Abstract
Modern α-proteobacteria are thought to be closely related to the ancient symbiont of eukaryotes, an ancestor of mitochondria. Respiratory complex I from α-proteobacteria and mitochondria is well conserved at the level of the 14 “core” subunits, consistent with that notion. Mitochondrial complex I contains the core subunits, present in all species, and up to 31 “supernumerary” subunits, generally thought to have originated only within eukaryotic lineages. However, the full protein composition of an α-proteobacterial complex I has not been established previously. Here, we report the first purification and characterization of complex I from the α-proteobacterium Paracoccus denitrificans. Single particle electron microscopy shows that the complex has a well defined L-shape. Unexpectedly, in addition to the 14 core subunits, the enzyme also contains homologues of three supernumerary mitochondrial subunits as follows: B17.2, AQDQ/18, and 13 kDa (bovine nomenclature). This finding suggests that evolution of complex I via addition of supernumerary or “accessory” subunits started before the original endosymbiotic event that led to the creation of the eukaryotic cell. It also provides further confirmation that α-proteobacteria are the closest extant relatives of mitochondria.
Keywords: Bioenergetics, Electron Microscopy (EM), Membrane Proteins, Mitochondria, Respiratory Chain
Introduction
Most eukaryotic cells contain mitochondria, which are thought to be the remnants of an ancient endosymbiotic event about 1.5–2 billion years ago, when a respiring bacterium entered a host proto-eukaryotic cell (1–3). Sequence comparisons suggest that the closest modern relatives of the proto-mitochondrion are α-proteobacteria, Gram-negative bacteria that can exist as a free-living bacteria, although the majority of members of this group live as symbionts or parasites of plants and animals (4). Members of the rickettsial subdivision of the α-proteobacteria, a group of intracellular parasites, are considered to be the closest known eubacterial relatives of mitochondria (5). Phylogenetic evidence suggests that all mitochondrial genomes descended from a common ancestor, implying that mitochondria originated only once in evolution (4). Energy production in the form of ATP is the major mitochondrial function and can be traced directly to the α-proteobacterial ancestor. The respiratory chain complexes of these bacteria are thus most closely related to the mitochondrial enzymes.
Complex I (NADH:ubiquinone oxidoreductase) is the first and the largest component of the respiratory chain, catalyzing transfer of two electrons from NADH to ubiquinone, coupled to the translocation of four protons (current consensus value) across the membrane (6–10). In doing so, it provides about 40% of the proton-motive force used by ATP synthase for the generation of ATP. Because of its central role in bioenergetics and the high energy demand in neuronal tissues, complex I is implicated in many human neurodegenerative diseases and possibly in aging (11). Bacterial complex I resides in the plasma membrane and is thought to consist of a minimal set of 14 different subunits of about 550 kDa in total. One known exception is complex I from Thermus thermophilus, which contains an additional frataxin-like subunit Nqo15 (12). The 14 “core” subunits are also present in the mitochondrial enzyme, which resides in the inner membrane and contains up to 31 additional subunits, bringing the total molecular mass to about 1 MDa (13). The core subunits are well conserved and contain all the redox centers of the complex (flavin mononucleotide and 8–9 Fe-S clusters). Furthermore, bacterial and mitochondrial enzymes have a similar distinctive L-shape and catalyze the same reaction. Therefore, bacterial complex I is a valid “minimal” model of the mitochondrial enzyme. Seven of the core subunits are hydrophilic and form the peripheral arm of the complex, containing all the redox centers, which are involved in the electron transfer from NADH to quinone. The other seven core subunits are highly hydrophobic and form the membrane arm of the enzyme, containing proton-translocating machinery. We have determined the crystal structures of both domains separately, as well as that of the entire complex (at the backbone level for the membrane domain), suggesting the likely mechanisms for electron transfer and proton translocation (14–16).
The additional, so-called “supernumerary” or “accessory,” subunits are generally thought to have been acquired by complex I during the evolution of eukaryotes (17). Their specific roles are not clear but are likely to include assistance in the assembly, stability, and regulation of the complex. Some supernumerary subunits may confer additional functions to the mitochondrial enzyme; the 39-kDa subunit (bovine nomenclature is used here for these subunits) contains tightly bound NADPH and is homologous to short chain dehydrogenases (18); the SDAP subunit is homologous to acyl-carrier proteins, and subunit B16.6 is identical to GRIM-19, an apoptosis-inducing factor (6, 7, 19). Previous bioinformatic analyses identified a group of 16 “extra” subunits and two assembly factors that are common to all eukaryotic species, likely representing supernumerary subunits of a primitive ancient eukaryotic complex I (20). After the diversification of eukaryotes, complex I in plants, fungi, and metazoans acquired different sets of yet more subunits. It was also noted that α-proteobacterial genomes contain proteins that are homologous to five supernumerary subunits (20). However, they were not expected to be incorporated into complex I, as, in the sequenced genomes, they never occur in operons with complex I proteins (20). Without direct characterization of the enzyme, it is not possible to establish whether these proteins were already components of complex I in the proto-mitochondrion or were recruited later in evolution into a primitive eukaryotic complex I.
Paracoccus denitrificans is a well characterized model α-proteobacterial soil organism with a fully sequenced genome (strain Pd 1222, img.jgi.doe.gov). Complex I from this species is relatively well studied in terms of activity, subunit topology and proximity, characteristics of Fe-S clusters and mutagenesis (9, 21). However, the intact complex was never purified previously due to its apparent fragility upon solubilization (22, 23). The nucleotide sequence of the nqo operon, encoding core complex I subunits Nqo1–14, also revealed the presence of six unidentified reading frames within the operon (24). However, their putative products, if any, are not related to known complex I subunits and were not expected to form part of the complex (23, 24). Clearly, to establish subunit composition, the intact complex has to be purified. One of the reasons to study P. denitrificans complex I is its high sequence similarity to the mammalian enzyme, which makes it a very good bacterial model to study, for example, pathological mutations in human complex I. Indeed, sequence identities to the human enzyme for many hydrophilic subunits are in the range of 60–70% for complex I from P. denitrificans but are only 30–40% for Escherichia coli or T. thermophilus (Table 1).
TABLE 1.
