Abstract
Transforming growth factor-β (TGFβ) binding to its receptor leads to intracellular phosphorylation of Smad2 and Smad3, which oligomerize with Smad4. These complexes accumulate in the nucleus and induce gene transcription. Here we describe mitogen- and stress-activated kinase 1 (MSK1) as an antagonist of TGFβ-induced cell death. Induction of MSK1 activity by TGFβ depends on Smad4 and p38 MAPK activation. Knockdown of GADD45, a Smad4-induced upstream regulator of p38 MAPK prevents TGFβ-induced p38 and MSK1 activity. MSK1 functionally regulates pro-apoptotic BH3-only BCL2 proteins, as MSK1 knockdown reduces Bad phosphorylation and enhances Noxa and Bim expression, leading to enhanced TGFβ-induced caspase-3 activity and cell death. This finding suggests that MSK1 represents a pro-survival pathway bifurcating downstream of p38 and antagonizes the established pro-apoptotic p38 MAPK function. Furthermore, EGF could reverse all the effects observed after MSK1 knockdown. Monitoring the status of MSK1 activity in cancer promises new therapeutic targets as inactivating both MSK1 and EGF signaling may (re)-sensitize cells to TGFβ-induced cell death.
Keywords: Cell Death, CREB, MAP Kinases (MAPKs), p38 MAPK, SMAD Transcription Factor, MSK1
Introduction
Transforming growth factor-β (TGFβ)2 is the prototypic member of the TGFβ family, which includes also activins and bone morphogenetic proteins (1). TGFβ exerts a broad range of biological activities, such as growth inhibition, differentiation, cell death, wound healing, and immune-modulation, and improper TGFβ signaling correlates with various diseases, such as cancer (2–5).
TGFβ mediates its pleiotropic effects by signaling through transmembrane serine/threonine kinase type I and type II receptors (4, 5). The activated type I receptor signals through phosphorylation of Smad2 and Smad3, which form heteromeric complexes with Smad4. These complexes accumulate in the nucleus, where they control gene expression in a cell type-specific and ligand dose-dependent manner (4, 5). In addition to the Smad pathway, TGFβ can also activate protein kinase pathways that in specific cases do not require Smad-dependent gene transcription (4, 6–9). p38 MAPK is a serine/threonine protein kinase that is activated in response to TGFβ but also by extracellular stress signals (9–13). p38 has an important role downstream of TGFβ signaling as it has been shown to be required for the induction of cell death. TGFβ can activate p38 through two distinct pathways. The first pathway involves GADD45, a Smad4-induced gene, which activates the upstream p38 activator MTK1 (12–14). The second pathway does not directly involve the Smads but instead involves TGFβ-induced activation of the upstream p38 activator TAK1 (TGFβ-activated kinase 1) by the ubiquitin ligase TRAF6 (11, 15). Whether the two mechanisms described leading to p38 activation interact or are completely independent of each other, remains to be explored in greater detail.
In contrast to the mechanistic detail available describing how TGFβ induces p38 activation, less is known about the downstream p38 substrates that mediate cell death (16). Cell death can be initiated by permeabilization of the mitochondrial outer membrane, which allows soluble proteins such as cytochrome c to diffuse from the mitochondrial inter-membrane space to the cytosol (17). Cytochrome c interacts with apoptotic protease activating factor-1 to form a caspase activating complex. Activated caspases subsequently cleave various cellular substrates, a process which ultimately results in cell death.
Permeability of the mitochondrial outer membrane is controlled by the BCL-2 family, which includes anti-apoptotic or pro-apoptotic members. Anti-apoptotic BCL-2 proteins contain four BCL-2 homology (BH) domains and they preserve mitochondrial membrane integrity by directly inhibiting pro-apoptotic BCL-2 proteins (17). Pro-apoptotic BCL-2 members can be classified into effector proteins and BH3 domain-only proteins. The effector proteins Bax and Bak homo-oligomerize upon activation and fuse with the mitochondrial outer membrane to induce mitochondrial outer membrane permeabilization. BH3-only proteins that are able to interact only with the anti-apoptotic BCL-2 family members are referred to as sensitizers or derepressors (and include BAD and Noxa). Bim and Bid are able to interact with the antiapoptotic BCL-2 proteins as well as the effector proteins, and can directly activate Bax and Bak, which are referred to as direct activators. BH3-only proteins can be regulated at both the transcriptional and post-transcriptional level.
TGFβ is well known to act on a cell type- and physiological context-dependent manner in exerting its biological effects (2). With respect to the apoptotic response, TGFβ has been described to induce cell death through the BH3-only direct activator Bim in a p38 MAPK-dependent manner (18). Others have shown that TGFβ-ERK1/2 MAPK-induced phosphorylation (and thus inactivation) of Bad, results in neuroprotection (19). Possibly TGFβ has the potential to alter the balance between the anti-apoptotic and the BH3-only BCL-2 family members depending on the cellular context.
Here we describe mitogen- and stress-activated kinase 1 (MSK1) as a novel factor regulating TGFβ-p38-dependent cell death. MSK1 is a multifunctional kinase that may function as an effector of ERK1/ERK2 or p38 MAPK in TGFβ signaling (20, 21). MSK1 is an 85 kDa nuclear protein closely related to the RSK family of protein kinases. MSK1 contains two separate kinase domains within a single polypeptide (21). Both N- and C-terminal kinase domains are required for full MSK1 activity. The molecular mechanism of MSK1 activation is complex and requires multi-site phosphorylation events by upstream kinases followed by autophosphorylation (21–23). MSK1 is known to phosphorylate a variety of downstream targets, which include the transcription factors CREB, ATF1, RelA, and Stat3 (21, 24–28).
We found that TGFβ can induce MSK1 activity in a p38-dependent manner, which in turn requires the mobilization of Smad4 and transcriptional induction of GADD45. We then present evidence that MSK1 unexpectedly plays a pro-survival role as its knockdown potentiated the activation of caspase 3, reduced Bad phosphorylation, and increased Noxa and Bim expression. With MSK1 we have identified a pro-survival kinase downstream of the p38 pathway that attenuates the extent of TGFβ-p38-induced cell death.
EXPERIMENTAL PROCEDURES
Inhibitors and Antibodies
All inhibitors were from Biomol (Hamburg Germany), except Gö6976 and Gö6983, which were from Calbiochem, PD184352 was a kind gift from J. Lennartson. All phosphospecific antibodies and antibodies against cleaved caspase 3 and Bad were from Cell Signaling Technology, except phospho-Smad2, which was homemade (29). As indicated on the provided datasheet the phospho-CREB antibody recognizes both phosphorylated CREB and phosphorylated ATF1. The Smad4 antibody was from Santa Cruz Biotechnology. MSK1 antibodies were from Santa Cruz Biotechnology and R&D Systems.
