Abstract
Elongation of transcription by mammalian RNA polymerase II (RNAPII) is regulated by specific factors, including transcription factor IIS (TFIIS) and positive transcription elongation factor b (P-TEFb). We show that the E3 ubiquitin ligase UBR5 associates with the CDK9 subunit of positive transcription elongation factor b to mediate its polyubiquitination in human cells. TFIIS also binds UBR5 to stimulate CDK9 polyubiquitination. Co-localization of UBR5, CDK9, and TFIIS along specific regions of the γ fibrinogen (γFBG) gene indicates that a ternary complex involving these factors participates in the transcriptional regulation of this gene. In support of this notion, overexpression of TFIIS not only modifies the ubiquitination pattern of CDK9 in vivo but also increases the association of CDK9 with various regions of the γFBG gene. Notably, the TFIIS-mediated increase in CDK9 loading is obtained during both basal and activated transcription of the γFBG gene. This increased CDK9 binding is paralleled by an increase in the recruitment of RNAPII along the γFBG gene and the phosphorylation of the C-terminal domain of the RNAPII largest subunit RPB1 on Ser-2, a known target of CDK9. Together, these results identify UBR5 as a novel E3 ligase that regulates transcription and define an additional function of TFIIS in the regulation of CDK9.
Keywords: Protein-Protein Interactions, RNA Polymerase II, Transcription Elongation Factors, Transcription Regulation, Ubiquitin Ligase
Introduction
Transcription factor IIS (TFIIS)3 is the first general transcription factor reported to associate with RNAPII (1). The functions of TFIIS in transcription have been extensively studied, and although early evidence pointed to a specific role in elongation, more recent studies have revealed the implication of TFIIS in early transcriptional stages such as formation of the preinitiation complex (2) and release of the polymerase from elongation complexes paused close to the promoter (3). The architecture of TFIIS is modular and comprises three domains with distinct functions: domain III is essential for the stimulation of elongation; domain II and the flexible linker between domains II and III are required for binding to RNAPII; and domain I is required for the interaction with the Mediator and other initiation factors (4–6). The most well characterized function of TFIIS in elongation consists of the rescue of backtracked RNAPII through stimulation of its intrinsic endonucleolytic activity (7). In support of the function of TFIIS in both early and late stages of transcription, chromatin immunoprecipitation studies in human cells (8) and lower eukaryotes (2, 9) have revealed the association of TFIIS along the entire sequence of tested genes, including promoters, and have shown that this factor is required for the recruitment of polymerase molecules at specific promoters.
TFIIS participates in important regulatory mechanisms that evolved from lower to higher eukaryotes. For instance, although yeast TFIIS is not essential for growth, TFIIS knock-out mice die during embryonic development (10). The regulation of post-initiation stages of transcription was generally believed to be involved in the control of a small subset of genes (e.g. heat shock response genes in Drosophila, c-Myc, c-Fos, and HIV-1) (11). However, genome-wide studies in different systems suggest that this regulatory mechanism is much more common than initially thought (12), given that paused polymerase molecules were identified along a majority of transcribed genes. Because TFIIS stimulates the release of polymerase from promoter-proximal paused complexes, the regulatory potential of TFIIS could be much wider than previously believed.
The transcription elongation factor P-TEFb also stimulates the release of promoter-proximal paused RNAPII (13–15). This activity is mediated by the kinase subunit of P-TEFb, CDK9, which phosphorylates multiple targets, including the C-terminal domain (CTD) of the Rpb1 subunit of RNAPII and the negative elongation factors DSIF and NELF (16). These phosphorylation events abolish the inhibitory effect of DSIF and NELF (14, 17) and allow the phosphorylated CTD to become a platform for the recruitment of stimulatory elongation factors (18). These concerted actions cause the release of the polymerase from its paused state and its entry into productive elongation. Notably, it has been shown that DSIF and NELF not only inhibit RNAPII but also negatively affect the activity of TFIIS in vitro, both negative factors being required (19). These data suggest that a self-regulatory loop could be formed with positive and negative elongation factors inhibiting each other; P-TEFb counteracts DSIF and NELF, which in turn inhibits TFIIS. Although DSIF and NELF have been shown to function as negative elongation factors only when both proteins are present, no direct link between TFIIS and P-TEFb has been reported yet.
In this article, we report on the identification of a new interaction partner for TFIIS and CDK9, namely, the E3 ubiquitin ligase UBR5. We show that UBR5 catalyzes the ubiquitination of CDK9 in vitro and that both UBR5 and TFIIS are essential for this ubiquitination event in human cells. We also show that overexpression of TFIIS increases the occupancy of CDK9 along the γFBG gene, an event that is accompanied by an increased loading of RNAPII and an increased phosphorylation of the Ser-2 residue of its RPB1 CTD. Moreover, we provide direct evidence that the effect of TFIIS on the ubiquitination of CDK9 and its occupancy level along the γFBG gene is dependent upon the UBR5 ubiquitin ligase.
EXPERIMENTAL PROCEDURES
Protein Affinity Purification Coupled with Mass Spectrometry
Human EcR 293 cells stably transfected were induced with 3 μm ponasterone A (Invitrogen) to express TAP-tagged versions of TFIIS and TFIIS.1. Whole cell extracts were subjected to TAP purification, and the eluates were run on SDS-PAGE gels, as described previously (20). Noninduced cells were used as TAP purification controls. After silver or Sypro Ruby (Bio-Rad) staining, the bands were excised and digested with trypsin. The tryptic peptides were purified and identified by tandem mass spectrometry, as described previously (21). To discriminate between specific and spurious interactions, we used a computational method that we have described previously (21, 22). This method uses an algorithm that assigns an interaction reliability score to each protein interaction based on parameters such as the strength of the MS score and the local topology of the network (e.g. number of common interaction partners and the identification of the interaction in reciprocal purifications). The graph in Fig. 1A shows the interaction partners of TFIIS with an interaction reliability score higher than 0.73. We estimate that above this threshold, the specificity is ∼88%, and the rate of false positives is lower than 12%. By setting this threshold value of interaction reliability, we evaluate that the sensitivity of our method to detect relevant interactions approaches 72%, which suggests a rate of 28% false negatives (see Ref. 22 for details on the computational method used to evaluate the confidence of our interaction networks).
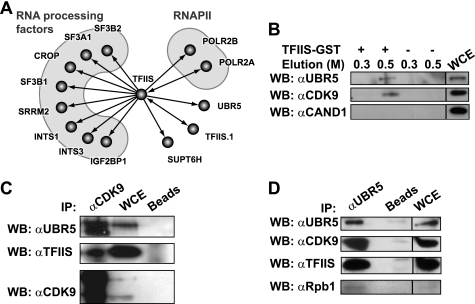
FIGURE 1.
TFIIS interacts with the E3 ligase UBR5 and the P-TEFb kinase CDK9. A, high confidence interaction network for human TFIIS. The graph shows part of a network of high confidence interactions obtained using affinity purification (in vivo pulldown) coupled with mass spectrometry, with 77 TAP-tagged baits targeting many transcription and RNA processing factors (22). B, in vitro pulldown using TFIIS-GST. In vitro pulldown experiments used HEK 293 whole cell extracts with TFIIS-GST bound to glutathione-Sepharose beads or with control glutathione-Sepharose beads. The eluates and input (5%) were immunoblotted using antibodies directed against UBR5, CDK9, or the control protein CAND1. The NaCl concentration (m) in the elution buffer is indicated. C and D, immunoprecipitation using anti-CDK9 (C) and anti-UBR5 (D) antibodies. Immunoprecipitation experiments used HEK 293 whole cell extracts with an antibody raised against CDK9 (C) or UBR5 (D) bound to protein A beads or with mock protein A beads. The eluates and input (5%) were run on SDS gels and immunoblotted using antibodies against UBR5, TFIIS, CDK9, and Rpb1. WB, Western blotting; IP, immunoprecipitation; WCE, whole cell extract.