Sequence comparisons of the 14 core complex I subunits from different species
Subunits are grouped according to complex I domain organization (8, 32). The names of subunits are in bovine and P. denitrificans nomenclature. Multiple sequence alignment of complex I subunits from human and species listed was performed with ClustalW2 (53). Sequence identity (and similarity in brackets) was calculated compared with human sequences.
| Domain | Subunit | Bovine | P. denitrificans | T. thermophilus | E. coli |
|---|---|---|---|---|---|
| % | % | % | % | ||
| Dehydrogenase | 51 kDa/Nqo1 | 97 (99) | 64 (77) | 43 (57) | 38 (56) |
| 24 kDa/Nqo2 | 96 (98) | 36 (52) | 33 (54) | 34 (56) | |
| 75 kDa/Nqo3 | 97 (99) | 52 (64) | 23 (39) | 16 (29) | |
| 49 kDa/Nqo4 | 95 (99) | 59 (77) | 44 (64) | 27 (42) | |
| 30 kDa/Nqo5 | 89 (94) | 51 (66) | 25 (38) | 8 (15) | |
| Connecting | PSST/Nqo6 | 86 (91) | 69 (81) | 47 (67) | 36 (51) |
| TYKY/Nqo9 | 93 (95) | 72 (82) | 31 (43) | 29 (41) | |
| ND3/Nqo7 | 74 (83) | 28 (50) | 21 (41) | 22 (41) | |
| Membrane | ND1/Nqo8 | 78 (88) | 35 (54) | 35 (51) | 35 (56) |
| ND6/Nqo10 | 62 (75) | 17 (33) | 15 (31) | 17 (36) | |
| ND4L/Nqo11 | 73 (90) | 24 (49) | 23 (42) | 19 (46) | |
| ND5/Nqo12 | 69 (81) | 28 (41) | 31 (46) | 28 (45) | |
| ND4/Nqo13 | 74 (85) | 26 (47) | 24 (40) | 26 (44) | |
| ND2/Nqo14 | 63 (81) | 14 (29) | 19 (31) | 16 (32) |
Here, we report the first purification and characterization of the intact complex I from P. denitrificans. Unexpectedly, in addition to the 14 core subunits, the complex contains three supernumerary subunits, homologues of bovine complex I subunits B17.2, AQDQ, and 13 kDa.
EXPERIMENTAL PROCEDURES
Materials
All detergents were purchased from Glycon (Luckenwalde, Germany). Chromatography columns and instrumentation were from GE Healthcare. E. coli total lipid extract was from Avanti Polar Lipids (Delfzyl, Netherlands). CompleteTM protease inhibitor mixture, EDTA-free, tablets were from Roche Diagnostics, and all other chemicals were from Sigma.
Bacterial Growth and Membrane Preparation
P. denitrificans strain DSM 413 was obtained as a freeze-dried culture from DSMZ and grown in 5 ml of BT media (2.5 g/liter peptone, 2.5 g/liter yeast extract, 2.5 g/liter casamino acids, 10 g/liter glucose, 10 g/liter K2HPO4, adjusted to pH 6.8 with H2SO4) (25). Samples were stored in 1-ml aliquots at −80 °C in 20% glycerol. Cells were grown from these subcultures in 50 ml of BTS media (BT media with the addition of 1 g/liter NH4Cl, 1.25 mm MgCl2, 1 mm citric acid, 1 ml/liter salts stock (0.1 m CaCl2, 90 mm FeCl2, 50 mm MnCl2, 25 mm ZnCl2, 10 mm CoCl2, 5 mm CuCl2, 5 mm H3BO3, 10 mm Na2MoO4, dissolved by addition of 1 m HCl), adjusted to pH 6.8 with H2SO4) for 16 h. The extra salts added are derived from the recipe in Ref. 26. 10 ml of this culture was added to 0.5 liters of BTS media for 8 h, and finally the resulting 0.5 liters of culture was added to 55 liters of BTS in a 70-liter Bio Clave fermentor (Applikon, Gloucestershire, UK) at 30 °C and 80% dissolved oxygen until late log phase (about 16 h). Cells were harvested by centrifugation at 18,300 × g (11,000 rpm in a Sorvall SLA-1500 rotor) for 20 min and then stored at −80 °C. The total yield was around 2.2 kg of cell paste. All subsequent steps were carried out at 4 °C. Thawed cells (350 g wet weight) were resuspended in 900 ml of RBP buffer (50 mm BisTris,3 pH 6.5, 0.002% phenylmethanesulfonyl fluoride (PMSF), and a CompleteTM protease inhibitor mixture, EDTA-free, tablet) using a glass-Teflon homogenizer and passed through a Z-plus 2.2 kilowatt disruptor once at 15,000 p.s.i. and three times at 30 000 p.s.i. Cell debris was removed by centrifugation at 18,300 × g (11,000 rpm in a Sorvall SLA-1500 rotor) for 45 min, followed by 31,900 × g (14,500 rpm in SLA-1500 rotor) for 1 h. The membrane fraction was collected by ultracentrifugation at 151,500 × g (44,000 rpm in Ti45 rotor) for 4 h, the supernatant discarded, and the pellet resuspended in 250 ml of RBP in a glass-Teflon homogenizer. The membranes were stored at −80 °C.
Purification of Complex I
Membranes made from 350 g of cells were used for a single purification. All steps were carried out at 4 °C. PMSF (0.002%), 100 mm NaCl, 10 mm CaCl2, a protease inhibitor cocktail-EDTA-free tablet, 10% glycerol, and 2.85% dodecyl maltoside (DDM) were added to membranes left under constant stirring for 4 h, and nonsolubilized material was removed by centrifugation at 151,500 × g for 1 h. The supernatant was passed through a 0.45-μm filter. The sample was purified using three ion exchange steps and a final gel filtration step using the ÄKTA Explorer chromatography system. The absorbance of the eluate was monitored at 280/420 nm to follow absorbance changes due to co-factors present in various proteins. All ion exchange columns were pre-equilibrated with buffer A (20 mm BisTris, pH 6.5, 10 mm CaCl2, 0.1% DDM, 20% glycerol, and 0.002% PMSF). The sample was applied to a HiLoad 26/10 Q-Sepharose column, washed with 10% buffer B (buffer A and 1 m NaCl), and then eluted with a 50-ml gradient from 10 to 15% followed by an 800-ml gradient from 15 to 25% buffer B. Fractions (9 ml) were collected, and those with high NADH:ferricyanide (FeCy) activity were pooled and diluted with an equal volume of buffer A. The diluted material was reapplied to the Q-Sepharose column and eluted with the same method. Fractions (9 ml) were collected, and those with NADH:FeCy activity were pooled, concentrated, and then diluted with 40 volumes of buffer A. Pooled, diluted material from the Q-Sepharose column was applied to a Mono S (HR 16/10) column. Protein was eluted with an 800-ml gradient from 0 to 30% of buffer B. Fractions (9 ml) were collected, assayed for NADH:FeCy activity, and analyzed by SDS-PAGE. Selected fractions were pooled and concentrated (Vivacell-20; 50,000-Da molecular mass cutoff) to ∼2 ml and applied to a HiLoad 16/60 Superdex 200 gel filtration column equilibrated in 20 mm BisTris, pH 6.5, 10 mm CaCl2, 150 mm NaCl2, 0.1% DDM, 10% glycerol, and 0.002% PMSF. Eluted fractions (1.5 ml) were assayed for NADH:FeCy activity and analyzed by SDS-PAGE. Selected fractions were pooled, and then glycerol and CaCl2 were added to give a final concentration of ∼20 mg/ml protein in 20 mm BisTris, pH 6.5, 100 mm NaCl, 10 mm CaCl2, and 25% glycerol. Protein was used immediately or stored under liquid nitrogen.