Cells and Cell Culture
NMuMG cells, clone 18 (30), were grown in DMEM containing 10% fetal bovine serum (FBS), antibiotics, and 5 mm l-glutamine. Smad4 knockdown cells were generated as described (30). Cells were maintained at 5% CO2 in a humidified atmosphere at 37 °C. For TGFβ stimulation, TGFβ3 was always used at a concentration of 10 ng/ml.
Plasmids
MSK1 plasmids were kindly provided by D. Alessi (21). The GADD45 short hairpin plasmids were from Sigma. The p38 plasmids were provided by E. Nishida.
Transfections
Transfections were performed using Lipofectamine 2000 (Invitrogen) and Dharmafect reagent 1 (Dharmacon). For transient overexpression of GADD45, plasmids were transfected and 24 h thereafter cells were selected overnight with 5 μg/ml puromycin. For siRNA transfection (MSK1 on-target Plus Smartpool, Smad4 on-target Smartpool from Dharmacon), cells were transfected with 100 nm siRNA.
Western Blotting and Detection
Cells were rinsed with ice-cold PBS before lysis in sample buffer. Samples were sonicated 3 × 30 s before boiling. Samples were separated on 10% SDS-polyacrylamide gels using a mini-gel system according to manufactures protocol (Bio-Rad) and transferred overnight to nitrocellulose membranes (Hybond-ECL, Amersham Biosciences) at 4 °C. Transfer efficiency was checked using Ponceau-S. Membranes were blocked with 5% skim milk in TBS-T for 1 h at room temperature. Primary antibodies were incubated for 3 h at room temperature or overnight at 4 °C. Secondary antibodies were incubated for 1 h at room temperature before detection with West-Dura Nano ECL substrate and Biomax films. Experiments were confirmed multiple times. Representative results out of at least three independent repeats are shown.
Viability Assay
Amount of viable cells was determined using an MTS assay according to the instructions of the manufacturer (Promega, Leiden, The Netherlands). Cells were serum-starved overnight before stimulation with TGFβ. After MTS detection cells were fixed with 4% formaldehyde in PBS and visualized using phase contrast microscopy. Digital images were adjusted for brightness and contrast. Experiments were performed in 5-fold and repeated at least four times.
Real-time PCR Analysis and Primers
NMuMG cells were treated as indicated in figures before extraction of RNA using RNeasy (Qiagen). Analysis of mRNA expression was performed as described earlier (31). The PCR primers used were: mouse MSK1, forward, 5′-AGCCACATGCACGATGTAGGA-3′, reverse, 5′-AGGCGTGCAAACCCAAAGT-3′; mouse TBP, forward, 5′-CCGCAGTGCCCAGCATCACT-3′, reverse, 5′-TGGGGAGGCCAAGCCCTGAG-3′; mouse Noxa, forward, 5′-ATAACTGTGGTTCTGGCGCAG-3′, reverse, 5′-TTGAGCACACTCGTCCTTCAA-3′; mouse Bim, forward, 5′-GTCCCGGTCCTCCAGTGGGT-3′, reverse, 5′-CGCAGCTCCTGTGCAATCCG-3′.
RESULTS
MSK1 Is Phosphorylated after TGFβ Stimulation
MSK1 can respond to both mitogen- and stress-induced MAPK pathways (21). Because TGFβ can activate the ERK1/2 and p38 MAPK pathways in mouse breast epithelial NMuMG cells (7, 9, 32), we investigated whether MSK1 is activated in this cell line.
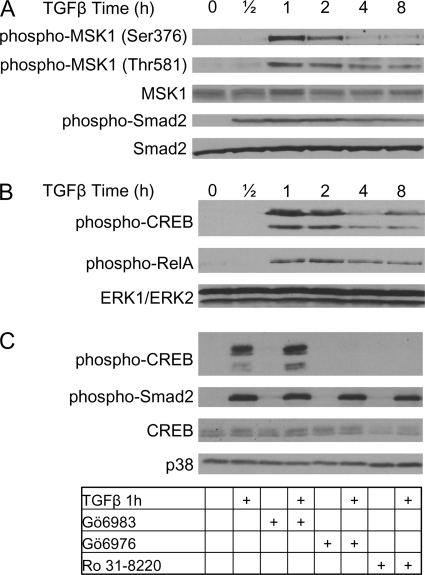
To establish the kinetics of MSK1 phosphorylation, NMuMG cells were stimulated with TGFβ for 0.5 up to 8 h (Fig. 1A). Subsequently, cell lysates were analyzed for MSK1 phosphorylation on residues Ser-376 and Thr-581. Phosphorylation of both Ser-376 and Thr-581 is required for MSK1 catalytic activity (22, 23). In non-stimulated controls (time point 0 h, Fig. 1A) basal MSK1 phosphorylation was very low. MSK1 phosphorylation remained unchanged after 0.5 h of TGFβ stimulation but increased dramatically after 1 h. After 4 h, phosphorylation of both Ser-376 and Thr-581 diminished and thereafter remained constant at lower levels for up to 8 h.
FIGURE 1.
TGFβ activates MSK1 in a p38-dependent manner. A, TGFβ induces MSK1 phosphorylation. NMuMG cells were seeded and grown for 24 h. Growth medium was replaced with serum-free medium, and cells were starved for 24 h before stimulation with TGFβ. TGFβ treatment resulted in the phosphorylation of MSK1 on Ser-376 and Thr-581 within 1 h. Smad2 phosphorylation is shown as a stimulation control and Smad2 total levels as a loading control. B, NMuMG cells were grown as in A. Lysates were probed with phospho-antibodies against CREB and RelA. TGFβ stimulation induces phosphorylation of CREB and RelA. The phospho-CREB antibody recognizes both phospho-CREB (upper band) and phospho-ATF1 (lower band). Total levels of ERK1 and ERK2 are shown as a loading control. Total levels of CREB and RelA remained unchanged during the experiment (data not shown). C, MSK1 inhibitors prevent TGFβ-induced CREB phosphorylation. Cells were grown as in A, but cells were preincubated with various inhibitors for 1 h prior to TGFβ treatment. Lysates were analyzed for phosphorylation of CREB and Smad2. Total levels of CREB and p38 are shown as loading controls. Gö6983 (10 μm) did not prevent CREB phosphorylation and had no effect on Smad2 phosphorylation. Gö6976 (10 μm) and Ro 31–8220 (10 μm) inhibited CREB phosphorylation and had no effect on Smad2 phosphorylation.
