102–112 [DOI] [PubMed] [Google Scholar]
- 6. Bernier J., Hall E. J., Giaccia A. (2004) Nat. Rev. Cancer 4, 737–747 [DOI] [PubMed] [Google Scholar]
- 7. Ding J., Miao Z. H., Meng L. H., Geng M. Y. (2006) Trends Pharmacol. Sci. 27, 338–344 [DOI] [PubMed] [Google Scholar]
- 8. Jackson S. P., Jeggo P. A. (1995) Trends Biochem. Sci. 20, 412–415 [DOI] [PubMed] [Google Scholar]
- 9. Pastwa E., Neumann R. D., Winters T. A. (2001) Nucleic Acids Res. 29, E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hopkins B. B., Paull T. T. (2008) Cell 135, 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Williams R. S., Moncalian G., Williams J. S., Yamada Y., Limbo O., Shin D. S., Groocock L. M., Cahill D., Hitomi C., Guenther G., Moiani D., Carney J. P., Russell P., Tainer J. A. (2008) Cell 135, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernstein K. A., Rothstein R. (2009) Cell 137, 807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Demple B., Harrison L. (1994) Annu. Rev. Biochem. 63, 915–948 [DOI] [PubMed] [Google Scholar]
- 14. Izumi T., Hazra T. K., Boldogh I., Tomkinson A. E., Park M. S., Ikeda S., Mitra S. (2000) Carcinogenesis 21, 1329–1334 [PubMed] [Google Scholar]
- 15. Parsons J. L., Dianova I. I., Dianov G. L. (2004) Nucleic Acids Res. 32, 3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen D. S., Herman T., Demple B. (1991) Nucleic Acids Res. 19, 5907–5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Georgakilas A. G., Bennett P. V., Wilson D. M., 3rd, Sutherland B. M. (2004) Nucleic Acids Res. 32, 5609–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sung J. S., Demple B. (2006) FEBS J. 273, 1620–1629 [DOI] [PubMed] [Google Scholar]
- 19. Demple B., DeMott M. S. (2002) Oncogene 21, 8926–8934 [DOI] [PubMed] [Google Scholar]
- 20. Sutherland B. M., Bennett P. V., Sidorkina O., Laval J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu L., Yan L., Donze J. R., Gerson S. L. (2003) Mol. Cancer Ther. 2, 1061–1066 [PubMed] [Google Scholar]
- 22. Liu L., Nakatsuru Y., Gerson S. L. (2002) Clin. Cancer Res. 8, 2985–2991 [PubMed] [Google Scholar]
- 23. Kawai H., Wiederschain D., Yuan Z. M. (2003) Mol. Cell. Biol. 23, 4939–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fung H., Demple B. (2005) Mol. Cell 17, 463–470 [DOI] [PubMed] [Google Scholar]
- 25. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 26. Ramotar D., Kim C., Lillis R., Demple B. (1993) J. Biol. Chem. 268, 20533–20539 [PubMed] [Google Scholar]
- 27. Franken N. A., Rodermond H. M., Stap J., Haveman J., van Bree C. (2006) Nat. Protoc. 1, 2315–2319 [DOI] [PubMed] [Google Scholar]
- 28. Skopek T. R., Liber H. L., Penman B. W., Thilly W. G. (1978) Biochem. Biophys. Res. Commun. 84, 411–416 [DOI] [PubMed] [Google Scholar]
- 29. Izumi T., Brown D. B., Naidu C. V., Bhakat K. K., Macinnes M. A., Saito H., Chen D. J., Mitra S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5739–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evans A. R., Limp-Foster M., Kelley M. R. (2000) Mutat. Res. 461, 83–108 [DOI] [PubMed] [Google Scholar]
- 31. Fadok V. A., Voelker D. R., Campbell P. A., Cohen J. J., Bratton D. L., Henson P. M. (1992) J. Immunol. 148, 2207–2216 [PubMed] [Google Scholar]
- 32. Singh N. P., Stephens R. E. (1997) Mutat. Res. 383, 167–175 [DOI] [PubMed] [Google Scholar]
- 33. Johnson R. E., Torres-Ramos C. A., Izumi T., Mitra S., Prakash S., Prakash L. (1998) Genes. Dev. 12, 3137–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choucroun P., Gillet D., Dorange G., Sawicki B., Dewitte J. D. (2001) Mutat. Res. 478, 89–96 [DOI] [PubMed] [Google Scholar]