Analytical Methods
Protein concentrations were measured using the bicinchoninic acid assay according to the manufacturer's instructions (Pierce). SDS-PAGE was performed with Novex Tris-glycine polyacrylamide gels containing a 4–20% acrylamide gradient (Invitrogen), according to the manufacturer's instructions. Protein bands were visualized with Coomassie Blue R-250 stain. Enzyme activity assays were performed at 30 °C in a Shimadzu UV-1601 spectrophotometer. NADH:FeCy reaction was followed at 340 nm in an assay buffer containing 50 mm MES, pH 6.5, 0.1% DDM, and 10 mm CaCl2. Reactions were started by the addition of ∼2 μg of protein to a 1-ml assay mixture containing 0.1 mm NADH and 1 mm FeCy. The oxidation of NADH in the presence of decyl-ubiquinone (DQ) was followed at 340 nm at 30 °C. The assay buffer contained 10 mm MES, pH 6.5, 25 mm NaCl, 2 mm CaCl2, 0.25 mg/ml E. coli total lipids, and 0.1% CHAPS. Reactions were initiated by the addition of 0.1 mm NADH after a 5-min preincubation of 2 ml of assay buffer with ∼5 μg of protein and 0.1 mm DQ.
Identification of Subunits
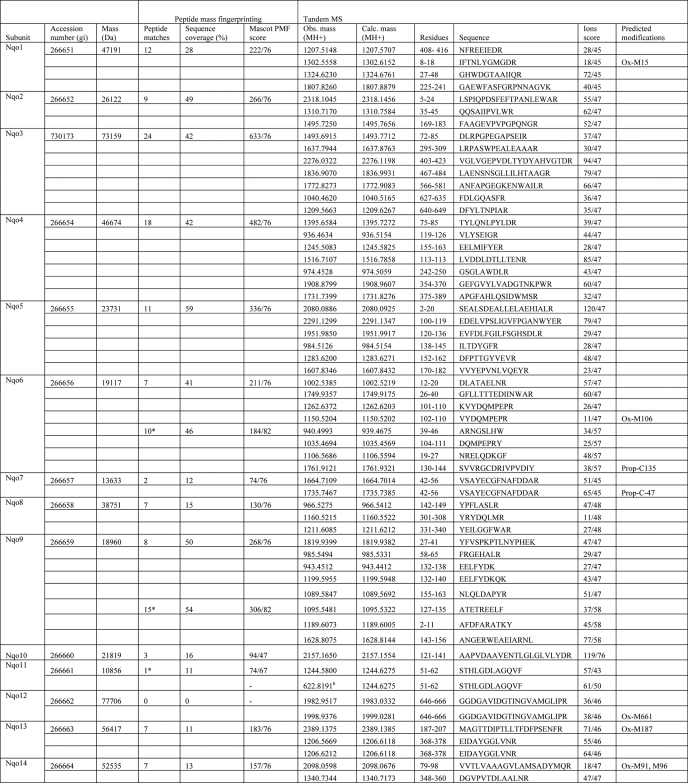
Subunits of the purified complex were separated by SDS-PAGE, stained with Coomassie Blue dye, excised from gels, and digested by “in gel” cleavage with either trypsin or chymotrypsin. The resultant peptides were analyzed by MALDI-TOF-TOF mass spectrometry (model 4800, Applied Biosystems) using α-cyanohydroxy-trans-cinnamic acid as the matrix. The instrument was calibrated with bovine trypsin autolysis products (m/z values 2163.057 and 2273.160) and a calcium-related matrix ion (m/z value, 1060.048). Some proteolytic digests were also analyzed by tandem MS using an LTQ OrbiTrap XL mass spectrometer (Thermo) following chromatography on a nanoscale reverse-phase column (75 μm inner diameter × 60 mm; Nanoseparations, Nieukoop, Netherlands) with an acetonitrile gradient 0.1% (v/v) formic acid; 250 nl/min). Proteins were identified by comparison of both peptide mass and peptide fragmentation data with National Center for Biotechnology Information (NCBI) sequence data base using the MASCOT algorithm (27).
Measurement of Molecular Masses
Molecular masses of subunits were determined by liquid chromatography mass spectrometry. Subunits of the purified complex were resolved by reverse-phase chromatography using a combination of chromatographic supports and solvent systems compatible with the separation and recovery of hydrophobic membrane proteins (28, 29). The eluate was introduced directly via an electrospray ionization (ESI) interface into a Quattro Ultima triple quadrupole mass spectrometer (Waters) operated in positive ion single mass spectrometry (MS) mode, tuned, and calibrated with a mixture of horse heart myoglobin and bovine trypsinogen. Molecular masses were determined from series of multiply charged ions using the component analysis function of the computer program MassLynx (Waters). In some cases mass measurements were made off-line to reverse-phase HPLC separations. Portions of fractions, collected according to the UV absorbance of the eluate, were introduced via a Rheodyne injector into a stream (3 μl/min) of 50% aqueous propanol and analyzed by ESI-MS.
Electron Microscopy
For single particle analysis, the protein was diluted to 5 μg/ml in buffer containing 10 mm MES, pH 6.5, 1 mm CaCl2, 5 mm NaCl, 0.1% DDM and applied to carbon-coated copper grids (glow-discharged in air). After a 2-min incubation, excess buffer was removed by blotting, and the grid was washed four times with the same buffer containing no detergent and then stained with 2% uranyl acetate for 10 s. Images were recorded with a Philips/FEI Tecnai 12 microscope operating at 120 kV and a calibrated magnification of ×40,580 (nominal 30,000), on an UltraScan 1000 2k × 2k camera (Gatan. Inc., Pleasanton, CA) with 14-μm pixel size (3.45 Å at the specimen level). Electron dose was ∼10 e−/Å2. A condenser aperture of 50 μm and an objective aperture of 150 μm were used. The best 157 images of well stained particles, with no drift or astigmatism, were selected for further image processing. The particles were manually selected and boxed (128 × 128 pixels) using Ximdisp from the MRC software suite (30). This dataset was processed using IMAGIC software (31). Boxed particles were bandpass-filtered to limit frequencies to a resolution range corresponding to 10–250 Å. The particles were then centered (in five iterations); for this, a translational alignment was used with a total sum of all particle images (rotationally averaged) as a reference. Centered particles were subjected to multivariate-statistical analysis and classified into 12 classes. The two best quality class averages (for “flip” and “flop” orientations) were used as references for a multireference alignment; the best class averages obtained were used as references for the next alignment. Class averages ceased to improve after four rounds of multireference alignment. For the final analysis, class averages of either four (each included from 51 to 33 particles) or two (flip and flop) classes were created.
RESULTS
Purification of Complex I from P. denitrificans
Bacterial complex I, especially that from proteobacteria (23), is notoriously unstable and sensitive to detergent. Well characterized preparations of intact bacterial complex I have so far been obtained from E. coli (32, 33), Aquifex aeolicus (34), and recently T. thermophilus (15). The P. denitrificans enzyme was previously isolated only either as a subcomplex (22) or in low yields as part of a supercomplex with respiratory complexes III and IV (35).
As a first step toward purification of P. denitrificans complex I in large yields, sufficient for structural studies, we optimized the cell growth conditions. Previously published media were adapted by a combination of rich media base (25) with trace metals and other salts (26, 36). The addition of salts approximately doubled the yield of cells, to about 50 g/liter in fermentor conditions under full (or 80% dissolved oxygen) aeration. In contrast to E. coli (33) and T. thermophilus (12), limiting oxygen supply did not increase expression of P. denitrificans complex I. However, the deamino-NADH:FeCy activity (specific for complex I, as other dehydrogenases cannot use dNADH) in the membranes (∼2 units/mg protein) was comparable with that achieved in other bacterial species. Specific activity was highest in the late log/early stationary phase, and therefore cells were grown in the fermentor until this phase.