TGFβ Induces Phosphorylation of CREB and RelA by MSK1
CREB and RelA are two transcription factors described to be downstream substrates of MSK1 in other cell types (21, 24, 27). Indeed, TGFβ induced the phosphorylation of CREB and RelA in a similar time frame as it induced the phosphorylation of MSK1 (Fig. 1B) suggesting that MSK1 may also regulate CREB and RelA in our cell model. The antibody used to detect phospho-CREB recognized two protein bands, the lower band is phospho-ATF1, whereas the upper band is phospho-CREB (see “Experimental Procedures”).
To confirm that the kinase activity of MSK1 is required for the phosphorylation of CREB and RelA in response to TGFβ, we preincubated NMuMG cells with different kinase inhibitors prior to TGFβ treatment. The inhibitors target several kinases, but all have in common that they potently inhibit MSK1 (Table 1) (33). The PKC inhibitor Gö6983, which does not act on MSK1, was used as a negative control, and did not prevent TGFβ-induced CREB phosphorylation, but consistently resulted in a slight increase in the phosphorylation of CREB and ATF1 (Fig. 1C). The inhibitors Gö6976 and Ro 31–8220 inhibited CREB phosphorylation. Because Gö6976 and Ro 31–8220, but not Gö6983 inhibit CREB phosphorylation, it is likely that MSK1 acts as the upstream CREB and RelA kinase.
TABLE 1.
Inhibitors used in this study and their target
| Inhibitor | Target |
|---|---|
| SB202190 | p38 |
| SB203580 | p38 |
| Ro 31–8220 | MSK1/PKC |
| Gö-6976 | MSK1/PKC |
| Gö-6983 | PKC |
| PD184352 | MEK1/2 |
| PD98059 | MEK1/2 |
| UO126 | MEK1/2 |
TGFβ-induced MSK1 Phosphorylation Is p38 MAPK-dependent
Having established that MSK1 is activated in response to TGFβ, we focused on the upstream pathway. It has been described that in certain cell types MSK1 is phosphorylated by ERK1/2 and p38. Therefore, we preincubated NMuMG cells with specific inhibitors to MEK1/2 and p38 prior to TGFβ stimulation.
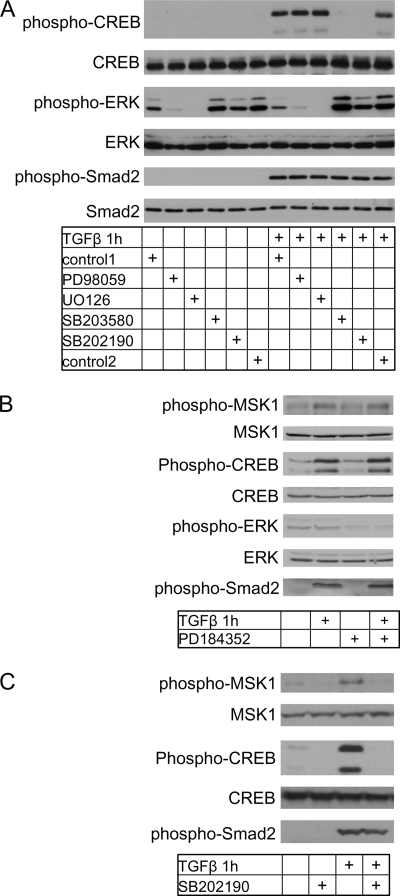
Two MEK1/2 inhibitors, UO126 or PD98059 both failed to prevent CREB phosphorylation (Fig. 2A). An additional MEK1/2 inhibitor PD184352 also did not prevent CREB phosphorylation nor did it prevent MSK1 phosphorylation (Fig. 2B). However, application of the p38 inhibitors SB202190 or SB203580 completely inhibited phosphorylation of CREB (Fig. 2A). SB202190 also prevented phosphorylation of MSK1 (Fig. 2C, SB203580 not shown). Phosphorylated Smad2 was included as a control and was not affected by these inhibitors as expected, whereas total p38 and CREB were used as controls to determine equal sample loading (Fig. 2A). These results suggest that p38 MAPK, but not ERK1/ERK2, plays a prominent role upstream of the MSK1 kinase in response to TGFβ stimulation.
FIGURE 2.
TGFβ activates MSK1 in a p38-dependent manner. A, p38 inhibitors prevent CREB/ATF1. Cells were grown as in Fig. 1C and treated with TGFβ for 1 h. Lysates were analyzed for phosphorylation of CREB. CREB and p38 were analyzed as loading controls. The MEK1/2 inhibitors PD98059 (50 μm) and UO126 (10 μm) did not prevent phosphorylation of MSK1, CREB, RelA, and Smad2. SB202190 (10 μm) and SB203580 (10 μm) did prevent phosphorylation of MSK1, CREB, RelA, but had no effect on Smad2. Control1 indicates the vehicle control for PD98059, control2 indicates vehicle control for U0126, SB202190, and SB203580. The changes in phosphorylation are not due to changes in total protein content as shown by CREB and p38 total levels. B, MEK1/2 inhibitor PD184352 did not prevent MSK1 or CREB phosphorylation. Cells were preincubated with PD184352 (100 nm) for 1 h before TGFβ treatment. C, p38 inhibitor SB202190 prevents MSK1 and CREB phosphorylation. Cells were preincubated with SB202190 (10 μm) for 1 h before TGFβ treatment.
TGFβ-induced p38-MSK1-CREB Phosphorylation Is TAK1-independent and Smad4-dependent
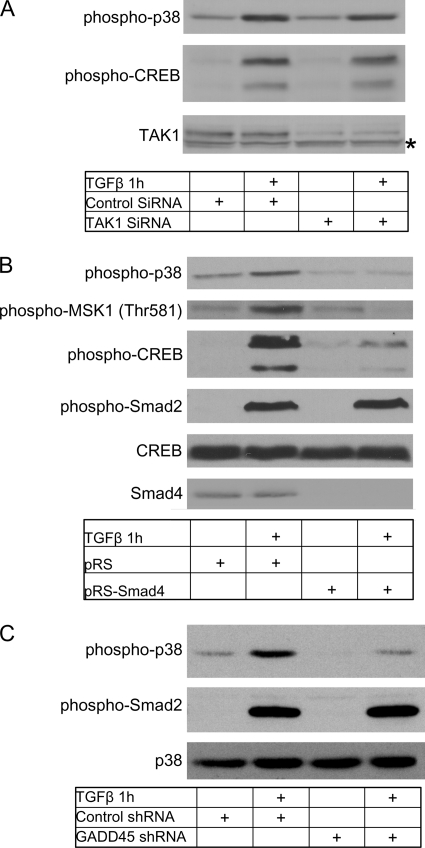
TGFβ-induced p38 activity has been described to depend on either Smad4-mediated transcription or TAK1 activity (11–13, 15). Therefore we knocked down either TAK1 or Smad4 and tested the ability of TGFβ to induce p38 phosphorylation. Transient knockdown of TAK1 with siRNA did not influence TGFβ-induced p38 or CREB phosphorylation (Fig. 3A), suggesting that they are activated in a TAK1-independent manner. To determine if p38-MSK1-CREB signaling requires Smad4, we analyzed NMuMG cells which stably express a short-hairpin RNA (shRNA) directed against Smad4. Treatment of these cells with TGFβ resulted in a greatly reduced phosphorylation of p38 MAPK, MSK1, and CREB. Smad4 knockdown had no effect on Smad2 phosphorylation, as expected, and indicating specificity toward the p38 pathway (Fig. 3B). Because Smad4 appears to be required for TGFβ-induced p38 activity, we presumed that a Smad4 target gene was required. One such candidate gene is Gadd45. We confirmed that all members of the Gadd45 gene family were expressed in NMuMG cells (supplemental Fig. S1) and were induced upon TGFβ treatment in a Smad4-dependent manner (data not shown). Knockdown of Gadd45 and subsequent TGFβ treatment significantly reduced TGFβ-induced p38 activity (Fig. 3C, knockdown efficiency supplemental Fig. S1), supporting a role for Gadd45 as a Smad4 target gene required for p38 activation.