Chemicals and Immunoreagents
The following antibodies were used: monoclonal anti-TFIIS (Transduction Laboratories), M2 monoclonal anti-FLAG (Sigma), polyclonal anti-CDK9 (C-20), anti-UBR5 (N-19), RNA Pol II (N-20 and C-21), anti-ubiquitin (FL-76), anti-CAND1 (A-13) (Santa Cruz Biotechnologies), monoclonal His6 tag (ab18184) (Abcam), and monoclonal anti Ser(P)-2 CTD Pol II (H5) (Convance). An UBR5 antiserum produced in rabbit was kindly provided by Robert L. Southerland (Sydney, Australia) and used for immunoprecipitation assays. Ubiquitin, ubiquitin-activating enzyme (UBE1), ubiquitin conjugating enzyme (UbcH5b), and ubiquitin aldehyde (Ub-H) were purchased from BostonBiochem; recombinant human IL-6 was purchased from Invitrogen; dissucinimidyl glutarate was purchased from Pierce; N-ethylmaleimide, AMP-PNP, and cycloheximide were purchased from Sigma; and lactacystin was purchased from Cayman Chemical.
Plasmid Constructions
PCR amplification was used to clone cDNAs for TFIIS, TFIIS.1, and UBR5 into the mammalian expression vector pMZI (23) carrying a TAP tag at its C terminus. From this template, we subcloned TFIIS into pCMV-3Tag-1A vector (Stratagene) encoding three copies of the FLAG epitope at the N terminus of the insert. GST-TFIIS was expressed from the pGEX-4T1 plasmid (GE Healthcare) in the Escherichia coli BL21 Star (DE3) strain (Invitrogen). The pCMV-Tag2B-UBR5 construct, which expresses UBR5 with a FLAG epitope at its N terminus, was obtained from Robert L. Southerland (Sydney, Australia). The cDNA for CDK9 was cloned by PCR amplification into pRLuc-C2 (h) and pRLuc-N2(h) vectors (BioSignal Packard) encoding the Renilla luciferase protein. As normalizing vector in luciferase assays, we used the pGL3 plasmid (Promega) encoding the firefly luciferase (Fluc). The pMT123 vector encoding HA-ubiquitin polypeptide was kindly provided by Sylvain Meloche (Institut de Recherche en Immunologie et Cancérologie, Université de Montréal, Montreal, Canada).
Cell Lines and Culture Conditions
Stable human embryonic kidney Ecr 293 cells (derived from HEK 293) were produced as described previously (20) and grown in DMEM supplemented with 10% FBS, 2 mm l-glutamine, 30 μg/ml BleocinTM (Calbiochem), and 300 μg/ml G418 (Invitrogen). Human HeLa and HepG2 cells were maintained in Eagle's minimal essential medium supplemented with 10% FBS and 2 mm l-glutamine. Stable HepG2 cell lines were obtained by transfection with the FuGENE reagent (Roche Applied Science) and a selection of stable clones with 600 μg/ml of G418 (Invitrogen). For IL-6 induction, the cells were serum-starved for at least 16 h, and IL-6 was added at a final concentration of 10 ng/ml for 30 min.
GST Pulldown and Immunoprecipitation Assays
TFIIS-GST was purified by affinity chromatography on glutathione-Sepharose 4B (GE Healthcare Biosciences) according to the manufacturer's instructions. Sepharose-bound TFIIS-GST was incubated with HEK 293 whole cell extracts (1 mg) prepared under native conditions (24). After thorough washing of the beads, the bound proteins were eluted at the indicated ionic strength, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies. For immunoprecipitation experiments, 0.5–1 mg of HEK 293 whole cell extracts prepared under native conditions were incubated with indicated antibodies, and the immunocomplexes were captured on protein A-Sepharose beads. After extensive washing with IPP100G (10 mm Tris, 100 mm NaCl, 0.1% Triton X-100), the bound proteins were eluted, resolved on SDS-PAGE, and immunoblotted with the indicated antibodies.
siRNA Knockdown Experiments
Pools of ON-TARGET plus Smartpool oligonucleotides (four oligonucleotides) targeting TFIIS, UBR5, and, as a negative control, a mix of four oligonucleotides that do not target any known human gene (Dharmacon) were used. HeLa and HepG2 cells cultured in six-well dishes were transfected using Oligofectamine (Invitrogen) and siPORT NeoFX transfection agent (Ambion), respectively, according to the manufacturers' instructions. For efficient knockdown, we used a double transfection procedure: after the first transfection, the cells were incubated at 37 °C for 24 h and then retransfected using the same protocol. To determine the knockdown efficiencies, the cell lysates obtained after 48, 72, and 96 h of culture were resolved on SDS-PAGE gels and tested by immunoblotting with relevant antibodies. Because the most efficient knockdown was obtained after 48 h (data not shown), these conditions were used for all siRNA knockdown experiments.
In Vitro Ubiquitination Assay
Full-length CDK9 carrying a C-terminal His6 tag was obtained by baculovirus expression along with cyclin T1 (1–290), followed by nickel-nitrilotriacetic acid and Mono S purification. UBR5 was purified by the TAP procedure from a human EcR 293 cell line stably expressing full-length UBR5 with a C-terminal TAP tag. After the second step of the TAP purification, the calmodulin beads containing immunoprecipitated UBR5 were washed twice with cold ubiquitination buffer (20 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm MgCl2, 1 mm DTT, 0.5% Nonidet P-40, and protease inhibitor mixture) and then washed twice with 50 mm Tris-HCl, pH 7.5, and 1 mm DTT. The reaction mixture (60 μl) for in vitro ubiquitination assays contained E1 (150 ng), E2 (UbcH5b, 500 ng), purified CDK9-His protein (2.5 μg), AMP-PNP (2 mm), ubiquitin aldehyde (5 μm), and 10% of the pellet containing affinity purified UBR5. The assay was performed in ubiquitination buffer: 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, and 1 mm DTT. Following preincubation at 37 °C for 5 min, ubiquitin (500 ng) was added to start the reaction. The mixtures were incubated for 55 min with gentle agitation every 10 min. The reaction was stopped by adding SDS loading buffer and boiling for 10 min. After the isolation of beads by centrifugation (1 min at 1000 rpm), the supernatant was loaded on 6% SDS-PAGE gels. The ubiquitination reactions were analyzed by Western blotting.
In Vivo Ubiquitination Assays
Transient transfection of HeLa cells with vectors driving the expression of TFIIS-FLAG or the empty vector, together with a ubiquitin-HA expression construct, was performed with calcium phosphate in 100-mm dishes. Lactacystin (12 μm) was added for 5 h. After 48 h of culture, the cells were lysed for 1 h at 4 °C in lysis buffer containing 1% Nonidet P-40, 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 10% glycerol, 2 mm N-ethylmaleimide, and protease inhibitor mixture (Roche Applied Science) and supplemented with 6 m urea. The soluble lysate was diluted to 0.3 m urea with lysis buffer supplemented with 1 mm DTT and immunoprecipitated at 4 °C overnight with a CDK9 antibody (10 μg). The immunocomplexes were collected with protein A-Sepharose beads, and after extensive washing, they were eluted with reducing SDS sample buffer. Eluates and a sample (5%) of the whole cell extracts were tested by immunoblotting with the indicated antibodies, using a standard procedure. The ubiquitination in HepG2 cells stably expressing UBR5-FLAG or TFIIS-FLAG was assessed using the same procedure.
To analyze the effect of TFIIS or UBR5 knockdown on the ubiquitination of CDK9, HeLa cells cultured in six-well plates were transfected with siRNAs and co-transfected with the DNA plasmids and siRNAs, as indicated in the figures, using Oligofectamine. After 24 h, the cells were transfected a second time with siRNAs. Final concentrations of siRNAs were 25 and 50 nm, as indicated. After a total culture time of 48 h, the cells were lysed and subjected to ubiquitination assays and to parallel verification of the knockdown efficiency by immunoblotting with relevant antibodies.
Luciferase Assay
HeLa cells were transfected directly into white 96-well plates with plasmids expressing N- or C-terminal CDK9-Renilla luciferase fusion proteins and with a normalizing plasmid expressing the Fluc using Oligofectamine (Invitrogen). UBR5-targeting and control siRNAs were co-transfected with the DNA plasmids, and after 24 h, a second transfection of siRNAs was performed. After a total of 48 h, the Renilla and firefly luciferase activities were measured with the Dual-Glo Luciferase system (Promega), according to the manufacturer's instructions. De novo protein synthesis was inhibited by adding cycloheximide at a final concentration of 100 μg/ml, and the luciferase substrates were added at the indicated time points.