To identify conditions where the solubilized complex remains stable, we used the apparent elution volume of dNADH:FeCy activity from the gel filtration column as an indicator. The size of the intact complex I with bound detergent micelle was expected to be ∼600–650 kDa. Initial attempts at purification roughly followed the procedure developed for the E. coli enzyme, using DDM as detergent, pH 6.0 buffers with 10% glycerol, and 2 mm CaCl2 as a stabilizing cation (33). When solubilized membranes were applied directly to a gel filtration column, most of complex appeared intact, eluting at ∼650 kDa, with only a minor fraction (∼20%) eluting as a peripheral arm (∼400 kDa) or smaller species. However, the complex dissociated after anion exchange chromatography. Therefore, a systematic search was performed for conditions where it remains stable under prolonged purification. For this assay, solubilized membranes were first applied to anion exchange spin Maxi-Q column and then applied to the gel filtration column. These tests indicated that the complex remains stable in the pH range 6–7 and gradually dissociates when the pH is decreased below 6 or increased above 7. The optimal pH was found to be 6.5. The addition of 10% glycerol during the solubilization of membranes was beneficial for stability. In these conditions, solubilization of membranes required high detergent to protein ratio to recover at least 80% of the complex I activity (2.85% DDM at ∼9–10 mg/ml, i.e. ∼3:1 detergent/protein). An increase of glycerol concentration in column buffers to 20% was also essential. Finally, we noted that the presence of EDTA had an extreme de-stabilizing effect, more severe than in E. coli or T. thermophilus, and therefore we increased the divalent cation concentration in buffers (10 mm CaCl2 was found to be optimal for stability).
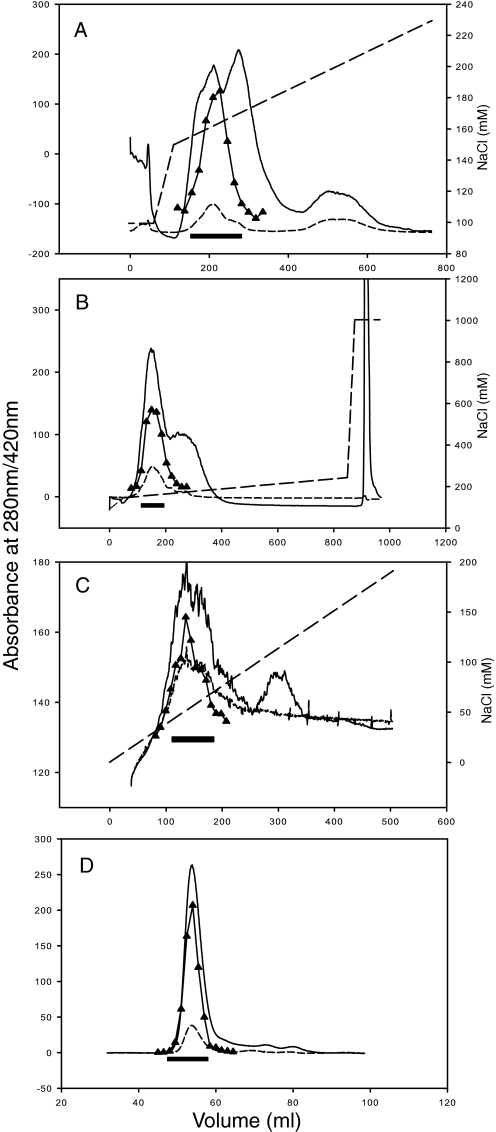
A combination of measures listed above allowed us to maintain complex integrity over the extensive four-step purification procedure. To achieve sufficient purity, a range of columns was explored. NADH:FeCy activity was used as a guide during purification, if an alternative dehydrogenase activity is present in P. denitrificans membranes, it does not appear to be significant. Although DEAE and ANX columns were useful, the best combination was found to be two similar Q-Sepharose steps, followed by Mono S and gel filtration columns (Table 2 and Fig. 1). The second Q-Sepharose step improved the purity of the sample due to a much lower protein load than in the first step. A peculiar property of complex I from E. coli, T. thermophilus, and P. denitrificans is that it binds to both anion exchange and cation exchange columns at the same pH. This appears to be a consequence of its bi-partite organization, and experiments with isolated domains (Refs. 12, 15 and data not shown) indicate that it binds to anion exchange columns mainly via the peripheral domain and to the cation exchange columns mainly via the membrane domain, presumably reflecting opposing surface charges of the domains. The significant loss of activity after purification on Mono S likely reflects the dissociation of a proportion of the enzyme, so that activity from the peripheral domain is found in the unbound fraction (as confirmed by gel filtration chromatography of the flow-through material, data not shown).
TABLE 2.
Representative purification of complex I from P. denitrificans
Starting material was 350 g of P. denitrificans cells.
| Purification step | Volume | Protein | Total activitya | Specific activity |
|---|---|---|---|---|
| ml | mg | μmol min−1 | μmol min−1mg−1 | |
| Membranes | 300 | 3000 | 4842 | 1.61 |
| Solubilized membranes | 780 | 2808 | 4064 | 1.45 |
| Q-Sepharose | 207 | 472 | 3185 | 6.75 |
| Q-Sepharose | 126 | 212 | 2331 | 11.01 |
| Mono S | 63 | 9 | 328 | 34.70 |
| Superdex 200 | 7.5 | 3.7 | 301 | 81.80 |
a The activity was measured by following the oxidation of NADH while FeCy was reduced at 30 °C. dNADH was used for membranes and solubilized membranes.
FIGURE 1.
Purification of complex I from P. denitrificans. A, Q-Sepharose 26/10 column chromatography; B, repeat of Q-Sepharose 26/10 chromatography; C, Mono S 16/10 column chromatography; and D, Superdex 200 16/60 column chromatography. The solid line represents the 280-nm absorbance; the short dashed line represents the 420-nm absorbance; the long dashed line represents the NaCl concentration; the black bar represents fractions used in subsequent steps; and triangles represent complex I activity (NADH:FeCy) in individual fractions.
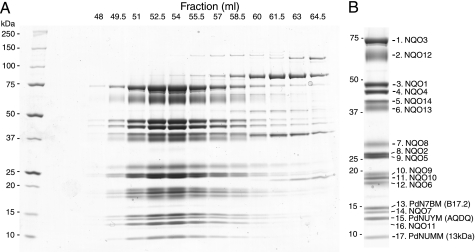
SDS-PAGE analysis of the fractions eluted from the last gel filtration column (Fig. 2A) shows that the preparation is highly pure, with 17 subunits of the complex (see below) all eluting in concert with the peak at about 53 ml (coincidental with thyroglobulin calibration standard, 669 kDa). Several protein impurities elute later, at 60–64 ml, and so only the first 5–6 fractions (∼48–56 ml, Fig. 2A) were routinely pooled as a pure enzyme. The final yield (4–5 mg) is sufficient for functional and, importantly, structural studies.
FIGURE 2.