FIGURE 3.
p38-MSK1 activation is Smad-dependent. A, knockdown of TAK1 does not affect CREB or p38 phosphorylation. Cells were plated and grown for 24 h before transfection with siRNA as described (“Experimental Procedures”). Cells were grown for an additional 48 h before transfection medium was replaced with serum-free medium. After 24 h, cells were treated with TGFβ for 1 h. An asterisk (*) shows a nonspecific protein band. B, knockdown of Smad4 inhibits CREB, MSK1, and p38 phosphorylation. Smad4 was knocked down using siRNA as described (“Experimental Procedures”). Cells stably overexpressing empty shRNA vector (pRS) or stably overexpressing Smad4 shRNA (pRS-Smad4) were treated with TGFβ for 1 h and analyzed for CREB, MSK1, p38, and Smad2 phosphorylation. C, knockdown of GADD45 prevents TGFβ-induced p38 phosphorylation. Cells were transfected with shRNA directed at GADD45 and grown for an additional 72 h before stimulation with TGFβ for 1 h. Lysates were analyzed for p38 and Smad2 phosphorylation. p38 was used as a loading control.
MSK1 Knockdown Potentiates TGFβ-induced Cell Death
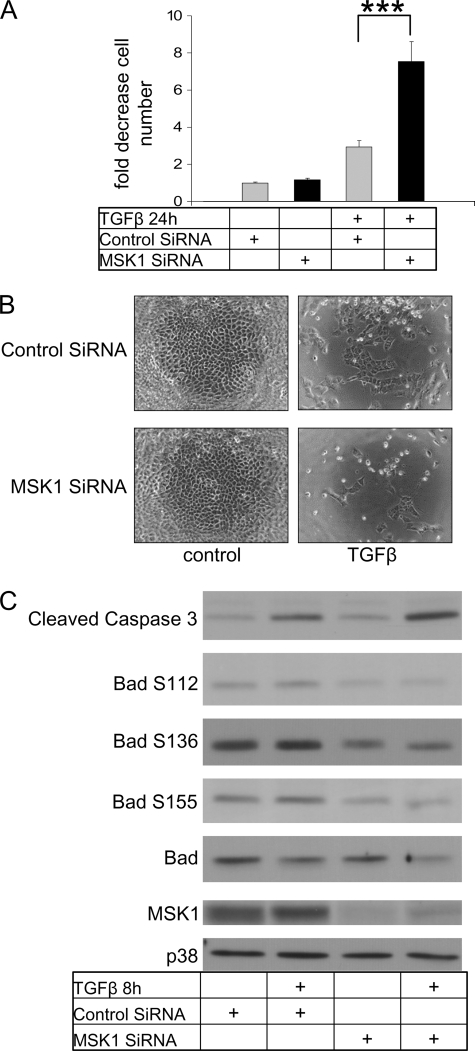
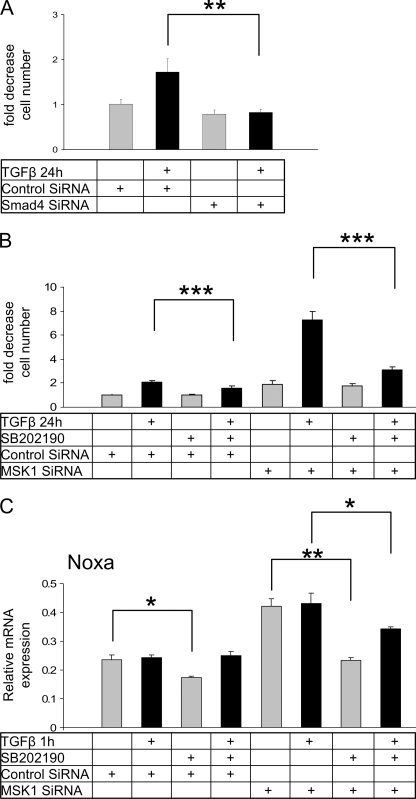
Because p38 activation by TGFβ is an important pathway leading to cell death, which has been previously described in NMuMG cells (18), we analyzed the effect of MSK1 knockdown on TGFβ-induced cell death. To measure viable cell number we used an MTS cell viability assay whereby signal intensity is directly correlated to the amount of viable cells. In controls, TGFβ enhanced cell death by 2.5-fold, whereas in the MSK1 siRNA-treated samples cell death was enhanced by 8-fold (Fig. 4A). The fold-induction reaffirms previously published data showing that TGFβ treatment induced cell death by 3-fold in this cell model (18). The results obtained on cell survival were also confirmed by microscopic inspection of cell numbers following formaldehyde fixation (Fig. 4B). The potentiation of TGFβ-induced cell death after MSK1 knockdown can be observed by the low amount of surviving cells after TGFβ treatment.
FIGURE 4.
MSK1 siRNA potentiates TGFβ-induced cell death. A, MSK1 siRNA potentiates TGFβ-induced cell death. Cells were transfected with siRNA, grown for an additional 72 h and then treated with TGFβ for 24 h and subsequently analyzed for cell death using an MTS assay as described (“Experimental Procedures”). The unstimulated control was set at value of 1. Bar graphs indicate changes relative to unstimulated control, and standard errors are derived from 5-fold determinations. ***, indicates p value < 0.001, as determined by Student's t test with unequal sample variance. B, MSK1 siRNA potentiates TGFβ-induced cell death. Cells were transfected with siRNA, grown for an additional 72 h and then treated with TGFβ for 24 h and subsequently fixed with 4% PFA. Cells were analyzed by phase-contrast microscopy. C, MSK1 knockdown induces caspase 3 cleavage. Cells were transfected with siRNA, grown for an additional 72 h and then treated with TGFβ for 8 h. Lysates were analyzed for cleaved caspase 3, Bad phosphorylation, total Bad, and total p38. MSK1 knockdown increases the amount of cleaved caspase 3, but reduces the amount of total Bad.