Chromatin Immunoprecipitation
Two-step ChIP assays were performed as described previously (25). HepG2 cells were cultured in 100-mm dishes, and 4–6 × 106 cells were cross-linked at room temperature in two steps. First, dissucinimidyl glutarate was added for 45 min for protein-protein cross-linking, and second, protein-DNA cross-linking was performed by adding formaldehyde for 10 min. After washing with PBS, the cells were lysed, and the nuclei were resuspended in SDS lysis buffer and sonicated on ice 12 times using 15-s pulses (Fisher Model 100 sonic dismembrator). The average length of DNA fragments was 350 bp. The immunoprecipitation of equal amounts of DNA was performed overnight at 4 °C in ChIP dilution buffer. We used 4 μg of the indicated antibodies or 10 μg in the case of the H5 antibody. The immunoprecipitated complexes were collected with protein A-Sepharose (GE Healthcare) for CDK9 (C20) and RNAPII (N20) antibodies, and with Dynabeads PanMouse IgG or rat anti-mouse IgM (Invitrogen) for anti-FLAG (M2) and anti-RNAPII Ser(P)-2-CTD form (H5) antibodies, respectively. After washing and elution, the cross-links were reversed at 65 °C for 4 h, and the DNA was purified by phenol/chloroform extraction. The quantification of the immunoprecipitated DNA fragments was performed by quantitative PCR, as described (20). The primers used are described elsewhere (26). For siRNA knockdowns coupled with ChIP experiments, HepG2 cells were cultured in 100-mm dishes, a double transfection of siRNAs was performed with siPORT NeoFX (Ambion), and after a total culture time of 48 h, the cells were subjected to the two-step ChIP assay as described above. For ChIP-re-ChIP experiments, the complexes were washed with washing buffer containing a protease inhibitor mixture (Roche Applied Science), eluted by incubation for 30 min at 37 °C in Re-ChIP buffer (0.5 mm DTT, 1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl, pH 8, protease inhibitor mixture), and subjected again to the ChIP procedure.
RESULTS
TFIIS Interacts with the E3 Ubiquitin Ligase UBR5 and CDK9
To map the space of the human interactome that contains the transcription elongation factor TFIIS (official gene symbol: TCEA1) and to identify novel interaction partners that may reveal new functions for TFIIS, we used affinity purification of TAP-tagged TFIIS coupled with mass spectrometry to indentify the interaction partners. High confidence interactions were selected computationally, as described previously (21, 22). As expected, TFIIS was found to interact with RNAPII and components of the RNA processing machinery (Fig. 1A). The interaction graph in Fig. 1A reveals the existence of two proteins that have not been previously reported to interact with TFIIS, namely, TFIIS.1 (official gene symbol: TCEA2) and UBR5. TFIIS.1 is a TFIIS-related protein that is expressed in specific tissues, with a high level of expression in the testis (27). Our functional analysis of TFIIS.1 revealed that, similarly to TFIIS, this protein stimulates transcriptional elongation in vitro and presents the same localization pattern in human cells (data not shown), suggesting similar roles in vivo. Based on these results, we decided not to proceed with further analysis of the TFIIS-TFIIS.1 interaction. UBR5 is a member of the HECT domain family of E3 ubiquitin ligases (28). The presence of UBR5 in the TFIIS-TAP eluate was confirmed by Western blotting (data not shown). To confirm the TFIIS-UBR5 interaction, we performed in vitro pulldown experiments using a TFIIS-GST fusion protein coupled with glutathione beads and a HEK 293 whole cell extract. Immunoblotting of the eluate with anti-UBR5 antibody revealed the presence of UBR5 (Fig. 1B).
According to the literature and our unpublished data, it was unlikely that TFIIS and RNAPII represent targets for UBR5 ubiquitination (data not shown). Recent reports, however, have indicated that CDK9, the kinase subunit of transcription elongation factor P-TEFb, which also interacts with RNAPII, is ubiquitinated in human cells (29, 30). We tested the hypothesis of UBR5 being involved in CDK9 ubiquitination, possibly through an interaction with TFIIS and/or RNAPII. We first assessed whether CDK9 can interact with TFIIS and UBR5 by probing the TFIIS-GST eluate for the presence of CDK9 by Western blotting. Fig. 1B shows that both UBR5 and CDK9, but not a control protein, are present in the TFIIS-GST pulldown eluate. CAND1 (cullin-associated and neddylation-dissociated 1) was used as a control because this protein was identified as an interaction partner of TFIIS.1 but not of TFIIS in our affinity purification experiments.
The constitutive interaction between these three proteins was further confirmed using immunoprecipitation with anti-CDK9 (Fig. 1C) and anti-UBR5 antibodies (Fig. 1D). These results indicate that TFIIS, UBR5, and CDK9 interact together to form a complex in vivo. Of note, the Rpb1 subunit of RNAPII is also found in the anti-UBR5 immunoprecipitate (Fig. 1D). This result suggests that UBR5 is indeed part of a complex also containing RNAPII, and confirms the result of our systematic affinity purification coupled with mass spectrometry analysis (Fig. 1A).
UBR5 Mediates the Ubiquitination of CDK9 in Vitro
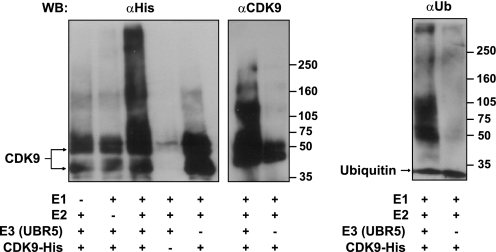
Because a previous report indicated that endogenous CDK9 molecules are ubiquitinated (30), it was legitimate to speculate that its interaction partner, UBR5, may be involved in this modification. To test this hypothesis, we first performed in vitro ubiquitination assays using TAP affinity purified UBR5 and CDK9 purified from insect cells as a substrate. When the complete set of components was used in the ubiquitination reaction, a robust polyubiquitination of CDK9 was observed, with the appearance of slower migrating high molecular weight species in lanes that contain the complete reaction mixture (Fig. 2). These forms contained ubiquitinated CDK9 molecules, because they were detected with both antibodies directed toward CDK9 (anti-tag and anti-CDK9), as well as with the anti-ubiquitin antibody. When one of the components was omitted (E1, E2, CDK9, or UBR5), no ubiquitination was observed. These results show that UBR5 directly catalyzes the ubiquitination of CDK9 in vitro.
FIGURE 2.
UBR5 ubiquitinates CDK9 in vitro. Ubiquitination of baculovirus-expressed CDK9 carrying a His tag was obtained in the presence of the enzymes E1, E2 (UbcH5b), and E3 (affinity purified UBR5). The reactions were incubated with ubiquitin (Ub), ubiquitin aldehyde, and AMP-PNP as described under “Experimental Procedures.” Higher molecular weight ubiquitinated forms of CDK9-His were detected by immunoblotting with anti-His or anti-CDK9 antibodies (left panel). To confirm that the higher molecular weight bands contained ubiquitinated forms of CDK9, replicate reactions were analyzed by immunoblotting with an anti-ubiquitin antibody (right panel). WB, Western blotting.