Subunit composition of complex I from P. denitrificans. A, SDS-PAGE analysis of Superdex 200 fractions. Protein was stained with Coomassie Blue. Molecular mass markers are in the 1st lane and are indicated at the left. B, purified enzyme, as used for MS analyses. Identified subunits are indicated on the right. Hydrophobic subunit Nqo11 is weakly stained by Coomassie Blue but is more clearly visible in silver-stained gels (not shown). Positions of Mr markers are shown on the left.
Activity and Stability of the Preparation
The activity of complex I in the membranes is similar to values reported previously (37) and is sensitive to piericidin A and rotenone, specific complex I inhibitors acting at the quinone-binding site. The purified enzyme shows high NADH:quinone oxidoreductase activity with DQ as an acceptor (Table 3). It corresponds to a turnover number of 140 s−1, similar to values observed for bovine complex I in the membranes (∼150 s−1 (38)) and achieved recently with the purified bovine enzyme (39). This activity is somewhat lower than can be achieved with E. coli enzyme under optimal conditions (33), possibly due to the use of E. coli rather P. denitrificans lipids in the assay. Similarly to E. coli enzyme (33), divalent cations promote NADH:DQ activity, and CaCl2 in 2 mm concentration was found to be optimal, with lower and higher concentrations inhibitory. The activity can be inhibited by piericidin A by more than 90%, indicating that the enzyme is functionally intact. The IC50 values for piericidin A and rotenone compare well to values determined for other bacterial species. However, rotenone is a less effective inhibitor of the P. denitrificans complex I than of the bovine enzyme, probably reflecting some differences in the quinone binding pocket.
TABLE 3.
Complex I catalytic activities in the membranes and after purification
| P. denitrificans sample | Reaction | Specific activity |
|---|---|---|
| μmol min−1mg−1 | ||
| Membranes | NADH:FeCy | 3.53 ± 0.230 |
| dNADH:FeCy | 2.06 ± 0.030 | |
| dNADH:O2 | 0.82 ± 0.069 | |
| dNADH:O2 + 20 μm piericidin A | 0.02 ± 0.003 | |
| dNADH:O2 + 20 μm rotenone | 0.05 ± 0.006 | |
| Purified complex I | NADH:FeCy | 88.29 ± 1.81 |
| NADH:DQ | 14.98 ± 0.44 | |
| NADH:DQ + 20 μm piericidin A | 1.05 ± 0.07 | |
| NADH:DQ + 20 μm rotenone | 2.05 ± 0.10 | |
| IC50 piericidin A | 0.10 μm | |
| IC50 rotenone | 0.17 μm |
A necessary condition for structural studies is that the complex should be stable over several days, as needed for crystallization. The stability was tested by incubating pure enzyme under a range of conditions and then checking for any dissociation of the complex by gel filtration chromatography. Analysis of the eluted fractions by SDS-PAGE indicated that complex is stable over 3–4 days at room temperature and at least a week at 4 °C, when kept at pH 6–6.5 and in the presence of about 15% glycerol (data not shown). The complex remained fully active under these conditions; at the end of storage NADH:DQ activity was about 85–90% of the initial activity and was inhibited by piericidin A by more than 90%. Therefore, the preparation fulfills the minimal requirements for structural studies. The addition of EDTA strongly destabilized the enzyme, as expected from initial trials on solubilized membranes.
Electron Microscopy
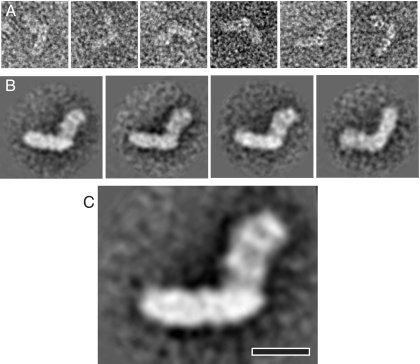
A low resolution EM reconstruction of the side view of the complex was produced by single particle analysis of negatively stained samples. Particles were picked on the basis of the L-shaped appearance in side views (Fig. 3A). These represented a majority of well defined particles, whereas many smaller fragments were also visible on grids, limiting the number of particles suitable for further processing to less than 200. Nevertheless, this was sufficient for the reconstruction and provided a clear outline of the molecule. Classification of images into four classes and averaging in IMAGIC (31) revealed three very similar classes in one orientation on the grid (flip) (40) and one class in the opposite orientation (flop) (Fig. 3B). This distribution is similar to that observed with the E. coli (40) and bovine enzymes (41) and likely reflects the asymmetrical nature of the molecule, so that it is absorbed to the carbon layer in a preferred orientation. Because all flip classes were very similar, only two classes (flip and flop) were used for the final analysis, and the best class average (flip), representing about 70% of total particles, is shown in Fig. 3C. The overall L-shape of the molecule is consistent with previous EM reconstructions (34, 40, 42), allowing us to assign the horizontal, strongly stain-excluding arm as the hydrophobic membrane domain. This domain is about 200 Å long and 60 Å thick, consistent with previous reconstructions and the recent crystal structure of the T. thermophilus enzyme (15). The length of the peripheral arm is about 140 Å, also consistent with available crystal structures (14, 15). The peripheral domain has features similar to the A. aeolicus reconstruction (34), with the stain-filled areas in the middle and at the tip. Thus, similarly to the case of the thermophile A. aeolicus, this reconstruction shows better defined features than similar reconstruction of the E. coli enzyme (33, 40), possibly reflecting higher stability of the complex.
FIGURE 3.
Electron microscopy and single particle analysis. A, representative raw images of side views of negatively stained complex I from P. denitrificans. B, class sums of such images obtained after multiple reference analysis and classification into four classes. Three class sums on the left are of particles in the flip orientation, and the fourth is for flop orientation. C, best side view EM reconstruction (flip orientation after classification into two classes). The membrane arm of the complex is horizontal. Scale bar, 10 nm.
Identification of Subunits
SDS-PAGE analysis of the preparation revealed 17 polypeptide bands (Fig. 2B), which were identified by mass spectrometry, using peptide mass mapping and tandem MS (Table 4). Fourteen polypeptides, including the highly hydrophobic membrane proteins, were identified as core conserved complex I subunits, encoded in the nqo operon. This analysis confirmed the purification of intact complex I, with all subunits present. This is important because distal subunits of the membrane domain (Nqo12, Nqo13, and Nqo8) are readily dissociated from the bacterial complex I (15, 43, 44).
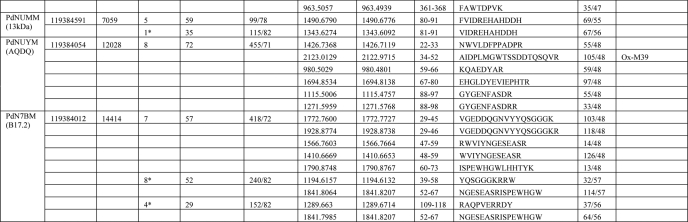
TABLE 4.