As a molecular corollary of cell death induced by TGFβ, we examined the total levels of active caspase 3, the total levels of Bad and of phosphorylated Bad on Ser-112, Ser-136, and Ser-155, as potential mediators of cell death (Fig. 4C). After 8 h of TGFβ treatment there was clearly an increase in the amount of cleaved caspase 3 (the antibody used is directed toward the active caspase 3 fragment). Although Bad phosphorylation appeared to be weakly induced, the total levels of the Bad protein were reduced upon stimulation with TGFβ. Total p38 levels were provided as a control to indicate equal loading and are not affected by TGFβ. The specific decrease in total Bad levels may reflect its cleavage by caspases and degradation as has been reported earlier (34). The cleavage of Bad should result in a Bad fragment which is extremely potent at inducing cell death. Unfortunately, the Bad antibody did not appear to recognize this cleaved Bad form. In agreement with the cell viability data of Fig. 4A, knockdown of MSK1 enhanced cleaved caspase 3 and further reduced Bad levels (Fig. 4C).
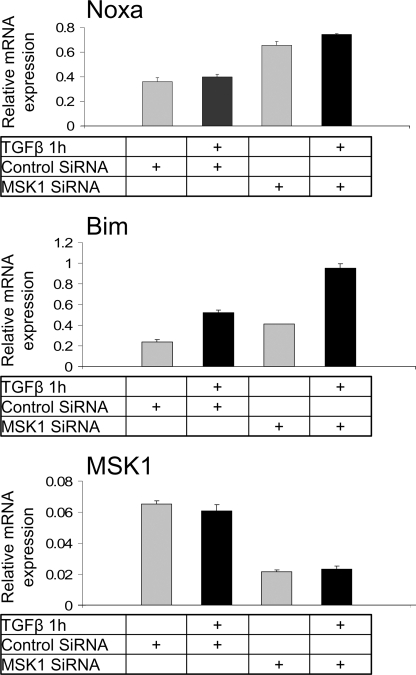
Because TGFβ affected Bad at the post-translational level we screened for other BCL-2 members at the transcriptional level and examined the effect of MSK1 knockdown on these transcripts. Both Noxa and Bim expression levels were increased after MSK1 knockdown, whereas only Bim expression was augmented after TGFβ stimulation (Fig. 5). In summary, MSK1 knockdown increases cell death presumably by decreasing Bad phosphorylation, increasing Noxa and Bim transcription and enhancing caspase 3 cleavage.
FIGURE 5.
MSK1 knockdown enhances the expression of BH3-only Bcl2 family pro-apoptotic members. Top, MSK1 knockdown induces Noxa expression. Cells were transfected with siRNA, grown for an additional 72 h and then treated with TGFβ for 1 h. Total RNA was analyzed for Noxa mRNA expression by quantitative real-time RT-PCR as described under “Experimental Procedures.” Relative mRNA levels normalized to expression of the housekeeping gene Tbp are plotted as averages from triplicate determinations with standard errors. Middle, MSK1 knockdown induces Bim expression. Experimental conditions as in top panel and analysis of Bim mRNA in the same samples as in top panel. Bottom panel, control MSK1 mRNA analysis in the same as above samples verifies the degree of MSK1 silencing.
MSK1 Knockdown Potentiates p38-mediated Cell Death
To gain more mechanistic insight into the role of MSK1 in TGFβ-induced cell death, we examined the role of its upstream regulators, Smad4 and p38. Transient Smad4 knock-down with siRNA revealed that TGFβ-induced cell death completely depended on the presence of Smad4 (Fig. 6A). Because p38 activity has been described to be crucial for TGFβ-induced cell death to occur, and MSK1 knockdown potentiates cell death instead of inhibiting it, we speculated that MSK1 inhibits a second pathway downstream of p38. To test this hypothesis we knocked down MSK1 and inhibited p38 with either SB202190 or SB203580 (Fig. 6B, SB203580 supplemental Fig. S2). Both p38 inhibitors prevented the potentiation of cell death induced by MSK1 knockdown, indicating that MSK1 counteracts a pro-apoptotic p38 pathway.
FIGURE 6.
MSK1 inhibits p38-mediated cell death. A, TGFβ-induced cell death is Smad4-dependent. Cells transfected with Smad4 siRNA were grown for 48 h before a 24 h serum-starvation period. Cells were subsequently treated with TGFβ for 24 h and analyzed for viability. Smad4 knockdown completely prevented the induction of cell death by TGFβ. ** indicates p value < 0.01 as determined by Student's t test with unequal sample variance. B, MSK1 inhibits p38-mediated cell death. Cells were transfected with siRNA targeting MSK1 and grown for an additional 48 h before starvation for 24 h and subsequent TGFβ treatment. One hour before TGFβ treatment cells were preincubated with SB202190. Inhibition of p38 MAPK with 10 μm SB202190 prevented the potentiation of cell death caused by MSK1 knockdown and TGFβ treatment. C, induction of Noxa mRNA after MSK1 knockdown is compromised by p38 MAPK inhibition. Cells were prepared exactly as in B, and total RNA was analyzed for Noxa mRNA expression by quantitative real-time RT-PCR as described in the legend to Fig. 4A.
Because p38 inhibition prevented cell death, we examined if the transcripts of Bim and Noxa were influenced by SB202190. Incubation of cells with SB202190 indeed reduced the transcript levels of Noxa (Fig. 6C), but not Bim (data not shown). SB202190 effectively reduced the enhanced expression of Noxa observed when MSK1 was knocked down, indicating that some BH3-only BCL-2 members are potentially sensitive to p38 inhibition.