TFIIS and UBR5 Are Required for the Ubiquitination of CDK9 in Vivo
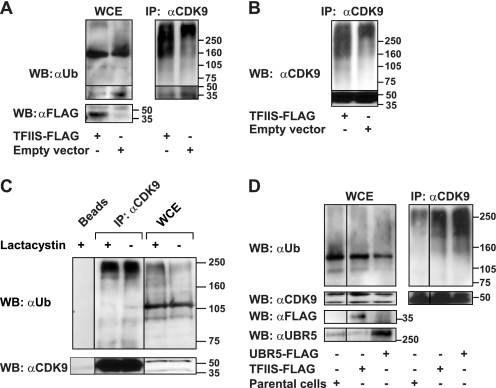
We next performed in vivo ubiquitination assays in extracts from cells in which either UBR5 or TFIIS were individually overexpressed or depleted. HeLa cells were co-transfected with constructs driving the expression of TFIIS carrying a FLAG epitope (TFIIS-FLAG) and an ubiquitin-HA peptide. Co-transfections of the empty vector and the ubiquitin-HA construct were used as controls. The cells were treated with the proteasome inhibitor lactacystin, and the whole cell extracts were incubated with an anti-CDK9 antibody. After CDK9 was isolated, the eluates were resolved on SDS-PAGE and immunoblotted with anti-ubiquitin, anti-CDK9, or anti-FLAG antibodies. The overexpression of TFIIS markedly increased the smear of high molecular weight proteins as compared with the controls (Fig. 3A, right panel). This effect was specific to CDK9 because no significant modification in the general ubiquitination pattern was observed in the whole cell extracts (Fig. 3A, left panel). Immunoblotting with an anti-CDK9 antibody shows that the high molecular weight species are polyubiquitinated CDK9 molecules (Fig. 3B). Moreover, these species appear specifically in the eluates from the CDK9 immunoprecipitates but not in the eluates from mock immunoprecipitation (Fig. 3C). Interestingly, proteasome inhibition by lactacystin does not affect the stability of the polyubiquitinated CDK9 species, whereas stabilization of the polyubiquitinated proteins is observed in the whole cell extracts using the same treatment (Fig. 3C).
FIGURE 3.
TFIIS and UBR5 overexpression affects the ubiquitination of CDK9 in vivo. A, HeLa cells transfected with a vector driving the expression of FLAG-tagged TFIIS or the empty vector, together with a construct driving the expression of ubiquitin-HA, were treated with the proteasome inhibitor lactacystin and lysed. The whole cell extract (5% input) (left panel) or eluates from anti-CDK9 immunoprecipitation with the C20 antibody (right panel) were run on SDS gels and immunoblotted with anti-ubiquitin or anti-FLAG antibodies (B) Immunodetection of CDK9 in the anti-CDK9 immunoprecipitate obtained as described in A. C, HeLa cells treated with lactacystin or mock-treated were lysed. The lysates were immunoprecipitated using anti-CDK9 antibody (C20) bound to protein A beads or with mock protein A beads. The eluates and the input (5%) were run on SDS gels and immunoblotted using anti-ubiquitin and anti-CDK9 antibodies. D, HepG2 cells stably transfected with constructs driving the expression of UBR5-FLAG or TFIIS-FLAG were subjected to in vivo ubiquitination as described above. Parental HepG2 cells treated in parallel were used as a control. The eluates of CDK9 immunoprecipitation (C20) (right panel) and the input (5%) (left panel) were immunoblotted using anti-ubiquitin, anti-CDK9, anti-FLAG, and anti-UBR5 antibodies. WB, Western blotting; IP, immunoprecipitation; WCE, whole cell extract.
To assess whether UBR5 is involved in the ubiquitination of CDK9 in vivo, we performed ubiquitination assays using extracts from HepG2 cells stably transfected with a construct expressing UBR5-FLAG. As controls, we used extracts prepared in parallel from a stable HepG2 cell line expressing TFIIS-FLAG or from parental HepG2 cells. Fig. 3D shows that overexpression of UBR5 specifically increased the smear of high molecular weight CDK9 species.
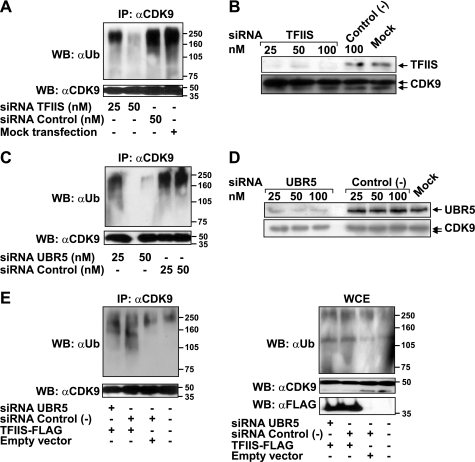
To confirm the role of UBR5 and TFIIS in CDK9 ubiquitination, siRNA treatments were used to knock down each protein individually in HeLa cells (Fig. 4, B and D, respectively), and the whole cell extracts were subjected to CDK9 immunoprecipitation and in vivo ubiquitination assays, as described above. The knockdown of each of TFIIS or UBR5 protein reduced the smear of polyubiquitinated CDK9 species in a dose-dependent fashion compared with control nontargeting siRNAs (Fig. 4, A and C, respectively). Together, these results indicate that both UBR5 and TFIIS are involved in CDK9 ubiquitination in vivo.
FIGURE 4.
TFIIS or UBR5 knockdown decreases the ubiquitination of CDK9 in vivo, and UBR5 is required for the effect of TFIIS on this ubiquitination event. A, HeLa cells were transfected with the indicated concentrations of siRNAs targeting TFIIS or control nontargeting siRNAs or mock transfected. The cells were treated with lactacystin and lysed, and the lysate was used in immunoprecipitation experiments with an anti-CDK9 antibody (C20). The eluates were run on SDS gels and immunoblotted with anti-ubiquitin or anti-CDK9 antibodies. B, comparison of knockdown efficiencies resulting from treatment with siRNAs targeting TFIIS or control nontargeting siRNAs. C, HeLa cells treated with the indicated concentrations of siRNAs targeting UBR5 or nontargeting siRNAs were assayed for in vivo ubiquitination (as in A). D, comparison of knockdown efficiencies resulting from treatment with siRNAs targeting UBR5 or control nontargeting siRNAs. E, HeLa cells treated with siRNAs targeting UBR5 or nontargeting siRNAs (50 nm) or mock-treated were transfected with a construct driving the expression of TFIIS-FLAG or with an empty vector. An in vivo ubiquitination assay (as in A) was performed with the transfected cells. The lysates were used for anti-CDK9 immunoprecipitation, and the eluates (left panel) or the input (5%) (right panel) were run on SDS gels and immunoblotted with anti-ubiquitin or anti-CDK9 antibodies. The expression of the FLAG-tagged protein was monitored by immunodetection with an anti-FLAG antibody. WB, Western blotting; IP, immunoprecipitation; WCE, whole cell extract.
UBR5 Is Required for the Effect of TFIIS on CDK9 Ubiquitination in Vivo
Because both TFIIS and P-TEFb stimulate transcriptional elongation by RNAPII, we hypothesized that TFIIS may potentiate the activity of P-TEFb by recruiting the E3 ubiquitin ligase UBR5 to facilitate CDK9 ubiquitination. To test this idea, we performed in vivo ubiquitination assays in HeLa cells co-transfected with constructs driving the expression of TFIIS-FLAG and siRNAs targeting UBR5. As shown in Fig. 4E, TFIIS overexpression increased the accumulation of high molecular weight CDK9 species as compared with transfection with the empty vector when a nontargeting control siRNA was co-transfected. However, co-transfection with the UBR5-targeting siRNA almost completely abolished this effect on CDK9 ubiquitination. This suggests that the effect of TFIIS on CDK9 ubiquitination is related, to a significant extent, to the activity of UBR5. The residual ubiquitination of CDK9 may be due to the incomplete depletion of UBR5.