Identification of subunits of complex I by mass spectrometrya
a For peptide mass fingerprinting, data were searched against the current NCBI nonredundant nucleotide database using Mascot software (Matrix science) with a tolerance of 70 ppm. Sequences from strain ATCC 13543 were used for the calculation of the subunit mass, except for the supernumerary subunits, for which sequences are available only from strain Pd 1222. Obs. mass, observed mass; Calc. mass, calculated mass; Ox, oxidized; Prop, propionate. Samples were trypsin-digested unless the number of peptide matches is marked with an asterisk, which indicates that this sample was chymotrypsin-digested. For supernumerary subunits, the bovine homologue name is shown in brackets.
b Sample was further analyzed using the Orbitrap mass spectrometer.
Unexpectedly, three additional proteins, homologous to the accessory subunits of eukaryotic complex I, were also identified in the preparation. These are P. denitrificans analogues of bovine subunits B17.2, AQDQ (also known as 18 kDa), and 13 kDa. We named them as PdN7BM (P. denitrificans N7BM), PdNUYM, and PdNUMM subunits, respectively, using the SwissProt nomenclature for genes of complex I subunits. This brings the total molecular mass of the complex to about 561 kDa, significantly larger than 522 kDa in T. thermophilus. None of the products of the six urfs present within the nqo operon were identified in these preparations, as was expected from mutagenesis studies and sequence comparisons (22, 23). Thus, unlike other bacterial complexes I, the P. denitrificans enzyme consists of 17, rather than 14, subunits and contains components not encoded by the nqo operon.
Manual inspection of the MS-MS fragmentation spectra from a sample of PdNUMM (analogue of 13-kDa subunit) suggested the presence of an N-terminal peptide TIPAPEIQTVTSWK (with removal of N-terminal methionine; m/z observed 1570.83, predicted 1570.80) and a tryptic peptide DTGWVECGYCDKR containing an internal disulfide bond between two conserved cysteines (indicated by −2 m/z mass difference; m/z observed 1529.61, predicted 1531.67). These assignments are consistent with measurements of peptide masses (given above) and the molecular mass of the intact subunit (Table 5).
TABLE 5.
Molecular masses of subunits of P. denitrificans complex I
Masses of entire subunits were measured by LC-ESI-MS, with the standard deviation shown in parentheses. No mass measurements were obtained for subunits Nqo7 and Nqo10. Calculated masses are for strains ATCC 13543 and, in brackets, Pd 1222. For supernumerary subunits, bovine homologue name is shown in parentheses.
| Subunit | Mass |
Mass difference | Modification | |
|---|---|---|---|---|
| Observed | Calculated | |||
| Da | ||||
| Nqo1 | 47100.9 (10.5) | 47191.28 (47219.33) | −90.4 (−118.43) | −Met +acetyl |
| Nqo2 | 26124.7 (2.2) | 26122.01 (26122.01) | +2.6 (+2.72) | None |
| Nqo3 | 73002.6 (30.6) | 73159.66 (73203.66) | −157.1 (−201.06) | ? |
| Nqo4 | 46631.1 (8.1) | 46674.63 (46724.64) | −43.5 (−93.51) | ? |
| Nqo5 | 23765.6 (8.3) | 23730.98 (23892.18) | +34.6 (−126.54) | ? |
| Nqo6 | 19527.6 (1.3) | 19177.04 (19504.63) | +350.5 (+22.93) | ? |
| Nqo7 | 13633.19 (13651.23) | |||
| Nqo8 | 38725.3 (16.5) | 38751.4 (38841.56) | −26 (−116.26) | ? |
| Nqo9 | 18765.0 (0.9) | 18959.70 (18900.62) | −194.6 (−135.58) | ? |
| Nqo10 | 21819.39 (21688.19) | |||
| Nqo11 | 10855.5 (0.7) | 10856.16 (10856.16) | −0.7 | None |
| Nqo12 | 77813.7 (21.7) | 77705.77 (77735.86) | +108 (+77.88) | ? |
| Nqo13 | 56572.9 (18.4) | 56417.48 (56484.55) | +155.4 (+88.36) | ? |
| Nqo14 | 52612.1 (15.7) | 52535.40 (52535.40) | +76.7 | ? |
| PdNUMM (13 kDa) | 6924.2 (1.0) | (7058.92) | −134.7 | −Met −disulfide |
| PdNUYM (AQDQ) | 12029.7 (0.7) | (12028.40) | +1.3 | None |
| PdN7BM (B17.2) | 14415.4 (0.6) | (14413.93) | +1.5 | None |
| 14443.3 (0.6) | +29.3 | +formyl | ||
Intact molecular masses of the subunits were measured by electrospray-mass spectrometry on samples resolved by reverse-phase chromatography (LC ESI-MS, see Table 5). Importantly, all three supernumerary subunits were clearly identified by these analyses. Approximate masses for most core subunits were also observed, but only Nqo1 (with N-terminal modification), Nqo2, and Nqo11 corresponded exactly to the expected mass. The reason for these discrepancies is that strain DSM 413 was used for the purification, while nqo sequences are available only for strains ATCC 13543 and Pd 1222. These sequences differ from each other (and probably from DSM 413) in a few amino acids in many subunits, resulting in slightly different masses (Table 5). All three supernumerary subunits could be identified unambiguously because their sequences do not differ between the strains.
DISCUSSION
We describe the first purification and characterization of the intact complex I from an α-proteobacterium, using P. denitrificans, a well characterized model organism. Despite many attempts by several research groups, intact proteobacterial complex I has not been isolated previously. The crucial parameters that allowed purification were appropriate choice of pH, the presence of glycerol throughout the procedure, and addition of divalent cations in relatively high concentrations. Although Ca2+ or Mg2+ stabilize complex I from E. coli (33) and T. thermophilus (15) in a concentration of about 2 mm, the effect appears to be more pronounced in P. denitrificans, and 10 mm CaCl2 was required for optimal stability.
The preparation shows robust NADH:decylubiquinone oxidoreductase activity, which is inhibited by piericidin-A and rotenone, classical indicators of complex I integrity. Single particle analysis of EM images produced a familiar L-shaped molecule, as observed for complexes I from all species studied. The features of the EM reconstruction are relatively well defined, and the enzyme is stable over several days, indicating that the preparation is likely to be stable enough for crystallization trials and further structural studies. All 14 core subunits of complex I, coded in the nqo operon, were identified in the preparation, further confirming the purification of intact enzyme.
The most unexpected result of this work is that complex I from P. denitrificans was found to consist of 17 subunits, containing 3 supernumerary subunits in addition to the 14 expected core subunits. These subunits are clearly homologous to the bovine complex I subunits B17.2 (PdN7BM), AQDQ (PdNUYM), and 13 kDa (PdNUMM) (Table 6). The sequence identity (∼30–35%) is lower than for most hydrophilic core subunits, but it is higher than for most membrane subunits. Database comparisons with these subunits using BLASTP show, as expected, the highest similarity scores for homologous proteins from various α-proteobacteria, such as Rhodobacter sphaeroides (Table 6). These are followed by homologous complex I subunits from eukaryotes, including mammals, plants, and fungi. However, there is no strict distinction in the degree of similarity between α-proteobacteria and eukaryotes, as some α-proteobacterial proteins (including NUMM and N7BM homologues from members of the rickettsial subdivision) show lower similarity scores to P. denitrificans subunits than some eukaryotes. Subunits from fungi show lower similarity than those from animals (Table 6). Furthermore, proteins closely related to these three subunits were not found in any bacteria other than α-proteobacteria.