EGF Rescues Cell Viability in the Absence of MSK1
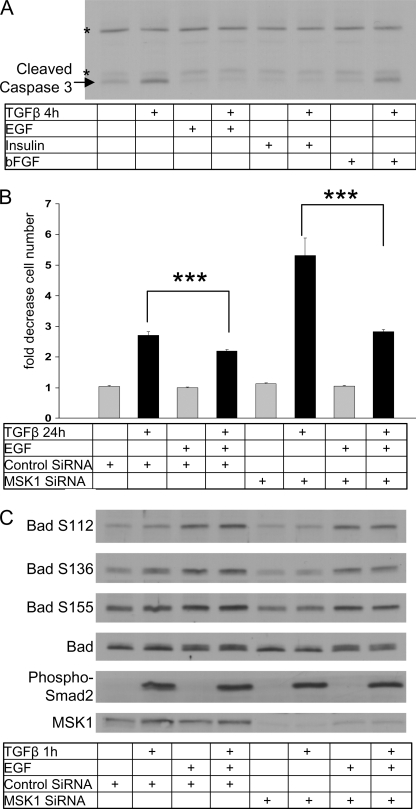
Because MSK1 appeared to provide the cell with a survival signal we investigated if specific growth factors could compensate for the loss of MSK1. We first tested the effect of EGF, insulin and bFGF on caspase 3 cleavage. Co-stimulation of TGFβ and EGF prevented caspase 3 cleavage (Fig. 7A) as did co-stimulation of TGFβ and insulin. bFGF did not prevent the induction of caspase 3 by TGFβ, despite the ability of bFGF to activate signaling in NMuMG cells (35).
FIGURE 7.
EGF rescues TGFβ- and p38-dependent cell death. A, EGF and insulin prevents TGFβ-induced caspase cleavage. Cells were seeded and grown for 24 h before starvation for an additional 24 h. Cells were subsequently treated with TGFβ or EGF for 4 h. After incubation cells were analyzed for the cleaved fragment of caspase 3. Asterisks (*) indicate background protein bands. B, EGF rescues MSK1 siRNA-potentiated cell death. Cells were plated and transfected with MSK1 siRNA after 24 h. Forty-eight hours after transfection cells were serum-starved for 24 h before treatment with TGFβ and EGF for 24 h. EGF rescued the potentiation of TGFβ-induced cell death induced by MSK1 knockdown. * indicates p value < 0.05, ** indicates p value < 0.01, *** indicates p value < 0.001, as determined by Student's t test with unequal sample variance. C, cells were treated as in B and processed for Western blotting. Samples were analyzed for Bad phosphorylation on indicated sites, total Bad levels, phospho-Smad2, and MSK1.
Next, we established that overnight incubation of cells with insulin or EGF slightly inhibited TGFβ-induced cell death (Fig. 7B and supplemental Fig. S3). Notably the effect of EGF and insulin on caspase 3 was measured after 4 h, considerably shorter than the time frame of the cell death assay (24 h). As observed before, knockdown of MSK1 potentiated TGFβ-induced cell death by 2-fold. Insulin and TGFβ co-stimulation resulted in a decrease of cell death, but insulin could not fully rescue the effect of MSK1 knockdown (supplemental Fig. S3). In contrast to insulin, EGF could fully rescue the effect of MSK1 knockdown on cell death as EGF application restored the amount of cell death to control levels.
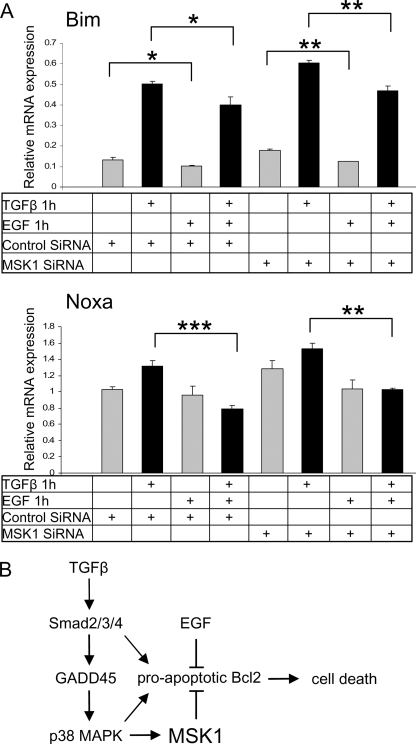
As we had observed a reduction in Bad phosphorylation and an increase in the transcript expression of Noxa and Bim after MSK1 knockdown, we investigated if EGF supplementation could revert this. Indeed EGF treatment resulted in a phosphorylation of Bad on Ser-112, Ser-136, and Ser-155. After MSK1 knockdown Bad phosphorylation on Ser-136 was reduced as observed before (Fig. 7C), but EGF fully restored phosphorylation on all phosphorylation sites. Additionally, EGF supplementation slightly repressed transcript levels of Bim and Noxa in control conditions and reversed the increases observed when MSK1 was knocked down (Fig. 8, A and B). These experiments show that EGF provides specific pro-survival signals that counteract the pro-apoptotic branch of the TGFβ pathway. They also establish new dynamic means by which the cellular response to TGFβ leading to cell death can be modulated by physiological stimuli within the cellular microenvironment.
FIGURE 8.
EGF suppresses the elevated levels of Noxa and Bim mRNA induced by MSK1 knockdown. A, EGF rescues MSK1 siRNA potentiation of Bim and Noxa expression. Cells were plated and transfected with MSK1 siRNA after 24 h. Forty-eight hours after transfection cells were serum starved for 24 before treatment with TGFβ and EGF for 1 h. * indicates p value < 0.05, ** indicates p value < 0.01, *** indicates p value < 0.001 as determined by Student's t test with unequal sample variance. B, TGFβ-induced MSK1 activity inhibits cell death. Model depicting MSK1 regulation by TGFβ based on experiments performed in this study and by others. TGFβ activates Smad-dependent transcription of GADD45. GADD45 induces the activity of p38. The activated p38 activates MSK1. MSK1 inhibits TGFβ-induced activation of BH3-only pro-apoptotic BCL2 members. In the absence of MSK1 EGF can fully compensate the loss of MSK1.
DISCUSSION
TGFβ-induced p38 MAPK activity has been shown to induce cell death, however, the downstream components linking p38 MAPK to the cell death machinery remain unclear. In addition, the upstream pathway that links TGFβ to the activation of p38 MAPK has so far appeared to be cell type-dependent, thus necessitating a careful examination of this mechanism. Here we demonstrate that the p38-activated kinase MSK1 modulates the extent of TGFβ-induced cell death by negatively regulating p38-dependent cell death, possibly by regulating the pro-apoptotic BH3-only members of the BCL-2 family (Fig. 8C). Additionally, with MSK1 we have identified a pro-survival kinase downstream of p38 MAPK, which is often considered pro-apoptotic. This novel role of MSK1 acting downstream of the TGFβ-p38 MAPK signaling branch provides an interesting nodal point that may explain mechanistically some of the context-dependent effects of TGFβ and the ability of different cell types to respond differentially to the pro-apoptotic signals of this cytokine. Overall, MSK1 appears to be an important hub in TGFβ signaling and a crucial component in controlling the extent of TGFβ-induced cell death.