The Knockdown of UBR5 Does Not Affect the Stability of CDK9
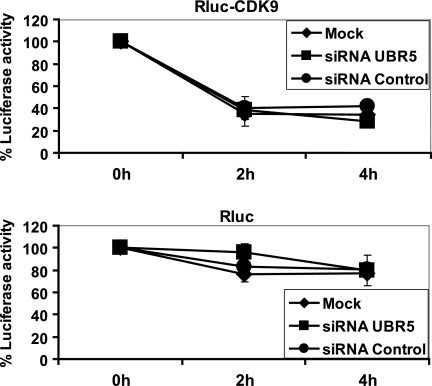
Although endogenous CDK9 is a long-lived protein, with a half-life (t½) between 4 and 7 h depending on the cell type, the exogenously overexpressed CDK9 fusion protein was reported to degrade faster, with a t½ dependent on the level of its overexpression (31). Because we have shown that the ubiquitination of CDK9 is dependent on UBR5, we reasoned that if the role of this modification is to target CDK9 for proteosomal degradation, then the knockdown of UBR5 should have a detectable effect on the stability of this kinase. We used a luciferase-based reporter assay combined with siRNA treatment to evaluate this hypothesis. Constructs encoding N- or C-terminal CDK9-Renilla luciferase fusion proteins were co-transfected into HeLa cells with a normalizing plasmid expressing the Fluc and with UBR5-targeting or control siRNAs. After a double transfection procedure, de novo protein synthesis was blocked by incubating the cells with cycloheximide for 2–4 h, and the luciferase activity was measured with a dual luciferase assay, as described by Archambault and co-workers (32). As a control, we also performed the same set of experiments with the Renilla luciferase plasmid with no attached CDK9 moiety. The results indicate that, in our conditions, the overexpressed CDK9 has a half-life of less than 2 h (Fig. 5, upper panel), although the control protein Renilla luciferase (Rluc) was relatively stable over the duration of the experiment (Fig. 5, lower panel). In cells in which UBR5 was knocked down, the kinetics of CDK9 degradation was similar to that of cells transfected with control nontargeting siRNA or mock-treated cells (Fig. 5, upper panel). More specifically, the luciferase activity of the CDK9-Rluc fusion protein, normalized to that of Fluc in each sample, presents similar values at all time points and for all tested conditions. Similar results were obtained when the RLuc fused to the N-terminal (Fig. 5, upper panel) or C-terminal (data not shown) ends of CDK9 was analyzed. These results suggest that UBR5 is not involved in regulating CDK9 stability and that the ubiquitination event mediated by this E3 ligase does not target CDK9 for degradation by the proteasome.
FIGURE 5.
UBR5 knockdown does not affect the stability of CDK9. HeLa cells were co-transfected with constructs driving the expression of RLuc fused to the N-terminal region of CDK9 and with Fluc as an internal normalization control, together with siRNAs targeting UBR5 or control nontargeting siRNAs. Cycloheximide was added to culture medium for 0, 2, and 4 h to block overall protein synthesis, and luciferase activities were measured (upper panel). Parallel control experiments using the Rluc construct alone were conducted (lower panel). Each value represents the average of four assays, including two independent experiments.
TFIIS Increases CDK9 Occupancy along the γFBG Gene in a UBR5-dependent Manner and Activates Its Transcription
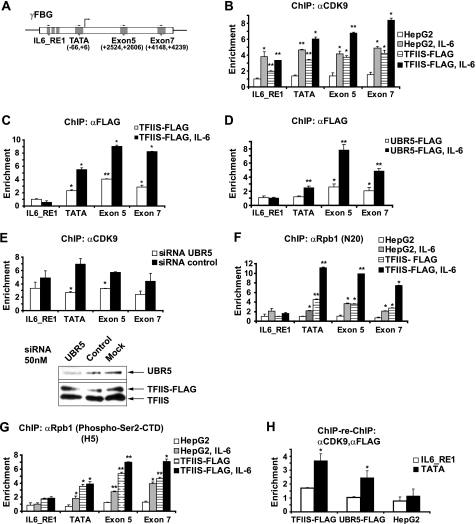
It was shown previously that the ubiquitination of CDK9 facilitates transactivation of HIV-1 genome transcription by Tat (30), suggesting that CDK9 ubiquitination may have a stimulatory effect on transcription. One possibility is that TFIIS promotes the activity of this kinase by mediating its ubiquitination by UBR5. To investigate this hypothesis, we used as a model the γFBG gene, which has been shown to be regulated through the recruitment of CDK9 upon IL-6 induction of HepG2 hepatocarcinoma cells (26). ChIP assays were performed in HepG2 cells with a two-step cross-linking procedure. Parental HepG2 cells or stable HepG2 cells expressing the TFIIS-FLAG protein were used for this ChIP procedure. The positions of the primers used to analyze the recruitment of different proteins along the γFBG gene are shown in Fig. 6A. Fig. 6B shows a significant increase in CDK9 occupancy levels along the γFBG gene upon IL-6 induction, as reported previously (26). Overexpression of TFIIS resulted in increased CDK9 occupancy levels along the sequence of this gene in basal conditions, with a further increase after IL-6 induction (Fig. 6B). This finding suggests that TFIIS stimulates the association of CDK9 with the γFBG gene chromatin.
FIGURE 6.
TFIIS, CDK9, and UBR5 are recruited to the γFBG gene, and TFIIS increases the occupancy of this gene by CDK9 in a UBR5-dependent manner. A, schematic representation of the γFBG gene and the position of primers used to amplify specific regions (IL6_RE1, TATA box, exon 5, and exon 7). B, parental or TFIIS-FLAG overexpressing HepG2 cells were subjected to a two-step ChIP assay prior to or following treatment with IL-6. After cross-linking, cell lysis, and sonication, the nucleoprotein complexes were immunoprecipitated with antibodies raised against CDK9 (C20). The occupancy levels were evaluated by quantitative PCR with primers along the γFBG gene, as indicated in A. C, TFIIS-FLAG-overexpressing HepG2 cells were used in ChIP experiments (as in B) prior to or following treatment with IL-6, with an anti-FLAG (M2) antibody at the immunoprecipitation step. D, UBR5-FLAG-overexpressing HepG2 cells were used in ChIP experiments (as in B), prior to or following treatment with IL-6, with the anti-FLAG (M2) antibody at the immunoprecipitation step. E, TFIIS-FLAG-overexpressing HepG2 cells were treated with either siRNAs targeting UBR5 or control nontargeting siRNAs. The cells treated with IL-6 were used in ChIP experiments with an anti-CDK9 antibody (C20) (upper panel). The knockdown efficiency is shown (lower panel). F, the N20 antibody that recognizes all forms of RNAPII (total) was used in ChIP experiments (as in B), prior to or following treatment with IL-6. G, an antibody that specifically recognizes the RNAPII Ser(P)-2-CTD form (H5) was used for the ChIP experiments. H, cells overexpressing TFIIS-FLAG, UBR5-FLAG, or parental HepG2 were treated with IL-6 and subjected to the ChIP-re-ChIP procedure with anti-CDK9 followed by anti-FLAG antibodies. The enrichment values for the IL6_RE1 and TATA box regions are shown. The data are presented as the means ± S.D. of at least three values including two independent experiments. Student's t tests were performed, and the levels of significance are indicated: *, p value <0.05; **, p value <0.01.
To assess whether this effect is due to the increased recruitment of TFIIS itself on the γFBG gene, we performed ChIP assays with an anti-FLAG antibody in the HepG2 cells stably expressing the FLAG-TFIIS protein. As shown in Fig. 6C, TFIIS is detected both in the region of the TATA box and in downstream transcribed regions of the γFBG gene, with higher enrichment values after IL-6 induction. The low TFIIS ChIP signals in the IL-6-RE1 region, both in basal conditions and after IL-6 induction, suggest that the effect of TFIIS on CDK9 occupancy in this region may be the result of modifications induced to CDK9 before its recruitment to the preinitiation complex. A similar set of experiments was performed in HepG2 cells stably expressing a form of UBR5 carrying a FLAG tag. The location profile of UBR5 along the γFBG gene was similar to that observed for TFIIS (Fig. 6D). This supports the idea that TFIIS mediates the effect of UBR5 on CDK9 via a physical association of all three proteins in the same genomic regions. As for TFIIS, the UBR5 signals observed in the IL-6-RE1 region were low (Fig. 6D), supporting the notion that the modifications of CDK9 by UBR5 may take place both before and after the recruitment of CDK9 to the chromatin.