TABLE 6.
Sequence comparisons of the three novel supernumerary subunits of complex I from P. denitrificans with other species
Multiple sequence alignment of complex I subunits from P. denitrificans and species listed was performed with ClustalW2 (53). The names of homologous bovine subunits are shown in parentheses. GI accession numbers for strain Pd 1222 are also shown. Sequence identity (and similarity in parentheses) was calculated compared with P. denitrificans sequences.
| P. denitrificans | R. sphaeroides | Human | Bovine | Yarrowia lipolytica | Neurospora crassa |
|---|---|---|---|---|---|
| % | % | % | % | % | |
| PdNUMM (13 kDa) gi 119384591 | 56 (73) | 27 (44) | 29 (44) | 21 (35) | 24 (37) |
| PdNUYM (AQDQ) gi 119384054 | 64 (77) | 35 (55) | 35 (54) | 29 (48) | 12 (25) |
| PdN7BM (B17.2) gi 119384012 | 69 (81) | 31 (45) | 31 (45) | 23 (37) | 21 (34) |
These findings suggest that a precursor of eukaryotic complex I, composed of at least 17 subunits, is present in modern α-proteobacteria. It was noted previously that sequences coding for N7BM, NUYM, and NUMM homologues, as well as for homologues of ACPM (bovine SDAP) and NUEM (39 kDa) subunits of complex I, are present in the α-proteobacterial genomes (20). It was not known, however, whether they actually became incorporated into complex I. The genomic context did not hint at their association with the complex because these genes never occur in the same operon as core subunits (20). Our data clearly show that three of these proteins form part of complex I in P. denitrificans. All three subunits are encoded in different parts of chromosome 1, far separated from the nqo operon. It cannot be excluded that in α-proteobacteria most closely related to eukaryotes, such as Rickettsia prowazekii (5), the ACPM and NUEM homologues are also incorporated into complex I.
The function of supernumerary subunits in complex I has not been established, apart from some similarities to proteins of known function, noted in the Introduction. The three subunits that we have identified in P. denitrificans enzyme are hydrophilic and are found in the peripheral arm of the bovine complex I (45, 46). Because of the small size of the subunits, a comparison of our EM reconstruction to other bacterial species, lacking these subunits, is not likely to reveal their exact location. It cannot be excluded that these three subunits, incorporated together into complex I, contact each other and form a subdomain within the complex. It remains to be established whether the apparent higher stability of the complex (as compared with the E. coli enzyme) is due to the presence of supernumerary subunits or due to intrinsic properties of the core subunits.
Subunit B17.2 is one of five complex I subunits identified with nitrated tyrosine residues when mitochondria are incubated with peroxynitrite (47). Subunit AQDQ was suggested to be phosphorylated in a cAMP-dependent manner (48), but the phosphorylated subunit was identified subsequently as ESSS (49).
Interestingly, subunit PdNUMM (13-kDa analogue) is listed in NCBI as a member of the zf-CHCC (PF10276 in Pfam data base) superfamily of small zinc finger proteins. The members of this superfamily contain a zinc finger motif CX8–9HX14–15CX2C, likely to bind zinc or other metals, fully conserved in homologues of 13-kDa subunit from all species, including human. This superfamily belongs to the rubredoxin clan (CL0045 in Pfam data base), which also includes subunit Vb of bovine cytochrome c oxidase. This small intramembranous subunit coordinates tightly bound zinc atom by four cysteines from its zinc finger motif, forming a β-barrel structure (50). This compact conformation did not suggest any specific physiological role for the bound zinc and zinc finger domain.
Bovine complex I also contains one tightly bound zinc atom (39), apparently coordinated by Cys and His residues (51). We have not identified any bound zinc atoms in the structure of the core hydrophilic subunits of complex I from T. thermophilus (14, 16). Several NMR structures of close α-proteobacterial homologues of PdNUMM protein were determined by the Northeast Structural Genomics consortium. These are from R. sphaeroides (PDB 2JVM), Silicibacter pomeroyi (PDB 2JRR), and with the bound zinc atom from Bartonella henselae (PDB 2JZ8). Because this protein can now be assigned as a component of complex I, these structures represent only the second known structure of the supernumerary subunit in complex I (the other being the NMR structure of subunit B8 (52)). In PDB 2JZ8, the zinc atom is coordinated by the three Cys and one His residue from the conserved zinc finger motif, in a similar conformation to oxidase subunit Vb. Therefore, it is likely that tightly bound zinc found in bovine complex I is coordinated by the 13-kDa subunit. Our MS data, mentioned above, show that upon protein denaturation and tryptic digestion, two neighboring coordinating cysteines are likely to make a disulfide bridge.
Thus, it appears that both complexes I and IV of the respiratory chain contain on their matrix side subunits with a similar zinc-coordinating fold. Zinc inhibits complex I in relatively high concentrations (53), a process that is not likely to involve the tightly bound zinc atom, as it remains bound in the presence of EDTA (39). It remains to be established whether the 13-kDa subunit, if it indeed coordinates zinc, has any regulatory role.
In summary, our findings suggest that the evolution of the respiratory complex I by the addition of supernumerary subunits began before the original symbiotic event and has not happened exclusively within eukaryotic lineages, as has been thought previously. Three of these subunits are found in complex I from P. denitrificans, from five possible candidates present in α-proteobacterial genomes. One reason why these subunits were never incorporated into nqo operons could be that they perform additional functions, not as part of complex I. Our findings also confirm that α-proteobacteria are the closest extant relatives of the proto-mitochondrion, in agreement with the endosymbiotic theory.
This work was supported by the Medical Research Council.
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank
- DDM
- dodecyl maltoside
- ESI
- electrospray ionization
- DQ
- decyl-ubiquinone.