p38 MAPK Activation Is Smad4-dependent
An important issue in TGFβ signaling is non-Smad versus Smad-dependent signaling (2). To elucidate the role of Smad4 (Smad-dependent) and TAK1 (non-Smad) in p38-MSK1 signaling, we investigated MSK1 signaling in cells where either Smad4 or TAK1 were silenced, and obtained evidence for a requirement for Smad4 in p38 activation (Fig. 3, A and B). Interestingly, it has been shown that p38 MAPK activation by TGFβ in NMuMG cells does not require Smad activation (9). This was shown by over-expressing a catalytically active mutant of the TGFβ type I receptor that does not bind Smads, but does retain its intrinsic kinase activity. Overexpression of this mutant receptor induced p38 MAPK activation and cell death in NMuMG cells, but it did not induce other physiological responses to TGFβ, such as epithelial-mesenchymal transition. Our results are in apparent contradiction to these earlier findings, but may be explained in three ways. First, overexpression of the mutant TGFβ receptor that has both constitutive kinase activity and lacks the binding site for Smad2 and Smad3, may direct endogenous signaling components toward a predominant pathway (e.g. p38 MAPK) bypassing the physiological capacity of endogenous wild-type receptors to activate multiple pathways. Second, Smad4 may act as a scaffold in the activation of p38 MAPK, without the requirement of Smad4 transcriptional activity. And thirdly, Smad4 may regulate a target or scaffold gene essential for TGFβ-induced p38 MAPK activity. The latter two options do not completely contradict the results obtained with the mutant type I receptor, because Smad4 was still present in those cells (9).
It has been suggested that rapid (within minutes) activation of p38 MAPK by TGFβ is a TAK1-dependent process, whereas delayed (hours) p38 MAPK activation is Smad-dependent, as it has been shown to depend on the Smad4-mediated induction of GADD45β (12). GADD45β subsequently binds to and activates the MAP kinase kinase MTK1, which functions as an upstream activator of p38 MAPK. The activation of MTK1 by GADD45β appears to be specific since other potential upstream activators of p38 MAPK, such as TAK1 or ASK, are not activated by GADD45β (12). C-terminal truncation mutants of Smad4, as well as antisense GADD45β could prevent activation of p38 MAPK by TGFβ, consistent with the requirement of Smad-dependent transcription in p38 MAPK activation (12). Because the induction of p38-MSK1 and CREB depended on the presence of Smad4 (Fig. 3B), we tested the requirement of GADD45 for the first time in our system. Consistent with a role of GADD45 during TGFβ-induced p38 activation, p38 phosphorylation was greatly reduced by GADD45 knockdown (Fig. 3C). Thus, the Smad4 dependence for p38 activation is explained by the Smad4-dependent induction of GADD45. This mechanism is fully compatible with the time frame of p38-MSK1 activation by TGFβ, which requires ∼1 h of signaling instead of a few minutes (Fig. 1A).
MSK1 Is Involved in the Regulation of TGFβ-induced Survival
The delayed activation of p38 MAPK by TGFβ-induced GADD45 has been mainly linked to the induction of cell death. Knockdown of GADD45 prevents the TGFβ-induced increase in cell death, whereas overexpression of GADD45 itself is enough to induce p38 MAPK activation and cell death (13). GADD45 knock-out cells display deficient p38 MAPK activation and diminished cell death in response to TGFβ treatment (13). However, knockdown of MSK1 and subsequent stimulation with TGFβ resulted in a dramatic loss of viable cells and this was accompanied by an increase in active caspase 3 (Fig. 4). We examined a second parameter involved in cell death by measuring total levels and phosphorylation of Bad (Fig. 4C). Phosphorylated Bad is sequestered in the cytoplasm by 14-3-3 proteins, whereas de-phosphorylation of Bad results in its translocation to mitochondria where it inactivates the anti-apoptotic proteins Bcl-XL and Bcl-2. However, we observed that the total levels of Bad decreased upon TGFβ treatment and this was further potentiated upon knockdown of MSK1. Possibly, TGFβ induces caspase-dependent cleavage of Bad, resulting in a truncated version as has been described (34). Notably, overexpression of the truncated Bad protein enhanced TGFβ-induced cell death, whereas a mutant Bad resistant to caspase 3 cleavage blocked TGFβ-induced cell death (34). Therefore, the reduction of Bad protein and cell death may be directly linked to the increase observed in active caspase 3 levels. A third parameter we looked at is the transcriptional regulation of Noxa and Bim, which are, besides Bad, two additional pro-apoptotic BH3-only BCL-2 members. MSK1 knockdown significantly increased the transcript levels of both Noxa and Bim, whereas only Bim appeared to be sensitive to TGFβ (Fig. 5). Both Bad and Noxa only function as sensitizers/depressors of cell death (17), therefore the transcriptional induction of Bim by TGFβ is of importance since this is a direct activator of apoptosis and may mediate in a more direct manner the TGFβ-dependent mechanism that establishes cell death in response to this cytokine.
MSK1 Inhibits TGFβ-induced p38-dependent Cell Death
To establish how MSK1 influences TGFβ-induced cell death, we first addressed the roles of its upstream activators, Smad4 and p38, in this process. Knockdown of Smad4 eliminated all TGFβ-induced cell death endorsing the importance of Smad4-mediated transcription in this system (Fig. 6A). Inhibition of p38 with chemical inhibitors had small pro-survival effects on TGFβ-induced cell death, however, they effectively blocked the potentiation of cell death observed after MSK1 knockdown (Fig. 6B). Possibly some BH3-only members require p38 MAPK activity as levels of the Noxa transcript were significantly reduced upon chemical inhibition of p38 MAPK (Fig. 5, bottom). Regulation of Bim and Bmf protein expression has been shown to require p38 MAPK activity, implying an important role for p38 MAPK in the activation/induction of pro-apoptotic BH3-only BCL-2 family members (18). Because MSK1 itself is activated downstream of p38 and MSK1 knockdown potentiates p38-dependent cell death, MSK1 represents a bifurcating node downstream of p38, balancing the TGFβ response between survival and cell death.
MSK1 Provides the Cell with an EGF-like Survival Signal
Both insulin and EGF are known stimulants of cellular survival as they are able to modulate intracellular survival pathways (36, 37). Interestingly, EGF stimulation could completely revert the effects observed after MSK1 knockdown, whereas insulin could not (Fig. 7, supplemental Fig. S3). EGF prevented caspase cleavage, restored Bad phosphorylation after MSK1 knockdown and reduced transcript levels of Noxa and Bim (Figs. 7 and 8). EGF and TGFβ may converge on the same intracellular pathway. The effects of EGF also imply that in order to efficiently kill the cell, a combinatorial method targeting both MSK1 and EGF signaling is required.