To further define whether the effect of TFIIS on CDK9 loading along the γFBG gene requires UBR5, we assessed the impact of UBR5 knockdown (Fig. 6E, lower panel) on CDK9 occupancy in cells that overexpress TFIIS. The results show a decreased CDK9 occupancy of the promoter and exon 5 regions upon UBR5 depletion, as compared with a control siRNA treatment (Fig. 6E, upper panel). The effect of UBR5 knockdown on the CDK9 occupancy at the TATA box and exon 5 regions cannot be explained by a lower loading of CDK9 in the upstream IL6_RE1, because no significant effect was observed in this upstream region. Rather, this finding suggests a local, on-chromatin event. This conclusion is supported by the presence of the three proteins, CDK9, TFIIS, and UBR5, on TATA box and exon 5 regions, where an effect of UBR5 knockdown on the CDK9 occupancy is observed, paralleled by the absence of TFIIS and UBR5 in the IL6_RE1 region where no significant effect of UBR5 knockdown is observed. Taken together, these results argue in favor of a model in which the E3 ubiquitin ligase UBR5 mediates the activity of TFIIS on CDK9 in a mechanism that stimulates the loading of CDK9 along transcribed regions.
To investigate whether the effect of TFIIS on the recruitment of CDK9 has a functional consequence on transcription, we performed ChIP assays with two RNAPII antibodies. One antibody recognizes all forms of RNAPII (referred to here as total RNAPII) (Fig. 6F), and one specifically recognizes Ser(P)-2 on the CTD of the large subunit (Fig. 6G). Ser-2 residues are targets of CDK9, and RNAPII containing Ser(P)-2 is found in the 3′ region of genes (33). We analyzed both the parental HepG2 and TFIIS overexpressing cells. Total RNAPII loading on the γFBG gene is significantly increased upon induction. Notably, the overexpression of TFIIS increased the recruitment of total RNAPII to the promoter both in basal and induced conditions, suggesting that TFIIS increases the de novo recruitment of RNAPII. This conclusion is supported by the finding that TFIIS and RNAPII have similar location patterns on the γFBG promoter in cells overexpressing TFIIS both in basal conditions and after IL-6 induction (Fig. 6, C and F). Fig. 6G shows that the occupancy level of Ser(P)-2-CTD RNAPII is increased both in promoter and downstream transcribed regions upon IL-6 induction of parental HepG2 cells, which is again in agreement with previous results (26). Of note, overexpression of TFIIS markedly increased the occupancy level of this form of polymerase along the promoter and downstream transcribed regions, and the induction with IL-6 further increased the enrichment values. These results support the notion that the effect of TFIIS on the association of CDK9 with γFBG gene is functionally relevant (see “Discussion”). Notably, neither TFIIS nor the Ser(P)-2-CTD RNAPII was detected in the IL6_RE1 region.
Finally, to determine whether TFIIS, UBR5, and CDK9 belong to the same protein complex on the γFBG gene, we performed ChIP-re-ChIP experiments in cells stably expressing FLAG-tagged TFIIS or UBR5 proteins, by using anti-CDK9 and anti-FLAG antibodies in sequential immunoprecipitations. Fig. 6H shows that, when promoter-bound CDK9 is immunoprecipitated, both TFIIS and UBR5 are co-immunoprecipitated, because a significantly increased enrichment is observed in cells overexpressing TFIIS-FLAG or UBR5-FLAG but not in parental HepG2 cells. Nevertheless, no significant enrichment was observed at the IL6_RE1, suggesting, on one hand, that TFIIS and UBR5 are not associated with CDK9 over this region, and, on the other hand, that the signal obtained on the promoter region is specific. No co-localization signal for the three proteins was observed in transcribed regions downstream from the promoter (data not shown).
DISCUSSION
In this study, we provide evidence that TFIIS is involved in the ubiquitination of CDK9 (the kinase subunit of P-TEFb) by recruiting the E3 ubiquitin ligase UBR5. Our results indicate that UBR5 catalyzes the polyubiquitination of CDK9, and this modification does not lead to the degradation of CDK9. Rather, TFIIS increases the loading of CDK9 over a cytokine inducible gene in a UBR5-dependent fashion.
Independent reports showing that TFIIS and P-TEFb are required for the entry of RNAPII into productive elongation (3, 13) are in agreement with our findings. Our study provides a link between these two factors and reveals a new function of TFIIS in recruiting the E3 ubiquitin ligase UBR5 and thereby potentiating the loading of CDK9 on chromatin. UBR5 is a HECT domain E3 ubiquitin ligase involved in various processes, including the transactivation of progesterone and miocardin-controlled transcription (34, 35), the DNA damage response (36), and, on a larger scale, the initiation, maintenance, and/or termination of cell proliferation (37). UBR5 amplification and overexpression was reported to be common in different types of cancers, such as ovarian, breast, and hepatocellular carcinoma (38, 39). Interestingly, UBR5 has been shown to interact with and activate the DNA damage checkpoint kinase CHK2 (36). Our finding that endogenous UBR5 interacts under normal conditions with both TFIIS and CDK9 (Fig. 1) suggests that UBR5 participates in general RNAPII transcription mechanisms. Indeed, the chromatin immunoprecipitation of a FLAG-tagged version of UBR5 shows its association with transcribed regions in normal conditions (Fig. 6), a finding that is further supported by the identification of RNAPII itself in immunoprecipitates of endogenous UBR5 (Fig. 1).
Previous studies also pointed to a possible link between TFIIS and P-TEFb. TFIIS-deficient yeast cells were shown to present an increased dependence on BUR1/BUR2 or CTK1, the yeast orthologues of P-TEFb (40). Also, by using a combination of defined and crude in vitro transcriptions systems, a study conducted in humans concluded in the existence of a factor facilitating the function of P-TEFb in transcriptional elongation (41). Notably, whereas TFIIS, a nonessential gene in yeast, evolved to gain essential functions in higher eukaryotes, UBR5 does not have a yeast homologue and appeared later during evolution.
CDK9 is known to be ubiquitinated in vivo, and the level of its expression was suggested to be a determinant for the fate of the modified protein. When overexpressed, CDK9 is ubiquitinated and rapidly degraded (29); however, the endogenous protein is long-lived (31), and its ubiquitination may rather modulate its function in transcription by RNAPII (29, 30). Our results show that TFIIS is involved in the ubiquitination of endogenous CDK9 by recruiting the E3 ubiquitin ligase UBR5. Four lines of evidence support this conclusion. First, the interaction partner of TFIIS, UBR5, directly catalyzes the ubiquitination of CDK9 in an in vitro ubiquitination assay (Fig. 2). Second, when overexpressed, both TFIIS and UBR5 increase the smear of polyubiquitinated CDK9 species in in vivo ubiquitination assays (Fig. 3). Third, the knockdown of each of these proteins decreases the levels of ubiquitination of endogenous CDK9 (Fig. 4). Fourth, the knockdown of UBR5 abolishes the capacity of TFIIS to increase the smear of polyubiquitinated CDK9 species (Fig. 4). The appearance of new bands of polyubiquitinated species of CDK9 upon overexpression of TFIIS or UBR5 may indicate the use of new sites of ubiquitination on CDK9, or alternatively, the generation of intermediate polyubiquitinated species on the same site. On the other hand, dual ubiquitin ligase/deubiquitilase activities have been reported, as, for example, in the case of A20, an essential enzyme for attenuating NF-κB signaling (35). Based on our results, we cannot exclude that, when overexpressed, UBR5 may rather act as a deubiquitylase, leading to the appearance of intermediate polyubiquitinated species of CDK9.
In a previous report, the E3 ubiquitin ligase SCFSKP2 (SKP1-Cul1-F-box-protein ubiquitin ligase, S phase kinase-associated protein 2) was shown to ubiquitinate CDK9 (30). Because we did not identify this ubiquitin ligase as a TFIIS interaction partner in our systematic affinity purification coupled with mass spectrometry analysis, it is possible that different ubiquitination pathways participate in the ubiquitination of CDK9. The context could specify the ubiquitination pathway; for example, SCFSKP2 was shown to mediate the ubiquitination of CDK9 in the context of Tat transactivation of HIV1 transcription (30) or to serve as a signal for proteosomal degradation when CDK9 is overexpressed (29). As a precedent, it has been shown that p27 is efficiently ubiquitinated by SCFSKP2 only when it forms a complex with a CDK but not when it is in the free, noncomplexed form (42). Our results showing that the ubiquitination of endogenous CDK9 is dependent upon TFIIS and UBR5 suggest a role for this modification as a downstream event in processes mediated by TFIIS.