- 35. Loeb L. A., Preston B. D. (1986) Annu. Rev. Genet. 20, 201–230 [DOI] [PubMed] [Google Scholar]
- 36. Chen C. Z., Sutherland J. C. (1989) Electrophoresis 10, 318–326 [DOI] [PubMed] [Google Scholar]
- 37. Little J. B., Nagasawa H., Keng P. C., Yu Y., Li C. Y. (1995) J. Biol. Chem. 270, 11033–11036 [DOI] [PubMed] [Google Scholar]
- 38. Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 39. Yu Y., Li C. Y., Little J. B. (1997) Oncogene 14, 1661–1667 [DOI] [PubMed] [Google Scholar]
- 40. Einhorn L. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4592–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Willers H., Dahm-Daphi J., Powell S. N. (2004) Br. J. Cancer 90, 1297–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park C. Y., Tseng D., Weissman I. L. (2009) Mol. Ther. 17, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ghotra V. P., Puigvert J. C., Danen E. H. (2009) Int. J. Radiat. Biol. 85, 955–962 [DOI] [PubMed] [Google Scholar]
- 44. Frosina G. (2010) J. Biomed. Biotechnol. 2010, 845396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robertson K. A., Bullock H. A., Xu Y., Tritt R., Zimmerman E., Ulbright T. M., Foster R. S., Einhorn L. H., Kelley M. R. (2001) Cancer Res. 61, 2220–2225 [PubMed] [Google Scholar]
- 46. Madhusudan S., Smart F., Shrimpton P., Parsons J. L., Gardiner L., Houlbrook S., Talbot D. C., Hammonds T., Freemont P. A., Sternberg M. J., Dianov G. L., Hickson I. D. (2005) Nucleic Acids Res. 33, 4711–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sutherland B. M., Bennett P. V., Sidorkina O., Laval J. (2000) Biochemistry 39, 8026–8031 [DOI] [PubMed] [Google Scholar]
- 48. Lomax M. E., Cunniffe S., O'Neill P. (2004) Biochemistry 43, 11017–11026 [DOI] [PubMed] [Google Scholar]
- 49. Wilson D. M., 3rd, Takeshita M., Grollman A. P., Demple B. (1995) J. Biol. Chem. 270, 16002–16007 [DOI] [PubMed] [Google Scholar]
- 50. Malyarchuk S., Castore R., Harrison L. (2009) DNA Repair 8, 1343–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kingma P. S., Osheroff N. (1997) J. Biol. Chem. 272, 1148–1155 [DOI] [PubMed] [Google Scholar]
- 52. Zhou W., Doetsch P. W. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 6601–6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wiederhold L., Leppard J. B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A. E., Izumi T., Prasad R., Wilson S. H., Mitra S., Hazra T. K. (2004) Mol. Cell 15, 209–220 [DOI] [PubMed] [Google Scholar]
- 54. Henner W. D., Rodriguez L. O., Hecht S. M., Haseltine W. A. (1983) J. Biol. Chem. 258, 711–713 [PubMed] [Google Scholar]
- 55. Suh D., Wilson D. M., 3rd, Povirk L. F. (1997) Nucleic Acids Res. 25, 2495–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chaudhry M. A., Dedon P. C., Wilson D. M., 3rd, Demple B., Weinfeld M. (1999) Biochem. Pharmacol. 57, 531–538 [DOI] [PubMed] [Google Scholar]
- 57. Xu Y. J., Kim E. Y., Demple B. (1998) J. Biol. Chem. 273, 28837–28844 [DOI] [PubMed] [Google Scholar]
- 58. Zhou T., Akopiants K., Mohapatra S., Lin P. S., Valerie K., Ramsden D. A., Lees-Miller S. P., Povirk L. F. (2009) DNA Repair 8, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Povirk L. F., Zhou T., Zhou R., Cowan M. J., Yannone S. M. (2007) J. Biol. Chem. 282, 3547–3558 [DOI] [PubMed] [Google Scholar]
- 60. Madhusudan S., Hickson I. D. (2005) Trends Mol. Med. 11, 503–511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.