In summary, we describe MSK1 as an important modulator of TGFβ signaling downstream of Smad4-induced p38 activity (Fig. 8C). By demonstrating the role of MSK1 as an anti-apoptotic factor for the first time, we have identified a functional bifurcation downstream of p38. It will be of great interest to explore possible involvement of TGFβ-induced MSK1 activation in diseases caused by perturbed TGFβ signaling such as cancer. Targeting of both the MSK1 and the EGF pathway may be considered as a method to re-sensitize cancer cells to TGFβ-induced cell death.
Supplementary Material
Acknowledgments
We thank Carl-Henrik Heldin and E-Jean Tan for critical reading of the manuscript.
This work was supported by the Atlantic Philanthropies/Ludwig Institute for Cancer Research Clinical Discovery Program and Cancerfonden (Contract Number: 09 0773).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- TGFβ
- transforming growth factor β
- BH
- Bcl2-homology
- MAPK
- mitogen-activated protein kinase
- MSK1
- mitogen and stress-activated kinase 1
- TAK1
- TGFβ-activated kinase 1.
REFERENCES
- 1. Moustakas A., Heldin C. H. (2009) Development 136, 3699–3714 [DOI] [PubMed] [Google Scholar]
- 2. Heldin C. H., Landström M., Moustakas A. (2009) Curr. Opin. Cell Biol. 21, 166–176 [DOI] [PubMed] [Google Scholar]
- 3. Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ten Dijke P., Goumans M. J., Itoh F., Itoh S. (2002) J. Cell. Physiol. 191, 1–16 [DOI] [PubMed] [Google Scholar]
- 5. ten Dijke P., Hill C. S. (2004) Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 6. Itoh S., Thorikay M., Kowanetz M., Moustakas A., Itoh F., Heldin C. H., ten Dijke P. (2003) J. Biol. Chem. 278, 3751–3761 [DOI] [PubMed] [Google Scholar]
- 7. Lee M. K., Pardoux C., Hall M. C., Lee P. S., Warburton D., Qing J., Smith S. M., Derynck R. (2007) EMBO J. 26, 3957–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moustakas A., Heldin C. H. (2005) J. Cell Sci. 118, 3573–3584 [DOI] [PubMed] [Google Scholar]
- 9. Yu L., Hébert M. C., Zhang Y. E. (2002) EMBO J. 21, 3749–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edlund S., Bu S., Schuster N., Aspenström P., Heuchel R., Heldin N. E., ten Dijke P., Heldin C. H., Landström M. (2003) Mol. Biol. Cell 14, 529–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sorrentino A., Thakur N., Grimsby S., Marcusson A., von Bulow V., Schuster N., Zhang S., Heldin C. H., Landström M. (2008) Nat. Cell Biol. 10, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 12. Takekawa M., Tatebayashi K., Itoh F., Adachi M., Imai K., Saito H. (2002) EMBO J. 21, 6473–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoo J., Ghiassi M., Jirmanova L., Balliet A. G., Hoffman B., Fornace A. J., Jr., Liebermann D. A., Bottinger E. P., Roberts A. B. (2003) J. Biol. Chem. 278, 43001–43007 [DOI] [PubMed] [Google Scholar]
- 14. Miyake Z., Takekawa M., Ge Q., Saito H. (2007) Mol. Cell. Biol. 27, 2765–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., Zhang Y. E. (2008) Mol. Cell 31, 918–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sánchez-Capelo A. (2005) Cytokine Growth Factor Rev. 16, 15–34 [DOI] [PubMed] [Google Scholar]
- 17. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramjaun A. R., Tomlinson S., Eddaoudi A., Downward J. (2007) Oncogene 26, 970–981 [DOI] [PubMed] [Google Scholar]
- 19. Zhu Y., Yang G. Y., Ahlemeyer B., Pang L., Che X. M., Culmsee C., Klumpp S., Krieglstein J. (2002) J. Neurosci. 22, 3898–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abécassis L., Rogier E., Vazquez A., Atfi A., Bourgeade M. F. (2004) J. Biol. Chem. 279, 30474–30479 [DOI] [PubMed] [Google Scholar]
- 21. Deak M., Clifton A. D., Lucocq L. M., Alessi D. R. (1998) EMBO J. 17, 4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCoy C. E., Campbell D. G., Deak M., Bloomberg G. B., Arthur J. S. (2005) Biochem. J. 387, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCoy C. E., macdonald A., Morrice N. A., Campbell D. G., Deak M., Toth R., McIlrath J., Arthur J. S. (2007) Biochem. J. 402, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arthur J. S., Cohen P. (2000) FEBS Lett. 482, 44–48 [DOI] [PubMed] [Google Scholar]
- 25. Olson C. M., Hedrick M. N., Izadi H., Bates T. C., Olivera E. R., Anguita J. (2007) Infect. Immun. 75, 270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schiller M., Böhm M., Dennler S., Ehrchen J. M., Mauviel A. (2006) Oncogene 25, 4449–4457 [DOI] [PubMed] [Google Scholar]
- 27. Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. (2002) Mol. Cell. Biol. 22, 2871–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y., Cho Y. Y., Petersen B. L., Bode A. M., Zhu F., Dong Z. (2003) J. Biol. Chem. 278, 12650–12659 [DOI] [PubMed] [Google Scholar]
- 29. Piek E., Moustakas A., Kurisaki A., Heldin C. H., ten Dijke P. (1999) J. Cell Sci. 112, 4557–4568 [DOI] [PubMed] [Google Scholar]
- 30. Deckers M., van Dinther M., Buijs J., Que I., Löwik C., van der Pluijm G., ten Dijke P. (2006) Cancer Res. 66, 2202–2209 [DOI] [PubMed] [Google Scholar]
- 31. Kowanetz M., Lönn P., Vanlandewijck M., Kowanetz K., Heldin C. H., Moustakas A. (2008) J. Cell Biol. 182, 655–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie L., Law B. K., Chytil A. M., Brown K. A., Aakre M. E., Moses H. L. (2004) Neoplasia 6, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim B. C., Mamura M., Choi K. S., Calabretta B., Kim S. J. (2002) Mol. Cell. Biol. 22, 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reiland J., Rapraeger A. C. (1993) J. Cell Sci. 105, 1085–1093 [DOI] [PubMed] [Google Scholar]
- 36. van der Heide L. P., Ramakers G. M., Smidt M. P. (2006) Prog. Neurobiol. 79, 205–221 [DOI] [PubMed] [Google Scholar]
- 37. Rosfjord E. C., Dickson R. B. (1999) J. Mammary Gland Biol. Neoplasia 4, 229–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.