We used a sensitive luciferase assay (32) to evaluate whether the ubiquitination of CDK9 mediated by UBR5 has the role of triggering its degradation. Our results show that this is not the case, because the knockdown of UBR5 has no effect on the kinetics of CDK9 degradation (Fig. 5), suggesting that the depletion of UBR5 does not stabilize CDK9. This finding further suggests that CDK9 ubiquitination by UBR5 does not act in targeting CDK9 for degradation when it is overexpressed. Moreover, the endogenous CDK9 protein was not stabilized by the knockdown of UBR5 or that of TFIIS (Fig. 4), implying that when CDK9 is expressed at physiological levels, its ubiquitination by UBR5 has functions other than triggering its degradation, as in the case of the overexpressed protein. In support of this notion, the inhibition of proteosomal degradation by lactacystin did not affect the level of polyubiquitinated species of CDK9, whereas a parallel stabilization of the general polyubiquitinated proteins is observed in the whole cell extracts from the same cells (Fig. 3). Based on these results, it is tempting to speculate that polyubiquitinated CDK9 is recycled by de-ubiquitination, which would be in agreement with endogenous CDK9 being a long-lived protein, despite its ubiquitination in normal conditions. Alternatively, the polyubiquitin chains attached to CDK9 molecules could be edited, leading to a later degradation, as has been reported for other proteins, such as Rip1 and IRAK1 (43).
Experiments using pharmacological inhibitors or depletion by a siRNA treatment showed that CDK9 is generally required for RNAPII transcription (33, 44, 45). Its inhibition may affect transcription in two different ways. First, an accumulation of initiated RNAPII molecules, but paused in the promoter-proximal region of the transcription unit, was detected in the case of the Hsp70 gene in Drosophila melanogaster (13). Second, the inability to initiate transcription was observed for several cytokine inducible genes, such as those induced by IL-6 and TNF (46).
We took advantage of an IL-6 inducible system to further investigate the effect of TFIIS and UBR5 on CDK9. Indeed, the activation of the γFBG gene by IL-6 involves STAT3 activation by the JAK/Tyk tyrosine kinase pathway (47), which provokes the nuclear translocation of STAT3, the formation of a complex with CDK9, and their recruitment to the IL-6 response elements of the γFBG gene (26). By using antibodies detecting either all forms of RNAPII (total RNAPII) or the Ser(P)-2 CTD form, we showed that the enzyme is located both on the promoter and downstream transcribed regions only after IL-6 induction (Fig. 6). This finding suggests that upon IL-6 treatment, the transcription machinery is recruited to the promoter of this gene with no RNAPII molecules engaged in transcription or paused in the promoter-proximal region in the basal state. Although we do not know whether the polymerase is prone to stall in the promoter-proximal regions of the γFBG gene upon IL-6 induction, our results indicate that TFIIS is recruited to these regions. Its occupancy profile, paralleling that of UBR5, shows a similar increase in the promoter and downstream transcribed regions upon IL-6 induction, suggesting that TFIIS plays a role in both the initiation and elongation stages of transcription of this gene (Fig. 6).
Our ChIP experiments show that TFIIS is able to enhance the expression of the γFBG gene in basal conditions. When TFIIS is overexpressed, the occupancy of CDK9 along the promoter and downstream transcribed regions presents a significant increase compared with parental uninduced cells, and this is paralleled by a significant increase of the total RNAPII and of its Ser(P)-2 CTD form (Fig. 6). This finding suggests that TFIIS somehow mimics the events that follow STAT3 activation by IL-6. Remarkably, the occupancy of CDK9 is significantly increased on the upstream IL6_RE1 element by TFIIS overexpression, even though TFIIS does not itself occupy this region. Moreover, the polymerase occupancy is not increased concomitantly on the IL6_RE1 region, an effect that mimics RNAPII occupancy upon IL-6 induction, arguing against a nonspecific effect of TFIIS on CDK9 occupancy. Based on this observation, we propose that TFIIS associates with CDK9 before its recruitment to DNA and induces modifications that promote the binding of CDK9 to the chromatin.
The ubiquitination of CDK9 was shown to have both activating and inhibitory effects depending on the transcriptional context. It can facilitate the formation of a ternary complex, comprising P-TEFb, the Tat activator, and the TAR RNA motif on the HIV-1 LTR (30), but it can also have a negative impact on transcription activated by CIITA (major histocompatibility complex class II transactivator) (29). Because an association between CDK9, TFIIS, and UBR5 was observed in the soluble fraction of cell extracts (Fig. 1) and because the ubiquitination of CDK9 is dependent upon these two proteins (Fig. 4), it is tempting to speculate that this modification promotes CDK9 recruitment to the chromatin template. However, the knockdown of UBR5 in cells overexpressing TFIIS does not lead to a significant decrease of CDK9 occupancy at the IL6_RE1 (Fig. 6E), which rather suggests that neither the association with UBR5 nor the ubiquitination are essential for the CDK9 recruitment step. Notwithstanding, a significant decrease of CDK9 occupancy upon UBR5 knockdown is observed on and downstream of the promoter, which is apparently not caused by a reduced recruitment on the upstream regions (Fig. 6E). In agreement with these results, ChIP-re-ChIP experiments show that TFIIS, UBR5, and CDK9 belong to the same protein complex and co-localize on the promoter but not on the upstream IL6_RE1 (Fig. 6H). Of note, the effect of UBR5 on CDK9 occupancy is lost further downstream of the promoter, despite the fact that UBR5 remains physically associated with chromatin all along the gene. Together, these observations suggest that UBR5 is important for the transcriptional stages following initiation, such as the transition from initiation to early elongation. We speculate that the recruitment of UBR5 potentiated by TFIIS and the subsequent modifications induced to CDK9 favor the stability of the CDK9-RNAPII elongating complex and thereby promote RNAPII phosphorylation on Ser-2 of the CTD of its Rpb1 subunit. This model is supported by the observation that TFIIS increases the loading along the promoter and downstream transcribed regions not only of total RNAPII and CDK9 but also of Ser(P)-2 CTD RNAPII.
CDK9 is widely implicated in regulating the elongation phase of transcription by RNAPII and plays a key role in a multitude of biological processes, such as cell differentiation, proliferation, immune response, and inflammation. Its activity is tightly regulated by mechanisms involving (i) a dynamic release from and capture by inhibitory (7SK/HEXIM) and activating (phosphorylated transcription factors) complexes; (ii) regulated expression of its two isoforms; and (iii) post-translational modifications, including phosphorylation and ubiquitination (16, 46, 48). Our results depict a new mechanism of CDK9 regulation by the general transcription factor TFIIS. We show that this mechanism involves the ubiquitination of CDK9, most likely in the soluble cellular fraction, through the recruitment of the E3 ubiquitin ligase UBR5. We propose that TFIIS coordinates the initiation of transcription with an increased stability of the early CDK9(P-TEFb)-RNAPII transcribing complexes, which is dependent upon UBR5.
Acknowledgments
We are grateful to the members of our laboratory, Jacques Archambault, and François Robert for helpful discussions and comments. We thank Denis Faubert (Institut de Recherches Cliniques de Montréal Proteomics Discovery Platform) for mass spectrometry analysis.
This work is supported by grants from the Canadian Institutes for Health Research, Genome Canada, Génome Québec, the Natural Sciences and Engineering Research Council of Canada, and the Canadian Foundation for Innovation.
- TFIIS
- transcription factor IIS
- RNAPII
- RNA polymerase II
- P-TEFb
- positive transcription elongation factor b
- γFBG
- γ fibrinogen
- CTD
- C-terminal domain
- AMP-PNP
- adenosine 5′-(β,γ-imido) triphosphate tetralithium
- Fluc
- firefly luciferase
- Rluc
- Renilla luciferase
- CDK
- cyclin-dependent kinase
- DSIF
- DRB sensitivity factor
- NELF
- negative elongation factor
- TAP
- tandem-affinity purification.