REFERENCES
- 1. Andersson S. G., Kurland C. G. (1999) Curr. Opin. Microbiol. 2, 535–541 [DOI] [PubMed] [Google Scholar]
- 2. Margulis L. (1970) Origin of Eukaryotic Cells, Yale University Press, New Haven, CT [Google Scholar]
- 3. John P., Whatley F. R. (1975) Nature 254, 495–498 [DOI] [PubMed] [Google Scholar]
- 4. Gray M. W., Burger G., Lang B. F. (1999) Science 283, 1476–1481 [DOI] [PubMed] [Google Scholar]
- 5. Andersson S. G., Zomorodipour A., Andersson J. O., Sicheritz-Pontén T., Alsmark U. C., Podowski R. M., Näslund A. K., Eriksson A. S., Winkler H. H., Kurland C. G. (1998) Nature 396, 133–140 [DOI] [PubMed] [Google Scholar]
- 6. Walker J. E. (1992) Q. Rev. Biophys. 25, 253–324 [DOI] [PubMed] [Google Scholar]
- 7. Brandt U. (2006) Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 8. Sazanov L. A. (2007) Biochemistry 46, 2275–2288 [DOI] [PubMed] [Google Scholar]
- 9. Yagi T., Matsuno-Yagi A. (2003) Biochemistry 42, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 10. Ohnishi T. (1998) Biochim. Biophys. Acta 1364, 186–206 [DOI] [PubMed] [Google Scholar]
- 11. Balaban R. S., Nemoto S., Finkel T. (2005) Cell 120, 483–495 [DOI] [PubMed] [Google Scholar]
- 12. Hinchliffe P., Carroll J., Sazanov L. A. (2006) Biochemistry 45, 4413–4420 [DOI] [PubMed] [Google Scholar]
- 13. Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. (2006) J. Biol. Chem. 281, 32724–32727 [DOI] [PubMed] [Google Scholar]
- 14. Sazanov L. A., Hinchliffe P. (2006) Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 15. Efremov R. G., Baradaran R., Sazanov L. A. (2010) Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 16. Berrisford J. M., Sazanov L. A. (2009) J. Biol. Chem. 284, 29773–29783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedrich T., Scheide D. (2000) FEBS Lett. 479, 1–5 [DOI] [PubMed] [Google Scholar]
- 18. Fearnley I. M., Walker J. E. (1992) Biochim. Biophys. Acta 1140, 105–134 [DOI] [PubMed] [Google Scholar]
- 19. Fearnley I. M., Carroll J., Shannon R. J., Runswick M. J., Walker J. E., Hirst J. (2001) J. Biol. Chem. 276, 38345–38348 [DOI] [PubMed] [Google Scholar]
- 20. Gabaldón T., Rainey D., Huynen M. A. (2005) J. Mol. Biol. 348, 857–870 [DOI] [PubMed] [Google Scholar]
- 21. Yagi T., Yano T., Matsuno-Yagi A. (1993) J. Bioenerg. Biomembr. 25, 339–345 [DOI] [PubMed] [Google Scholar]
- 22. Yagi T. (1986) Arch. Biochem. Biophys. 250, 302–311 [DOI] [PubMed] [Google Scholar]
- 23. Dupuis A., Chevallet M., Darrouzet E., Duborjal H., Lunardi J., Issartel J. P. (1998) Biochim. Biophys. Acta 1364, 147–165 [DOI] [PubMed] [Google Scholar]
- 24. Xu X., Matsuno-Yagi A., Yagi T. (1993) Biochemistry 32, 968–981 [DOI] [PubMed] [Google Scholar]
- 25. Berry E. A., Trumpower B. L. (1985) J. Biol. Chem. 260, 2458–2467 [PubMed] [Google Scholar]
- 26. Ludwig B. (1986) Methods Enzymol. 126, 153–159 [DOI] [PubMed] [Google Scholar]
- 27. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 28. Carroll J., Fearnley I. M., Wang Q., Walker J. E. (2009) Anal. Biochem. 395, 249–255 [DOI] [PubMed] [Google Scholar]
- 29. Carroll J., Fearnley I. M., Walker J. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16170–16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith J. M. (1999) J. Struct. Biol. 125, 223–228 [DOI] [PubMed] [Google Scholar]
- 31. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 32. Leif H., Sled V. D., Ohnishi T., Weiss H., Friedrich T. (1995) Eur. J. Biochem. 230, 538–548 [DOI] [PubMed] [Google Scholar]
- 33. Sazanov L. A., Carroll J., Holt P., Toime L., Fearnley I. M. (2003) J. Biol. Chem. 278, 19483–19491 [DOI] [PubMed] [Google Scholar]
- 34. Peng G., Fritzsch G., Zickermann V., Schägger H., Mentele R., Lottspeich F., Bostina M., Radermacher M., Huber R., Stetter K. O., Michel H. (2003) Biochemistry 42, 3032–3039 [DOI] [PubMed] [Google Scholar]
- 35. Stroh A., Anderka O., Pfeiffer K., Yagi T., Finel M., Ludwig B., Schägger H. (2004) J. Biol. Chem. 279, 5000–5007 [DOI] [PubMed] [Google Scholar]
- 36. Iwata S., Ostermeier C., Ludwig B., Michel H. (1995) Nature 376, 660–669 [DOI] [PubMed] [Google Scholar]
- 37. Grivennikova V. G., Roth R., Zakharova N. V., Hägerhäll C., Vinogradov A. D. (2003) Biochim. Biophys. Acta 1607, 79–90 [DOI] [PubMed] [Google Scholar]
- 38. Vinogradov A. D. (1998) Biochim. Biophys. Acta 1364, 169–185 [DOI] [PubMed] [Google Scholar]
- 39. Shinzawa-Itoh K., Seiyama J., Terada H., Nakatsubo R., Naoki K., Nakashima Y., Yoshikawa S. (2010) Biochemistry 49, 487–492 [DOI] [PubMed] [Google Scholar]
- 40. Mamedova A. A., Holt P. J., Carroll J., Sazanov L. A. (2004) J. Biol. Chem. 279, 23830–23836 [DOI] [PubMed] [Google Scholar]
- 41. Morgan D. J., Sazanov L. A. (2008) Biochim. Biophys. Acta 1777, 711–718 [DOI] [PubMed] [Google Scholar]
- 42. Guénebaut V., Schlitt A., Weiss H., Leonard K., Friedrich T. (1998) J. Mol. Biol. 276, 105–112 [DOI] [PubMed] [Google Scholar]
- 43. Baranova E. A., Holt P. J., Sazanov L. A. (2007) J. Mol. Biol. 366, 140–154 [DOI] [PubMed] [Google Scholar]
- 44. Holt P. J., Morgan D. J., Sazanov L. A. (2003) J. Biol. Chem. 278, 43114–43120 [DOI] [PubMed] [Google Scholar]
- 45. Sazanov L. A., Peak-Chew S. Y., Fearnley I. M., Walker J. E. (2000) Biochemistry 39, 7229–7235 [DOI] [PubMed] [Google Scholar]
- 46. Hirst J., Carroll J., Fearnley I. M., Shannon R. J., Walker J. E. (2003) Biochim. Biophys. Acta 1604, 135–150 [DOI] [PubMed] [Google Scholar]
- 47. Murray J., Taylor S. W., Zhang B., Ghosh S. S., Capaldi R. A. (2003) J. Biol. Chem. 278, 37223–37230 [DOI] [PubMed] [Google Scholar]
- 48. Scacco S., Vergari R., Scarpulla R. C., Technikova-Dobrova Z., Sardanelli A., Lambo R., Lorusso V., Papa S. (2000) J. Biol. Chem. 275, 17578–17582 [DOI] [PubMed] [Google Scholar]
- 49. Chen R., Fearnley I. M., Peak-Chew S. Y., Walker J. E. (2004) J. Biol. Chem. 279, 26036–26045 [DOI] [PubMed] [Google Scholar]
- 50. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1996) Science 272, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 51. Giachini L., Francia F., Boscherini F., Pacelli C., Cocco T., Papa S., Venturoli G. (2007) FEBS Lett. 581, 5645–5648 [DOI] [PubMed] [Google Scholar]
- 52. Brockmann C., Diehl A., Rehbein K., Strauss H., Schmieder P., Korn B., Kühne R., Oschkinat H. (2004) Structure 12, 1645–1654 [DOI] [PubMed] [Google Scholar]
- 53. Sharpley M. S., Hirst J. (2006) J. Biol. Chem. 281, 34803–34809 [DOI] [PubMed] [Google Scholar]