REFERENCES
- 1. Sekimizu K., Kobayashi N., Mizuno D., Natori S. (1976) Biochemistry 15, 5064–5070 [DOI] [PubMed] [Google Scholar]
- 2. Kim B., Nesvizhskii A. I., Rani P. G., Hahn S., Aebersold R., Ranish J. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16068–16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adelman K., Marr M. T., Werner J., Saunders A., Ni Z., Andrulis E. D., Lis J. T. (2005) Mol. Cell 17, 103–112 [DOI] [PubMed] [Google Scholar]
- 4. Awrey D. E., Shimasaki N., Koth C., Weilbaecher R., Olmsted V., Kazanis S., Shan X., Arellano J., Arrowsmith C. H., Kane C. M., Edwards A. M. (1998) J. Biol. Chem. 273, 22595–22605 [DOI] [PubMed] [Google Scholar]
- 5. Wery M., Shematorova E., Van Driessche B., Vandenhaute J., Thuriaux P., Van Mullem V. (2004) EMBO J. 23, 4232–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kettenberger H., Armache K. J., Cramer P. (2003) Cell 114, 347–357 [DOI] [PubMed] [Google Scholar]
- 7. Nickels B. E., Hochschild A. (2004) Cell 118, 281–284 [DOI] [PubMed] [Google Scholar]
- 8. Cojocaru M., Jeronimo C., Forget D., Bouchard A., Bergeron D., Côte P., Poirier G. G., Greenblatt J., Coulombe B. (2008) Biochem. J. 409, 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prather D. M., Larschan E., Winston F. (2005) Mol. Cell. Biol. 25, 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ito T., Arimitsu N., Takeuchi M., Kawamura N., Nagata M., Saso K., Akimitsu N., Hamamoto H., Natori S., Miyajima A., Sekimizu K. (2006) Mol. Cell. Biol. 26, 3194–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saunders A., Core L. J., Lis J. T. (2006) Nat. Rev. Mol. Cell Biol. 7, 557–567 [DOI] [PubMed] [Google Scholar]
- 12. Margaritis T., Holstege F. C. (2008) Cell 133, 581–584 [DOI] [PubMed] [Google Scholar]
- 13. Ni Z., Saunders A., Fuda N. J., Yao J., Suarez J. R., Webb W. W., Lis J. T. (2008) Mol. Cell. Biol. 28, 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujinaga K., Irwin D., Huang Y., Taube R., Kurosu T., Peterlin B. M. (2004) Mol. Cell. Biol. 24, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H. (2006) Mol. Cell 21, 227–237 [DOI] [PubMed] [Google Scholar]
- 16. Peterlin B. M., Price D. H. (2006) Mol. Cell 23, 297–305 [DOI] [PubMed] [Google Scholar]
- 17. Ping Y. H., Rana T. M. (2001) J. Biol. Chem. 276, 12951–12958 [DOI] [PubMed] [Google Scholar]
- 18. Egloff S., Murphy S. (2008) Trends Genet. 24, 280–288 [DOI] [PubMed] [Google Scholar]
- 19. Palangat M., Renner D. B., Price D. H., Landick R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15036–15041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeronimo C., Langelier M. F., Zeghouf M., Cojocaru M., Bergeron D., Baali D., Forget D., Mnaimneh S., Davierwala A. P., Pootoolal J., Chandy M., Canadien V., Beattie B. K., Richards D. P., Workman J. L., Hughes T. R., Greenblatt J., Coulombe B. (2004) Mol. Cell. Biol. 24, 7043–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., Thérien C., Bergeron D., Bourassa S., Greenblatt J., Chabot B., Poirier G. G., Hughes T. R., Blanchette M., Price D. H., Coulombe B. (2007) Mol. Cell 27, 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cloutier P., Al-Khoury R., Lavallée-Adam M., Faubert D., Jiang H., Poitras C., Bouchard A., Forget D., Blanchette M., Coulombe B. (2009) Methods 48, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeghouf M., Li J., Butland G., Borkowska A., Canadien V., Richards D., Beattie B., Emili A., Greenblatt J. F. (2004) J. Proteome. Res. 3, 463–468 [DOI] [PubMed] [Google Scholar]
- 24. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 25. Nowak D. E., Tian B., Brasier A. R. (2005) BioTechniques 39, 715–725 [DOI] [PubMed] [Google Scholar]
- 26. Hou T., Ray S., Brasier A. R. (2007) J. Biol. Chem. 282, 37091–37102 [DOI] [PubMed] [Google Scholar]
- 27. Umehara T., Kida S., Yamamoto T., Horikoshi M. (1995) Gene 167, 297–302 [DOI] [PubMed] [Google Scholar]
- 28. Callaghan M. J., Russell A. J., Woollatt E., Sutherland G. R., Sutherland R. L., Watts C. K. (1998) Oncogene 17, 3479–3491 [DOI] [PubMed] [Google Scholar]
- 29. Kiernan R. E., Emiliani S., Nakayama K., Castro A., Labbé J. C., Lorca T., Nakayama K. K., Benkirane M. (2001) Mol. Cell. Biol. 21, 7956–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barboric M., Zhang F., Besenicar M., Plemenitas A., Peterlin B. M. (2005) J. Virol. 79, 11135–11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garriga J., Bhattacharya S., Calbó J., Marshall R. M., Truongcao M., Haines D. S., Graña X. (2003) Mol. Cell. Biol. 23, 5165–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gagnon D., Joubert S., Sénéchal H., Fradet-Turcotte A., Torre S., Archambault J. (2009) J. Virol. 83, 4127–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., Young R. A. (2010) Cell 141, 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henderson M. J., Russell A. J., Hird S., Muñoz M., Clancy J. L., Lehrbach G. M., Calanni S. T., Jans D. A., Sutherland R. L., Watts C. K. (2002) J. Biol. Chem. 277, 26468–26478 [DOI] [PubMed] [Google Scholar]
- 35. Hymowitz S. G., Wertz I. E. (2010) Nat. Rev. Cancer 10, 332–341 [DOI] [PubMed] [Google Scholar]
- 36. Henderson M. J., Munoz M. A., Saunders D. N., Clancy J. L., Russell A. J., Williams B., Pappin D., Khanna K. K., Jackson S. P., Sutherland R. L., Watts C. K. (2006) J. Biol. Chem. 281, 39990–40000 [DOI] [PubMed] [Google Scholar]
- 37. Mansfield E., Hersperger E., Biggs J., Shearn A. (1994) Dev. Biol. 165, 507–526 [DOI] [PubMed] [Google Scholar]
- 38. Clancy J. L., Henderson M. J., Russell A. J., Anderson D. W., Bova R. J., Campbell I. G., Choong D. Y., Macdonald G. A., Mann G. J., Nolan T., Brady G., Olopade O. I., Woollatt E., Davies M. J., Segara D., Hacker N. F., Henshall S. M., Sutherland R. L., Watts C. K. (2003) Oncogene 22, 5070–5081 [DOI] [PubMed] [Google Scholar]
- 39. Chin S. F., Teschendorff A. E., Marioni J. C., Wang Y., Barbosa-Morais N. L., Thorne N. P., Costa J. L., Pinder S. E., van de Wiel M. A., Green A. R., Ellis I. O., Porter P. L., Tavaré S., Brenton J. D., Ylstra B., Caldas C. (2007) Genome Biol. 8, R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lindstrom D. L., Hartzog G. A. (2001) Genetics 159, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng B., Price D. H. (2007) J. Biol. Chem. 282, 21901–21912 [DOI] [PubMed] [Google Scholar]
- 42. Lacy E. R., Filippov I., Lewis W. S., Otieno S., Xiao L., Weiss S., Hengst L., Kriwacki R. W. (2004) Nat. Struct. Mol. Biol. 11, 358–364 [DOI] [PubMed] [Google Scholar]
- 43. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 44. Chao S. H., Price D. H. (2001) J. Biol. Chem. 276, 31793–31799 [DOI] [PubMed] [Google Scholar]
- 45. Shim E. Y., Walker A. K., Shi Y., Blackwell T. K. (2002) Genes Dev. 16, 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brasier A. R. (2008) Cell Cycle 7, 2661–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heinrich P. C., Behrmann I., Müller-Newen G., Schaper F., Graeve L. (1998) Biochem. J. 334, 297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S., Fischer P. M. (2008) Trends Pharmacol. Sci. 29, 302–313 [DOI] [PubMed] [Google Scholar]